Introduction
Fluoropyrimidine-based chemotherapy plus antibody
therapy is currently the standard first-line treatment for
metastatic colorectal cancer (mCRC). Infusional 5-fluorouracil
(5-FU) and leucovorin (LV) or capecitabine with either oxaliplatin
(FOLFOX/XELOX) or irinotecan (FOLFIRI), both of which were shown to
have manageable toxicity profiles and to be able to improve
treatment efficacy (1–3), are administered as the chemotherapy
backbones in such treatment. Due to their proven anticancer
activity, the monoclonal antibodies (mAbs) cetuximab, panitumumab
and bevacizumab have also been approved for use as first-line
chemotherapy in mCRC patients in combination with FOLFOX/XELOX
and/or FOLFIRI.
Progress has been achieved with drugs targeting the
vascular endothelial growth factor (3) or the epidermal growth factor receptor
(EGFR) (4). The EGFR, a receptor
tyrosine kinase, triggers a downstream signaling cascade through
mechanisms such as the RAS/RAF/MAPK and PI3K/AKT pathways, which
are involved in cell proliferation, survival and motility. Based on
our knowledge of this cascade, the administration of cetuximab and
panitumumab, two mAbs targeting EGFR, was established as a novel
treatment option for mCRC patients.
Among the predictive biomarkers used to identify the
mCRC patients most likely to benefit from cetuximab and panitumumab
treatment, the best established is the KRAS gene. Mutations
in KRAS produce a constitutively active RAS protein, leading
in turn to EGFR-independent activation of the RAS/RAF/MAPK pathway
(5). The identification of this
phenomenon has led to the compelling hypothesis that the activation
of KRAS mutations may preclude response to anti-EGFR mAb
therapy, a hypothesis supported by earlier clinical observations
(6,7). Between 2007 and 2008, 6 randomized
clinical trials were conducted, in which the KRAS status was
retrospectively assessed in tumor samples from mCRC patients who
had been randomly assigned to receive panitumumab (8,9) or
cetuximab treatment (10–13).
The activation of mutations of BRAF, another
component of the EGFR/MAPK signal transduction pathway, is also
prevalent among mCRC patients. As with the KRAS mutation, it
is plausible that BRAF mutations may confer resistance to
anti-EGFR therapy, although their lower prevalence makes this
hypothesis more difficult to test clinically. However, two previous
studies reported that BRAF mutation at codon 600 (V600E),
resulting in strong activation of the BRAF protein downstream of
KRAS, is associated with a shorter progression-free survival
(PFS) and overall survival in mCRC chemorefractory patients treated
with anti-EGFR mAb therapy (14,15).
Tumor-derived mutant PI3K was shown to
stimulate the AKT pathway and promote cell growth in several types
of cancer, including CRC. Tumors with PIK3CA mutations have
been associated with poor prognosis, with mutations in the
PIK3CA gene found to significantly impair the response to
anti-EGFR mAb treatment in mCRC patients. In support of these
findings, a large-scale European study reported that acquiring
knowledge regarding the combined KRAS, BRAF,
PIK3CA and NRAS mutation status may improve the
sensitivity of prediction of the response to anti-EGFR mAb therapy
(16).
In order to counsel mCRC patients harbouring
KRAS mutations (and possibly other gene mutations), we need
to establish whether they may still benefit from standard
chemotherapeutic options. Therefore, this study pursued three
objectives: to evaluate the efficacy of the currently available
chemotherapeutic protocols for the treatment of mCRC patients, to
investigate the value of predictive biomarkers in the
personalization of 5-FU-based chemotherapy and to test the
hypothesis that mutations in several of the targeted oncogenes are
correlated with treatment outcomes in patients receiving different
first-line regimens. To test this hypothesis, we first evaluated
the predictive significance of KRAS, BRAF,
PIK3CA, NRAS and AKT1 mutations in a cohort of
mCRC patients who had undergone 5-FU-based chemotherapy;
subsequently, using a uniform catalog of retrospective but detailed
clinical data, we determined the predictive value of these
mutations regarding patient outcomes following completion of the
most common therapeutic regimens. This analysis allowed for
assessment of the predictive significance of KRAS,
BRAF, PIK3CA, NRAS and AKT1 mutations
independent of anti-EGFR therapy and their predictive value
regarding benefit from oxaliplatin- or irinotecan-based
therapy.
Materials and methods
Patients and treatment methods
This study was approved by the Ethics Committee of
Tohoku University School of Medicine and included a total of 194
mCRC patients who had received various forms of first-line
chemotherapy at the study site between February, 2005 and October,
2010.
The mFOLFOX6 regimen consisted of a 2-h infusion of
85 mg/m2 of oxaliplatin on day 1, a 2-h infusion of 200
mg/m2 of LV on day 1, a bolus of 400 mg/m2 of
5-FU on day 1 and a 46-h infusion of 2,400 mg/m2 of
5-FU/day on days 1–2. The FOLFIRI regimen consisted of a 1.5-h
infusion of 150 mg/m2 of irinotecan on day 1, a 2-h
infusion of 200 mg/m2 of LV on day 1, a bolus of 400
mg/m2 of 5-FU on day 1 and a 46-h infusion of 2,400
mg/m2 of 5-FU/day on days 1–2. The treatments had been
administered on day 1 and repeated on day 2 of a 14-day treatment
cycle. The IRIS regimen consisted of continuous administration of
150 mg/m2 of irinotecan for 90 min on day 1, followed by
twice-daily administration of S-1 for a 2-week period on days 3–16.
The administered dose of S-1 had been determined as follows: for a
body surface area (BSA) of <1.25 m2, 80 mg/day; for a
BSA of 1.25–1.5 m2, 100 mg/day; and for a BSA of >1.5
m2, 120 mg/day as a 3-week course.
Tumor collection and processing
Formalin-fixed, paraffin-embedded (FFPE) samples of
tumor tissue from archival specimens that had been collected at the
time of diagnosis and stored at Tohoku University Hospital were
investigated. The assays of the tissue samples for KRAS,
BRAF, PIK3CA, NRAS and AKT1 mutations
were performed at the Department of Clinical Oncology, Institute of
Development, Aging and Cancer, Tohoku University. All samples were
screened for KRAS mutations in codons 12, 13 and 61;
BRAF V600E; PIK3CA mutations in exons 9 and 20;
NRAS mutations in codons 12, 13 and 61; and AKT1
E17K. All samples were also classified as mutant or wild-type.
Nucleotide sequence analysis
Mutation analyses of KRAS, BRAF,
PIK3CA, NRAS and AKT1 were performed by
extraction of genomic DNA from FFPE tissue slides or sections. DNA
was extracted using the QIAamp DNA FFPE tissue kit (Qiagen, Hilden,
Germany) according to the manufacturer’s instructions. Analyses of
the DNA sequences were performed using the automated CEQ2000XL DNA
analysis system (Beckman Coulter, Fullerton, CA, USA) under
specific cycle and temperature conditions. The PCR products were
analyzed by 1.0% agarose gel electrophoresis. Appropriate positive
and negative controls were included for the KRAS,
BRAF, PIK3CA, NRAS and AKT1 analyses.
To minimize bias, all researchers who performed the mutation
analyses were blinded to the clinical outcomes.
Statistical analysis
All patients for whom data regarding KRAS,
BRAF, PIK3CA, NRAS and AKT1 mutation
status were available, were included in the analysis. The response
rate (RR) was determined according to the Response Evaluation
Criteria in Solid Tumors (RECIST), version 1.0. According to
RECIST, the patients were categorized as responders if they
achieved complete response (CR) or partial response (PR) and as
non-responders if they exhibited stable disease (SD) or progressive
disease (PD). The associations between treatment response or
patient characteristics and mutational status were assessed using
the χ2 test. PFS was defined as the time interval
between the initiation of chemotherapy and the first objective
evidence of disease progression or death from any cause. The PFS
was determined using the Kaplan-Meier method and compared using the
log-rank test. Statistical significance was set at a level of
P<0.05 for a bilateral test. Through such means, the KRAS
mutational status was investigated and the hypothesis that PFS
varies according to the type of first-line regimen (oxaliplatin- or
irinotecan-based) was tested.
Results
Study objective
This retrospective study investigated the efficacy
of first-line chemotherapeutic protocols in 194 mCRC patients
according to gene status and its association with several patient
clinical characteristics. As tumor samples and complete end-point
data were available for all patients, RR and PFS were determined
for the entire patient sample (100%).
Treatment regimens
Combination chemotherapy was administered as
first-line treatment to 174 patients (89.7%), either with or
without mAb supplementation (Table
I). 5-FU was administered as the only cytotoxic agent to 17
patients (8.8%). A total of 109 patients (56.2%) were treated with
oxaliplatin in the first-line setting. The oxaliplatin-containing
regimen consisted of only the FOLFOX regimen (infusion and bolus
5-FU plus oxaliplatin). A total of 65 patients (33.5%) were treated
with irinotecan in the first-line setting. The
irinotecan-containing regimen consisted of the FOLFIRI regimen
(infusion and bolus 5-FU with irinotecan) for 43 patients and S-1
plus irinotecan for 22 patients. As first-line treatment,
bevacizumab was administered as part of an oxaliplatin-containing
regimen to 27 patients (13.9%) and with irinotecan to 26 patients
(13.4%). As bevacizumab was not approved until 2007 in Japan, the
percentage of mCRC patients who received bevacizumab in this study
was low (27.3%).
 | Table I.First-line treatment regimens used in
this retrospective study (n=194). |
Table I.
First-line treatment regimens used in
this retrospective study (n=194).
| Regimens | No. (%) |
|---|
| FOLFOX +
bevacizumab | 27 (13.9) |
| FOLFOX | 82 (42.3) |
| FOLFIRI +
bevacizumab | 17 (8.8) |
| FOLFIRI | 26 (13.4) |
| IRIS +
bevacizumab | 9 (4.6) |
| IRIS | 13 (6.7) |
| 5-Fluorouracil
only | 17 (8.8) |
| No treatment | 3 (1.5) |
| Oxaliplatin-based
treatment | 109 (56.2) |
| Irinotecan-based
treatment | 65 (33.5) |
Mutation analyses of KRAS, BRAF, PIK3CA,
NRAS and AKT1
Table II lists the
mutations detected by direct sequencing. A relatively rare mutation
in codon 61 was analyzed in addition to the common mutations in
codons 12 and 13 in order to increase the sensitivity of mutation
detection. KRAS mutations at codons 12, 13 and 61 were
observed in 78 (40.2%) of the tumor samples. Of the 78 detected
mutations in codons 12 and 13, the most frequent was G12D (12.9%),
followed by G13D (11.3%), G12V (10.3%), G12C (1.5%), G12A (1.0%),
G12R (0.5%) and G13C (0.5%). In codon 61, Q61H and Q61R were
detected in 4 samples (2.0%). Three common KRAS mutations,
G12D, G13D and G12V, were also frequently detected. V600E was
detected in 10 samples (5.2%), all of which harboured wild-type
KRAS. PIK3CA mutations in exon 9 (E542K, E545K, E545G and
Q546K) were detected in 17 samples (8.8%) and PIK3CA
mutations in exon 20 (H1047R, H1047L and H1047Y) in 6 (3.1%).
Mutations in KRAS and PIK3CA were detected in 9
samples (4.7%). NRAS mutations at codons 12, 13 and 61 were
detected in 3 samples (1.5%); and an AKT1 mutation at codon
17 (E17K) was detected in two samples (1.0%).
 | Table II.KRAS, BRAF,
PIK3CA, NRAS and AKT1 mutation frequencies
(n=194). |
Table II.
KRAS, BRAF,
PIK3CA, NRAS and AKT1 mutation frequencies
(n=194).
| Gene | Codon | Nucleotide
substitution | Amino acid
substitution | No. (%) | Total (%) |
|---|
| KRAS | 12 | GGT→CGT | G12R | 1 (0.5) | 78 (40.2) |
| GGT→TGT | G12C | 3 (1.5) |
| GGT→GAT | G12D | 25 (12.9) |
| GGT→GCT | G12A | 2 (1.0) |
| GGT→GTT | G12V | 20 (10.3) |
| 13 | GGC→TGC | G13C | 1 (0.5) |
| GGC→GAC | G13D | 22 (11.3) |
| 61 | CAA→CGA | Q61R | 1 (0.5) |
| CAA→CAC | Q61H | 2 (1.0) |
| CAA→CAT | Q61H | 1 (0.5) |
| BRAF | 600 | GTG→GAG | V600E | 10 (5.2) | 10 (5.2) |
| PIK3CA | 542 | GAA→AAA | E542K | 4 (2.1) | 23 (11.9) |
| 545 | GAG→AAG | E545K | 4 (2.1) |
| GAG→GGG | E545K | 7 (3.6) |
| 546 | CAG→AAG | Q546K | 2 (1.0) |
| 1047 | CAT→TAT | H1047Y | 1 (0.5) |
| CAT→CTT | H1047L | 1 (0.5) |
| CAT→CGT | H1047R | 4 (2.1) |
| NRAS | 12 | GGT→GAT | G12D | 3 (1.5) | 3 (1.5) |
| AKT1 | 17 | GAG→AAG | E17K | 2 (1.0) | 2 (1.0) |
| KRAS and
PIK3CA | | | | 9 (4.7) | |
Patient characteristics
The characteristics of the 194 mCRC patients (median
age, 63 years; range, 16–82 years) from whom primary tumor tissue
samples had been collected were retrospectively analyzed (Table III). The most frequent type of
tumor was tumor of the rectum (75 patients; 38.7%), followed by
tumor of the ascending and sigmoid colon (42 patients; 21.6%),
transverse colon (15 patients; 7.7%) and descending colon and cecum
(10 patients; 5.2%). The most frequent site of metastasis was the
liver (114 patients; 58.8%), followed by the lungs (100 patients;
51.5%), intra-abdominal lymph nodes (72 patients; 37.1%) and the
peritoneum (36 patients; 18.6%).
 | Table III.Patient characteristics. |
Table III.
Patient characteristics.
|
Characteristics | All | KRAS
wild-type | KRAS
mutant | P-value |
|---|
| Total number of
patients | 194 | 116 | 78 | |
| Median age, [years
(range)] | 63 (16–82) | 62 (16–82) | 65 (37–81) | |
| Gender | | | | |
| Male | 111 | 73 | 38 | 0.0498 |
| Female | 83 | 43 | 40 | |
| Primary tumor | | | | |
| Cecum | 10 | 4 | 6 | 0.70 |
| Ascending
colon | 42 | 23 | 19 | |
| Transverse
colon | 15 | 9 | 6 | |
| Descending
colon | 10 | 6 | 4 | |
| Sigmoid
colon | 42 | 25 | 17 | |
| Rectum | 75 | 49 | 26 | |
| Metastatic
sites | | | | |
| Liver | 114 | 64 | 50 | 0.10 |
| Lung | 100 | 52 | 48 | |
| Intra-abdominal
lymph nodes | 72 | 51 | 21 | |
| Peritoneum | 36 | 16 | 20 | |
| Bone | 10 | 5 | 5 | |
| Others | 22 | 12 | 10 | |
Effect of mutation status on the outcome
of first-line chemotherapy
Tables IV and
V show the results of the analysis
of the association between clinical response in terms of RR and
median PFS (mPFS) and the presence or absence of gene mutations.
There were no significant differences in RR or mPFS between the
wild-type and mutant KRAS subgroups who had received
oxaliplatin- or irinotecan-based treatment as first-line therapy.
Furthermore, there was no significant difference in RR or mPFS
between the wild-type KRAS, BRAF, PIK3CA,
NRAS and AKT1 subgroups and the respective mutant
subgroups in any of the 5 genes.
 | Table IV.Response to oxaliplatin-based
treatment according to the presence or absence of gene mutations
(n=109). |
Table IV.
Response to oxaliplatin-based
treatment according to the presence or absence of gene mutations
(n=109).
| Tumor response | KRAS
status | Genetic status of
KRAS, BRAF, PIK3CA, NRAS and
AKT1 | Total (%) |
|---|
|
|
|---|
| Mutant (%) | Wild-type (%) | Mutant of any genes
(%) | Wild-type of all
genes (%) |
|---|
| Total | 43 (100) | 66 (100) | 58 (100) | 51 (100) | 109 (100) |
| Bevacizumab
use | 11 (25.6) | 16 (24.2) | 15 (25.9) | 12 (23.5) | 27 (24.8) |
| CR | 1 (2.3) | 1 (1.5) | 1 (1.7) | 1 (2.0) | 2 (1.8) |
| PR | 20 (46.5) | 33 (50.0) | 25 (43.1) | 28 (54.9) | 53 (48.6) |
| SD | 14 (32.6) | 25 (37.9) | 22 (37.9) | 17 (33.3) | 39 (35.8) |
| PD | 8 (18.6) | 7 (10.6) | 10 (17.2) | 5 (9.8) | 15 (13.8) |
| RR (%) | 48.8 | 51.5 | 44.8 | 56.9 | 50.5 |
| DCR (%) | 81.4 | 89.4 | 82.8 | 90.2 | 86.2 |
| mPFS (months) | 6.8 | 8.6 | 7.2 | 9.7 | 8.1 |
 | Table V.Response to irinotecan-based
treatment according to the presence or absence of gene mutations
(n=65). |
Table V.
Response to irinotecan-based
treatment according to the presence or absence of gene mutations
(n=65).
| Tumor response | KRAS
status | Genetic status of
KRAS, BRAF, PIK3CA, NRAS and
AKT1 | Total (%) |
|---|
|
|
|---|
| Mutant (%) | Wild-type (%) | Mutant of any genes
(%) | Wild-type of all
genes (%) |
|---|
| Total | 26 (100) | 39 (100) | 33 (100) | 32 (100) | 65 (100) |
| Bevacizumab
use | 9 (34.6) | 17 (43.6) | 12 (36.4) | 14 (43.8) | 26 (40.0) |
| CR | 2 (7.7) | 1 (2.6) | 2 (6.1) | 1 (3.1) | 3 (4.6) |
| PR | 12 (46.2) | 19 (48.7) | 15 (45.5) | 16 (50.0) | 31 (47.7) |
| SD | 8 (30.8) | 14 (35.9) | 12 (36.4) | 10 (31.3) | 22 (33.8) |
| PD | 4 (15.4) | 5 (12.8) | 4 (12.1) | 5 (15.6) | 9 (13.8) |
| RR (%) | 53.8 | 51.3 | 51.5 | 53.1 | 52.3 |
| DCR (%) | 84.6 | 87.2 | 87.9 | 84.4 | 86.2 |
| mPFS (months) | 9.7 | 7.7 | 10.0 | 7.1 | 9.1 |
The mPFS of the wild-type and mutant KRAS
subgroups who had received oxaliplatin-based treatment was 8.6
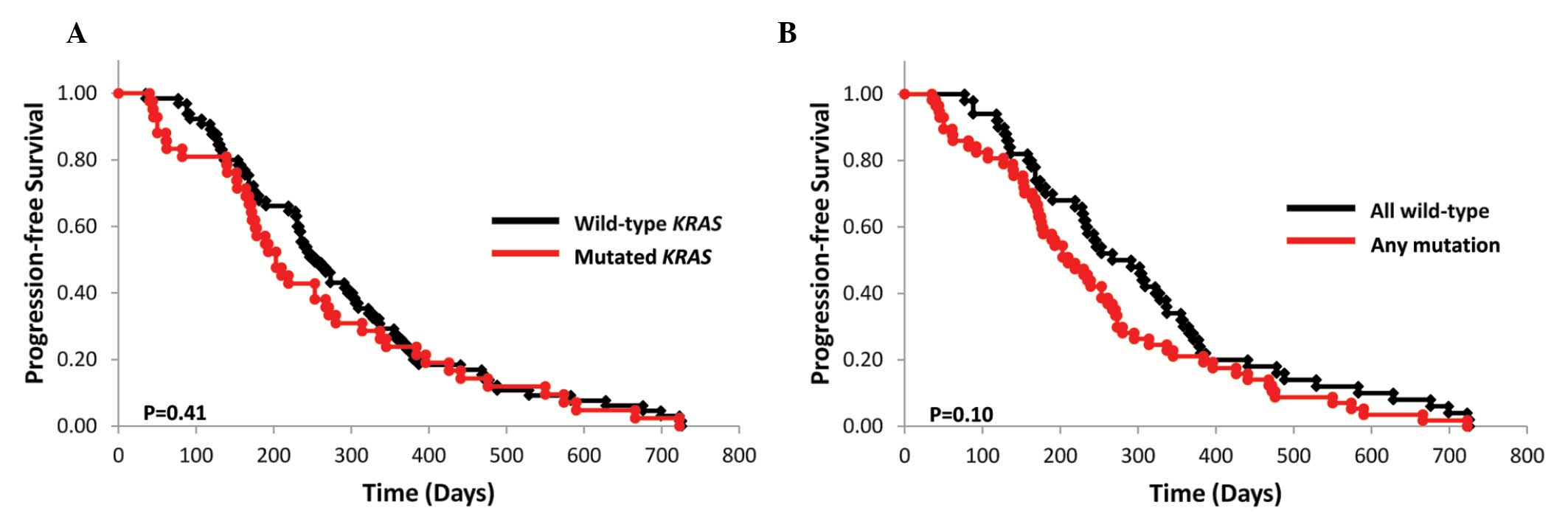
(n=66) and 6.8 months (n=43), respectively (P=0.41; Fig. 1A). Of the 109 assessed patients, 16
of the 66 (24.2%) patients in the wild-type subgroup and 11 of the
43 (25.6%) patients in the mutant KRAS subgroup had received
bevacizumab in combination with FOLFOX (P=0.87). The mPFS of the
wild-type KRAS, BRAF, PIK3CA, NRAS and
AKT1 subgroups and that of their respective mutant subgroups
was 9.7 (n=51) and 7.2 months (n=58), respectively (P=0.10;
Fig. 1B).
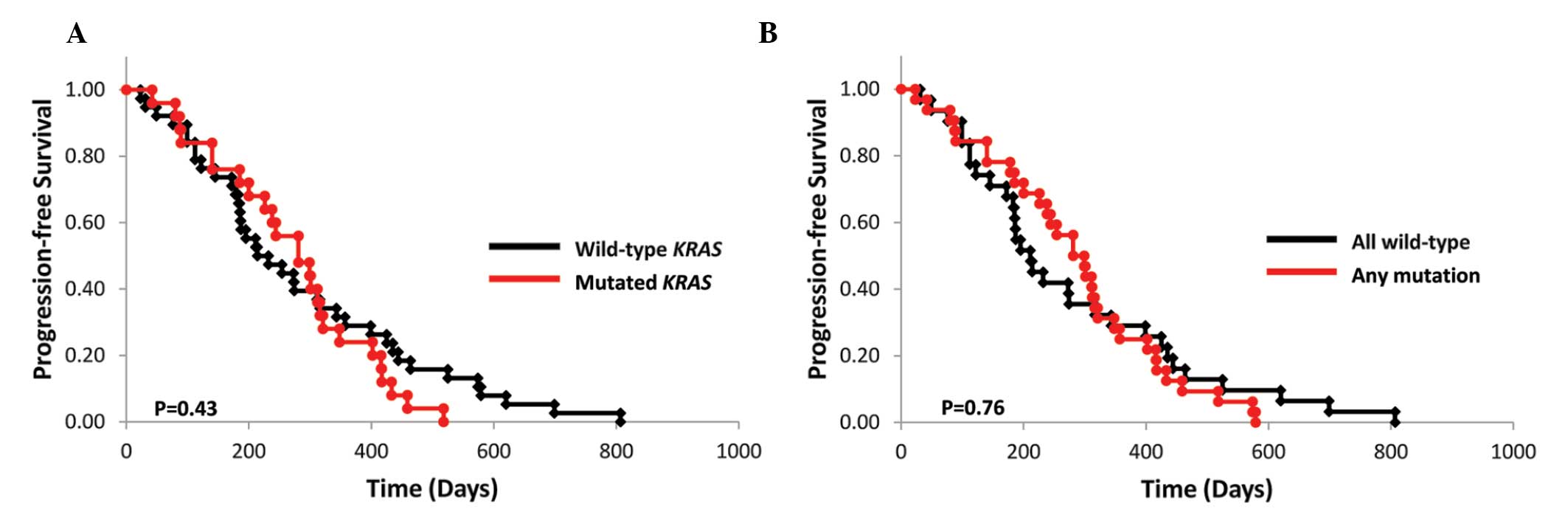
The mPFS of the wild-type and mutant KRAS
subgroups who had received irinotecan-based treatments was 7.7
(n=39) and 9.7 months (n=26), respectively (P=0.43; Fig. 2A). Of the 65 assessed patients, 17
of the 39 (43.6%) patients in the wild-type subgroup and 9 of the
26 (34.6%) patients in the mutant KRAS subgroup had received
bevacizumab in combination with FOLFIRI or IRIS treatment (P=
0.47). The mPFS of the wild-type KRAS, BRAF,
PIK3CA, NRAS and AKT1 subgroups and that of
their respective mutant subgroups were 7.1 (n=32) and 10.0 months
(n=33), respectively (P=0.76; Fig.
2B).
Discussion
This analysis of various mutations of the
KRAS, BRAF, PIK3CA, NRAS and
AKT1 genes in 194 Japanese mCRC patients resulted in the
detection of KRAS mutations at a frequency (78/194; 40.2%)
similar to that described in a previous study of Japanese patients
(17). As the pattern of
KRAS mutations was also found to be similar to that reported
in a previous study on Caucasian patients (18,19),
the results indicated that KRAS mutations do not differ
significantly between Japanese and Caucasian populations in terms
of frequency and mutation spectrum. The prevalence of PIK3CA
mutation (23/194; 11.9%) was also found to be similar to that
reported by previous studies (10–20%) (16). By contrast, the prevalence of
BRAF mutations (10/194; 5.2%) was found to be lower compared
to that reported in studies on Caucasian patients (20), possibly reflecting the genetic
differences between the populations. Of the mutations detected,
E542K, E545K and H1047R were identified as hotspot mutations,
whereas the E545G mutation was rarely detected (16,21).
A large-scale analysis is required to elucidate whether this
discrepancy in the mutation spectrum is the result of genetic
differences among different populations. Previous studies
identified mutations in the NRAS and AKT1 genes in
2.6% (16) and 5.9% (22) of mCRC patients, respectively. In
this study, NRAS and AKT1 mutations were detected in
1.5% (3 patients) and 1.0% (2 patients) of the sample,
respectively.
Similar to previous investigations, the present
study analyzed the association between gene mutations and patient
characteristics in order to determine whether such associations may
predict the efficacy of a first-line regimen. Sartore-Bianchi et
al (23) reported that
KRAS mutations were significantly more prevalent among
females compared with males, whereas PIK3CA mutations were
not found to be significantly associated with gender. In accordance
with Watanabe et al (17),
who reported a higher prevalence of KRAS mutations among
Japanese female (40.9%) compared to male mCRC patients (35.5%;
P=0.001), a higher prevalence of KRAS mutations was detected
among the samples obtained from female (48.2%) compared with those
obtained from male patients (34.2%, P=0.050) in the present
study.
The individualization of drug therapy for mCRC
patients is becoming increasingly feasible. Studies on patients
receiving first-line and subsequent lines of treatment demonstrated
that those with KRAS mutations do not respond to or
experience any survival benefit from treatment with anti-EGFR mAb
therapy. Based on this finding, all mCRC patients are currently
offered KRAS testing to determine whether their tumor is
wild-type KRAS and, if so, counseled that they would likely
benefit from anti-EGFR mAb therapy. As such, it is crucial to
establish whether the mutation may affect the ability to benefit
from anti-EGFR mAb therapy (or any other form of therapy), or
whether prognosis is independent of treatment. Retrospective
analyses of KRAS mutations in mCRC patients treated with
bevacizumab plus chemotherapy revealed that the clinical benefit of
bevacizumab is independent of the KRAS status (24,25).
Other studies investigated the association between KRAS
status (as well as other gene statuses) and clinical benefit from
oxaliplatin- or irinotecan-based treatment in the first-line
setting (26,27). Those studies reported that the
clinical benefit of oxaliplatin- or irinotecan-based treatment is
independent of the KRAS mutational status.
In this study, the patients who had received
oxaliplatin treatment exhibited longer mPFS in the wild-type
KRAS alleles. By contrast, the patients who had received
irinotecan treatment exhibited longer mPFS in the mutant
KRAS alleles. Although there were no statistically
significant differences in the distinct KRAS status between
the oxaliplatin- and irinotecan-based treatment groups, the
KRAS status is likely to affect the outcome of these
treatments in some of the patients. The results of the present
study indicated that mCRC patients with activation of KRAS
mutations, even those treated with oxaliplatin- and
irinotecan-based regimens as first-line treatments, may benefit
from cytotoxic drug therapy. We also provided evidence that both
the wild-type and mutant KRAS subgroups of mCRC patients may
benefit from oxaliplatin- and irinotecan-based therapy.
Acknowledgements
This study was supported in part by
grants-in-aid from the Ministry of Education, Science, Sports and
Culture of Japan. We would like to thank Eri Yokota for assisting
with the mutational analysis, Hiroyoshi Suzuki for preparing the
samples at the Sendai Medical Center and Yayoi Takahashi for
preparing the samples at the Tohoku University Hospital.
References
|
1.
|
Van Cutsem E, Kohne CH, Hitre E, et al:
Cetuximab and chemo-therapy as initial treatment for metastatic
colorectal cancer. N Engl J Med. 360:1408–1417. 2009.PubMed/NCBI
|
|
2.
|
Bokemeyer C, Bondarenko I, Makhson A, et
al: Fluorouracil, leucovorin, and oxaliplatin with and without
cetuximab in the first-line treatment of metastatic colorectal
cancer. J Clin Oncol. 27:663–671. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Hurwitz H, Fehrenbacher L, Novotny W, et
al: Bevacizumab plus irinotecan, fluorouracil, and leucovorin for
metastatic colorectal cancer. N Engl J Med. 350:2335–2342. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Cunningham D, Humblet Y, Siena S, et al:
Cetuximab mono-therapy and cetuximab plus irinotecan in
irinotecan-refractory metastatic colorectal cancer. N Engl J Med.
351:337–345. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Bos JL, Fearon ER, Hamilton SR, et al:
Prevalence of ras gene mutations in human colorectal cancers.
Nature. 327:293–297. 1987. View
Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Benvenuti S, Sartore-Bianchi A, Di
Nicolantonio F, et al: Oncogenic activation of the RAS/RAF
signaling pathway impairs the response of metastatic colorectal
cancers to anti-epidermal growth factor receptor antibody
therapies. Cancer Res. 67:2643–2648. 2007. View Article : Google Scholar
|
|
7.
|
Di Fiore F, Blanchard F, Charbonnier F, et
al: Clinical relevance of KRAS mutation detection in metastatic
colorectal cancer treated by cetuximab plus chemotherapy. Br J
Cancer. 96:1166–1169. 2007.PubMed/NCBI
|
|
8.
|
Amado RG, Wolf M, Peeters M, et al:
Wild-type KRAS is required for panitumumab efficacy in patients
with metastatic colorectal cancer. J Clin Oncol. 26:1626–1634.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Hecht JR, Mitchell E, Chidiac T, et al: A
randomized phase IIIB trial of chemotherapy, bevacizumab, and
panitumumab compared with chemotherapy and bevacizumab alone for
metastatic colorectal cancer. J Clin Oncol. 27:672–680. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Van Cutsem E, Lang I, D’haens G, et al:
KRAS status and efficacy in the first-line treatment of patients
with metastatic colorectal cancer (mCRC) treated with FOLFIRI with
or without cetuximab: the CRYSTAL experience. J Clin Oncol.
26:abstract 2. 2008.PubMed/NCBI
|
|
11.
|
Punt CJ, Tol J, Rodenburg CJ, et al:
Randomized phase III study of capecitabine, oxaliplatin, and
bevacizumab with or without cetuximab in advanced colorectal cancer
(ACC), the CAIRO2 study of the Dutch Colorectal Cancer Group
(DCCG). J Clin Oncol. 26:abstract LBA4011. 2008.PubMed/NCBI
|
|
12.
|
Bokemeyer C, Bondarenko I, Hartmann JT, et
al: KRAS status and efficacy of first-line treatment of patients
with metastatic colorectal cancer (mCRC) with FOLFOX with or
without cetuximab: the OPUS experience. J Clin Oncol. 26:abstract
4000. 2008.
|
|
13.
|
Karapetis CS, Khambata-Ford S, Jonker DJ,
et al: K-ras mutations and benefit from cetuximab in advanced
colorectal cancer. N Engl J Med. 359:1757–1765. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Di Nicolantonio F, Martini M, Molinari F,
et al: Wild-type BRAF is required for response to panitumumab or
cetuximab in meta-static colorectal cancer. J Clin Oncol.
26:5705–5712. 2008.
|
|
15.
|
Tol J, Nagtegaal ID and Punt CJ: BRAF
mutation in metastatic colorectal cancer. N Engl J Med. 361:98–99.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
De Roock W, Claes B, Bernasconi D, et al:
Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy
of cetuximab plus chemotherapy in chemotherapy-refractory
meta-static colorectal cancer: a retrospective consortium analysis.
Lancet Oncol. 11:753–762. 2010.PubMed/NCBI
|
|
17.
|
Watanabe T, Yoshino T, Uetake H, et al:
KRAS mutational status in Japanese patients with colorectal cancer:
results from a nationwide, multicenter, cross-sectional study. Jpn
J Clin Oncol. 43(7): 706–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Andreyev HJ, Norman AR, Cunningham D, et
al: Kirsten ras mutations in patients with colorectal cancer: the
multicenter ‘RASCAL’ study. J Natl Cancer Inst. 90:675–684.
1998.
|
|
19.
|
Andreyev HJ, Norman AR, Cunningham D, et
al: Kirsten ras mutations in patients with colorectal cancer: the
‘RASCAL II’ study. Br J Cancer. 85:692–696. 2001.
|
|
20.
|
Barault L, Veyrie N, Jooste V, et al:
Mutations in the RAS-MAPK, PI(3)K (phosphatidylinositol-3-OH
kinase) signaling network correlate with poor survival in a
population-based series of colon cancers. Int J Cancer.
122:2255–2259. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Ogino S, Nosho K, Kirkner GJ, et al:
PIK3CA mutation is associated with poor prognosis among patients
with curatively resected colon cancer. J Clin Oncol. 27:1477–1484.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Carpten JD, Faber AL, Horn C, et al: A
transforming mutation in the pleckstrin homology domain of AKT1 in
cancer. Nature. 448:439–444. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Sartore-Bianchi A, Martini M, Molinari F,
et al: PIK3CA mutations in colorectal cancer are associated with
clinical resistance to EGFR-targeted monoclonal antibodies. Cancer
Research. 69:1851–1857. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Ince WL, Jubb AM, Holden SN, et al:
Association of k-ras, b-raf, and p53 status with the treatment
effect of bevacizumab. J Natl Cancer Inst. 97:981–989. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Hurwitz HI, Yi J, Ince W, et al: The
clinical benefit of bevacizumab in metastatic colorectal cancer is
independent of K-ras mutation status: analysis of a phase III study
of bevacizumab with chemotherapy in previously untreated metastatic
colorectal cancer. Oncologist. 14:22–28. 2009. View Article : Google Scholar
|
|
26.
|
Richman SD, Seymour MT, Chambers P, et al:
KRAS and BRAF mutations in advanced colorectal cancer are
associated with poor prognosis but do not preclude benefit from
oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin
Oncol. 27:5931–5937. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Souglakos J, Philips J, Wang R, et al:
Prognostic and predictive value of common mutations for treatment
response and survival in patients with metastatic colorectal
cancer. Br J Cancer. 101:465–472. 2009. View Article : Google Scholar : PubMed/NCBI
|