Introduction
5-Fluorouracil (5-FU) is an anticancer agent widely
used in the treatment of several types of cancer, including
gastrointestinal, mammary, head and neck, non-small-cell lung and
oral cancers. The effects of 5-FU are closely associated with the
activity of its metabolic enzymes. The 5-FU-related enzymes, namely
thymidylate synthase (TS), dihydropyrimidine dehydrogenase (DPD),
orotate phosphoribosyl transferase (OPRT) and thymidine
phosphorylase (TP), play a significant role in the anticancer
effects of 5-FU (1–15). Therefore, evaluation of the mRNA
expression levels of these enzymes may determine the desirable
efficiency and concomitant side effects of 5-FU. From this point of
view, the correlation between enzyme expression and anticancer
effects has been extensively investigated (3,4,8,9,11,16).
Although there are controversial points, it was suggested that the
high expression of DPD in tumor tissues may reduce the anticancer
effects of 5-FU (8–10,17).
Cigarette smoking is a major risk factor for the
development of squamous cell carcinoma of the head and neck.
Additionally, alcohol consumption has long been known as a risk
factor for head and neck cancers (18). Previous evidence indicates that
patients with head and neck cancers, including oral cancers, are
highly likely to lead such lifestyles. Furthermore, it was reported
that lifestyle, particularly smoking and/or alcohol consumption,
may modulate drug metabolism through enzyme induction and/or
inhibition (19–21). However, there is currently no
available report regarding the effects of these lifestyle habits on
the status of 5-FU-related metabolic enzymes. In the present study,
in order to elucidate the effects of smoking and alcohol
consumption on the 5-FU-related metabolic enzymes, the expression
status of TS, DPD, TP and OPRT was investigated in oral squamous
cell carcinoma (OSCC) and adjacent normal tissues.
Materials and methods
Patient population
A total of 29 patients who underwent surgical
resection of primary OSCC at the Department of Oral and
Maxillofacial Surgery, Gifu University Hospital (Gifu, Japan)
between March, 2002 and October, 2004 were included in the present
study (Table I). In total, 15
patients were smokers and their average Brinkman index, calculated
by the numbers of cigarettes smoked per day multiplied by the
smoking years, was 784.5 (range, 140–2,100). Additionally, 13
patients had a habit of alcohol consumption and their average
number of drinking years was 41.2 (12–63 years).
 | Table I.Clinicopathological characteristics of
29 OSCC patients. |
Table I.
Clinicopathological characteristics of
29 OSCC patients.
| Characteristics | No. (%) |
|---|
| Age, years | |
| Mean ± SD | 68.8 ± 13.4 |
| Range | 39–91 |
| Gender | |
| Male | 16 (55.2%) |
| Female | 13 (44.8%) |
| T | |
| 1 | 5 (17.2%) |
| 2 | 13 (44.8%) |
| 3 | 5 (17.2%) |
| 4 | 6 (20.8%) |
| N | |
| 0 | 24 (82.8%) |
| 1 | 4 (13.8%) |
| 2 | 1 (3.4%) |
| Stage | |
| I | 5 (17.2%) |
| II | 10 (34.4%) |
| III | 8 (27.6%) |
| IV | 6 (20.8%) |
| Histopathological
grading | |
| G1 | 24 (82.8%) |
| G2 | 5 (17.2%) |
| Anneroth’s
classification | |
| I | 4 (13.8%) |
| II | 17 (58.6%) |
| III | 8 (27.6%) |
| Tumor location | |
| Buccal mucosa | 5 |
| Upper gum | 3 |
| Lower gum | 8 |
| Tongue | 7 |
| Floor of mouth | 6 |
| Smoking | |
| No | 14 (48.3%) |
| Yes | 15 (51.7%) |
| Brinkman index | |
| Mean ± SD | 784.5 ± 523.4 |
| Range | 140–2,100 |
| Drinking | |
| No | 16 (55.2%) |
| Yes | 13 (44.8%) |
| Drinking years | |
| Mean ± SD | 41.2 ± 16.6 |
| Range | 12–63 |
This study was approved by the Institutional Review
Board of Gifu University and all the participants provided written
informed consent.
RNA extraction and cDNA synthesis
Two tissue specimens, weighing ∼3 mg and measuring 3
mm in length each, were obtained from tumor and adjacent normal
tissues resected during surgery. Efforts were made to minimize the
time interval between surgery and freezing of the samples.
Total RNA was isolated using an RNeasy mini kit
(Qiagen, Inc., Chatsworth, CA, USA), and DNase treatment was
performed with the RNase-Free DNase set (Qiagen, Inc.) according to
the manufacturer’s instructions. Following RNA isolation, cDNA was
prepared from each sample using Super-Script II Reverse
Transcriptase (Invitrogen, Co., Carlsbad, CA, USA). Reverse
transcription with ≤4 μg of total RNA was performed in a
total volume of 1 μl containing 500 μg/ml oligo(dT)
in 250 mM Tris-HCl (pH 8.3), 375 mM KCl, 15 mM MgCl2,
0.1 M dithiothreitol and 1 μl deoxyribo-nucleotide
triphosphates. Initially, the total RNA solution and oligo(dT)
mixture was heated at 65°C for 5 min and immediately chilled on
ice, followed by the addition of the other reagents. First strand
cDNAs were obtained and dissolved in 20 μl distilled
water.
Quantitative reverse
transcription-polymerase chain reaction (qRT-PCR)
The mRNA levels of TS, DPD, OPRT and TP were
evaluated by qRT-PCR (TaqMan PCR) using an ABI Prism 7700 sequence
detector (Perkin-Elmer Applied Biosystems, Foster City, CA, USA).
The β-actin gene was used as an endogenous control gene. The
primers and TaqMan probes for each gene were designed based on the
nucleotide sequence of human TS, DPD, OPRT and TP (Table II). The PCR mixture contained 10
μl of each appropriately diluted cDNA sample (standard curve
points and patient samples), 200 nM forward primer, 200 nM reverse
primer, 100 nM TaqMan probe and 12.5 μl TaqMan Universal PCR
Master Mix (Perkin-Elmer Applied Biosystems) in a final volume of
25 μl. The PCR profile consisted of incubation at 94°C for 2
min, followed by incubation at 95°C for 10 min and 40 cycles of
amplification at 95°C for 15 s and at 60°C for 1 min. The
expression of the β-actin gene was used as an internal standard and
the relative gene expression was calculated by determining the
ratio between the amount of the PCR product of the 5-FU-related
metabolic enzyme genes and the β-actin gene.
 | Table II.Sequence of quantitative RT-PCR
primers and sequence-specific probes for target genes. |
Table II.
Sequence of quantitative RT-PCR
primers and sequence-specific probes for target genes.
| Target gene | Primer sequence
(5’→3’) |
|---|
| TS | |
| Sense |
ATTTACCTGAATCACATCGAGCC |
| Antisense |
TCGAAGAATCCTGAGCTTTGG |
| TaqManProbe |
FAM-AAAATTCAGCTTCAGCGAGAACCCAGACC-TAMRA |
| DPD | |
| Sense |
TCCCCAAAAGGCCTATTCCT |
| Antisense |
TGCTCAATTCACCAAATGTTCC |
| TaqManProbe |
FAM-CCATCAAGGATGTAATAGGAAAAGCACTGCAG-TAMRA |
| TP | |
| Sense |
GAGTCTATTCCTGGATTCAATGTCATC |
| Antisense |
CCCACGATACAGCAGCCC |
| TaqManProbe |
FAM-CAGATGCAAGTGCTGCTGGACCAGG-TAMRA |
| OPRT | |
| Sense |
ACTACACTAGAGCAGCGGTTAGAATG |
| Antisense |
AACTGAACTCCTGGAGTCAAGTGA |
| TaqManProbe |
FAM-TTCTGGCTCCCGAGTAAGCATGAAACC-TAMRA |
Statistical analysis
The association between smoking and drinking and the
mRNA levels of the 5-FU-related metabolic enzymes were compared
using the Kruskal-Wallis rank test. The mRNA levels of the
5-FU-related metabolic enzymes were compared by the Brinkman
index/drinking years using Pearson’s correlation coefficient.
P<0.05 was considered to indicate a statistically significant
difference for each analysis.
Results
Effect of smoking on mRNA expression of
5-FU-related meta- bolic enzymes
The P-values reflecting the correlation between the
Brinkman index and mRNA expression of the 5-FU-related metabolic
enzymes in tumor and adjacent normal tissues are summarized in
Table III. There was no
significant difference between the mRNA expression levels of TS and
TP in tumor and adjacent normal tissues, although there was a
significant positive correlation between the Brinkman index and the
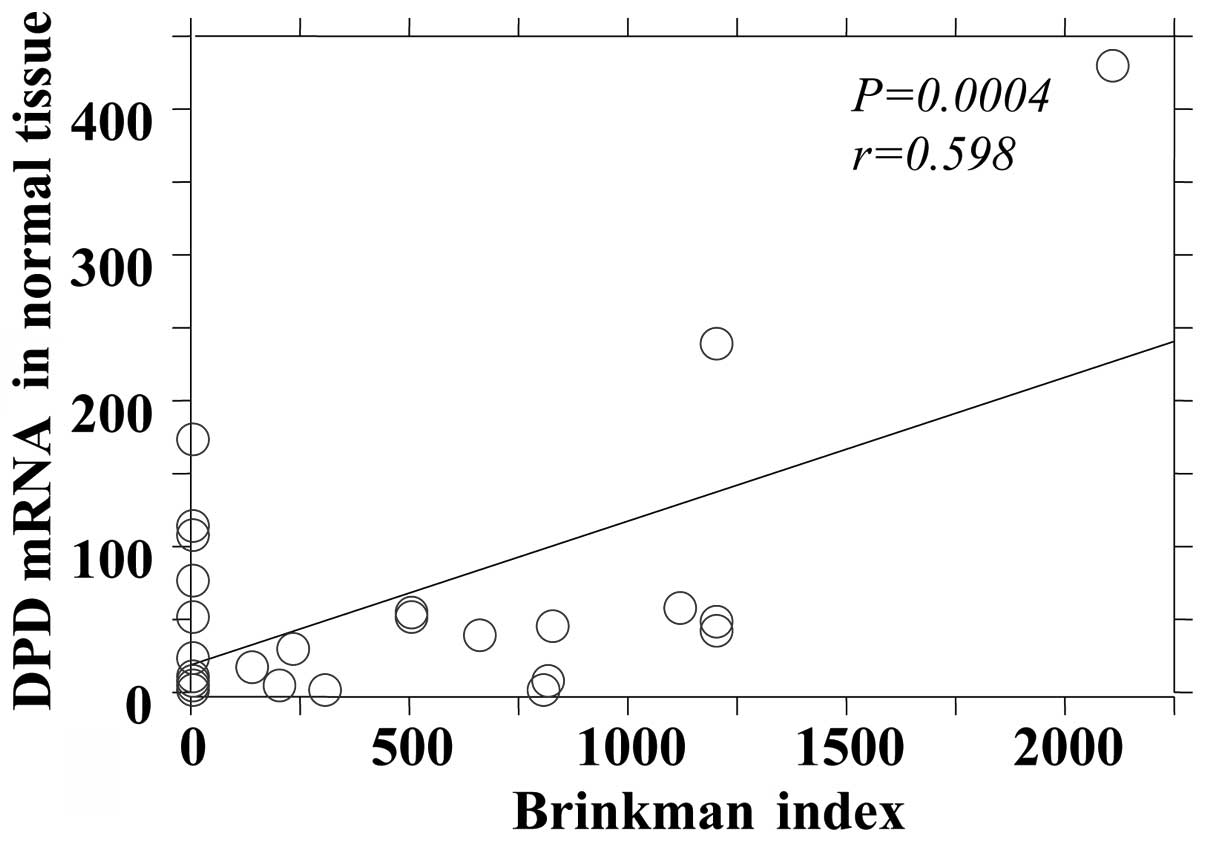
DPD mRNA expression in tumor and adjacent normal tissues (Figs. 1 and 2). The OPRT mRNA expression was found to
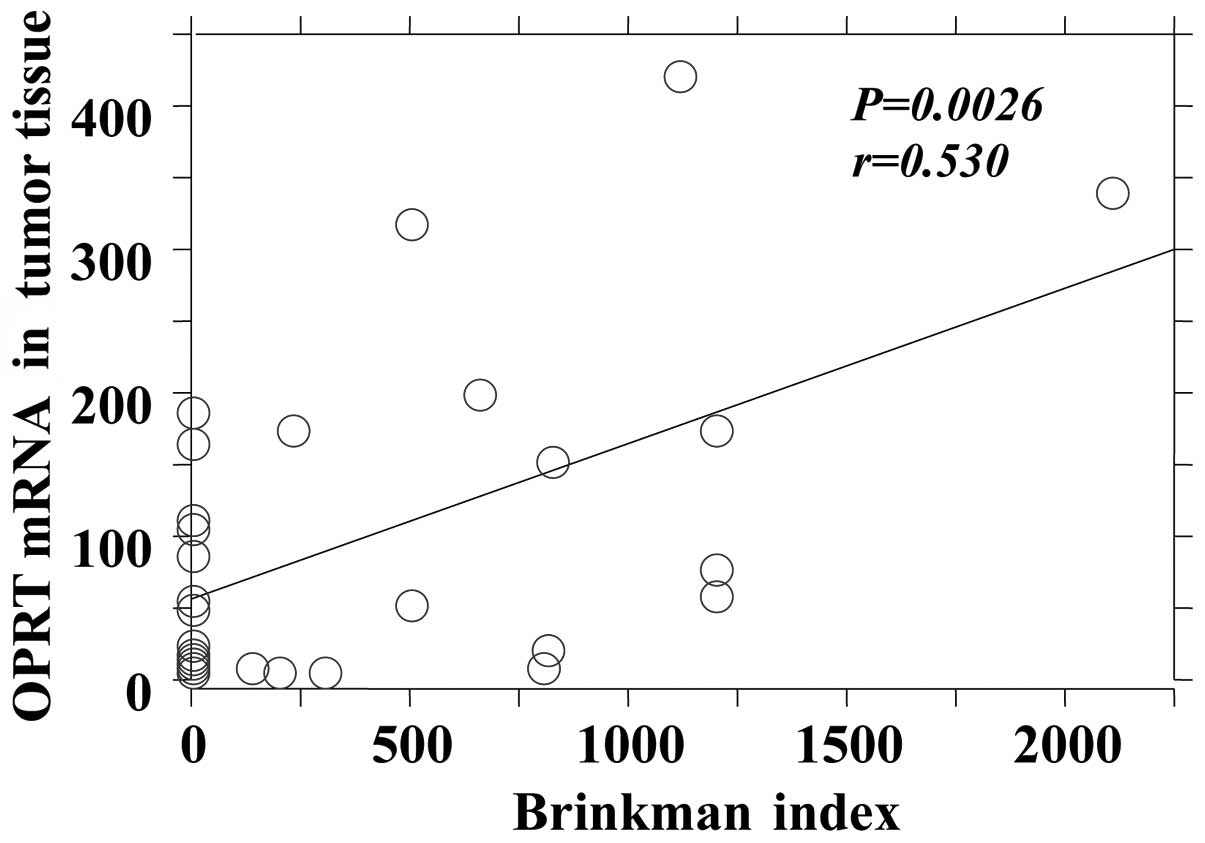
be significantly correlated with the Brinkman index in tumor
tissues only (Fig. 3).
 | Table III.P-values and correlation coefficients
between the Brinkman index and the mRNA expression of 5-FU-related
metabolic enzymes. |
Table III.
P-values and correlation coefficients
between the Brinkman index and the mRNA expression of 5-FU-related
metabolic enzymes.
| Enzymes | P-value | Correlation
coefficient |
|---|
| TS | | |
| Tumor | 0.5289 | 0.123 |
| Normal | 0.8253 | 0.048 |
| DPD | | |
| Tumor | 0.0005 | 0.593 |
| Normal | 0.0004 | 0.598 |
| OPRT | | |
| Tumor | 0.0026 | 0.530 |
| Normal | 0.097 | 0.314 |
| TP | | |
| Tumor | 0.3338 | 0.187 |
| Normal | 0.5064 | 0.130 |
Effect of alcohol consumption on mRNA
expression of 5-FU-related metabolic enzymes
The P-values reflecting the correlation between
drinking years and the mRNA expression of the 5-FU-related
metabolic enzymes in tumor and adjacent normal tissues are
summarized in Table IV. There was
no significant difference between the mRNA expression levels of TS,
OPRT and TP in tumor and adjacent normal tissues, although there
was a significant positive correlation between drinking years and
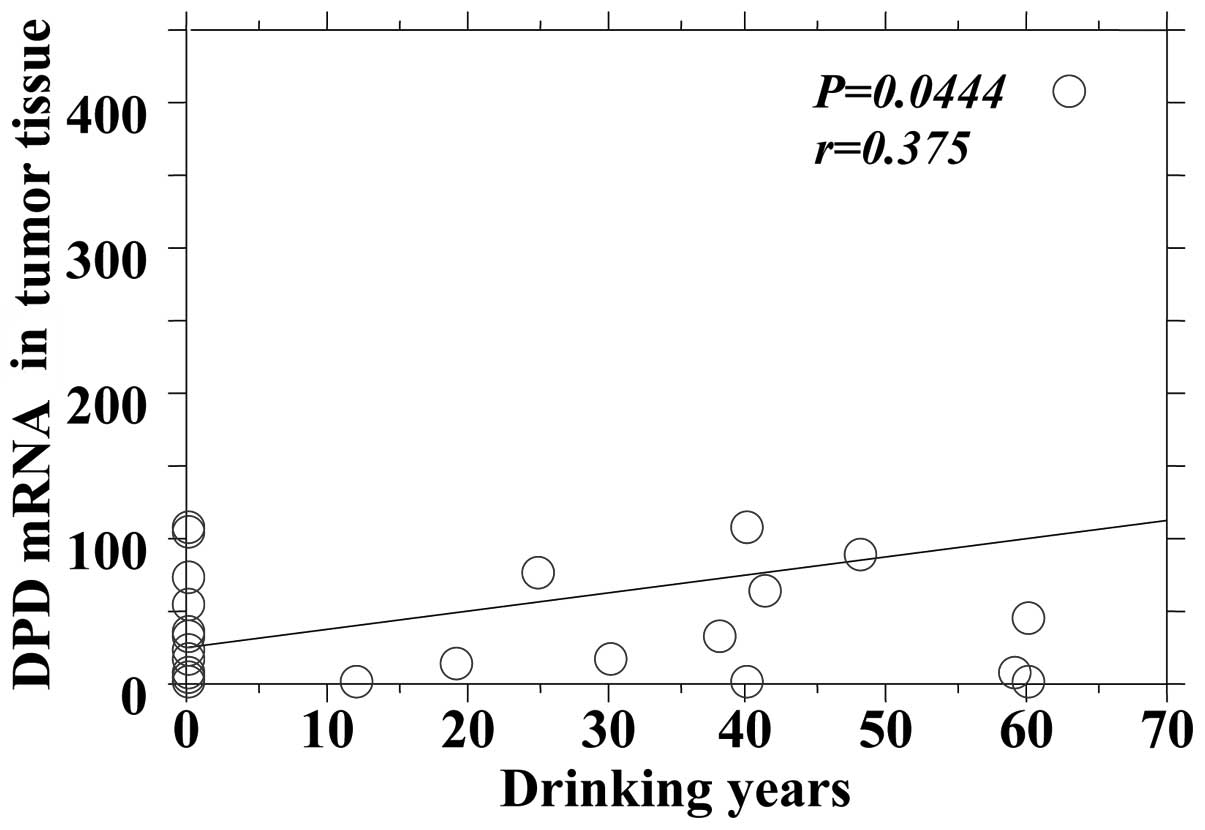
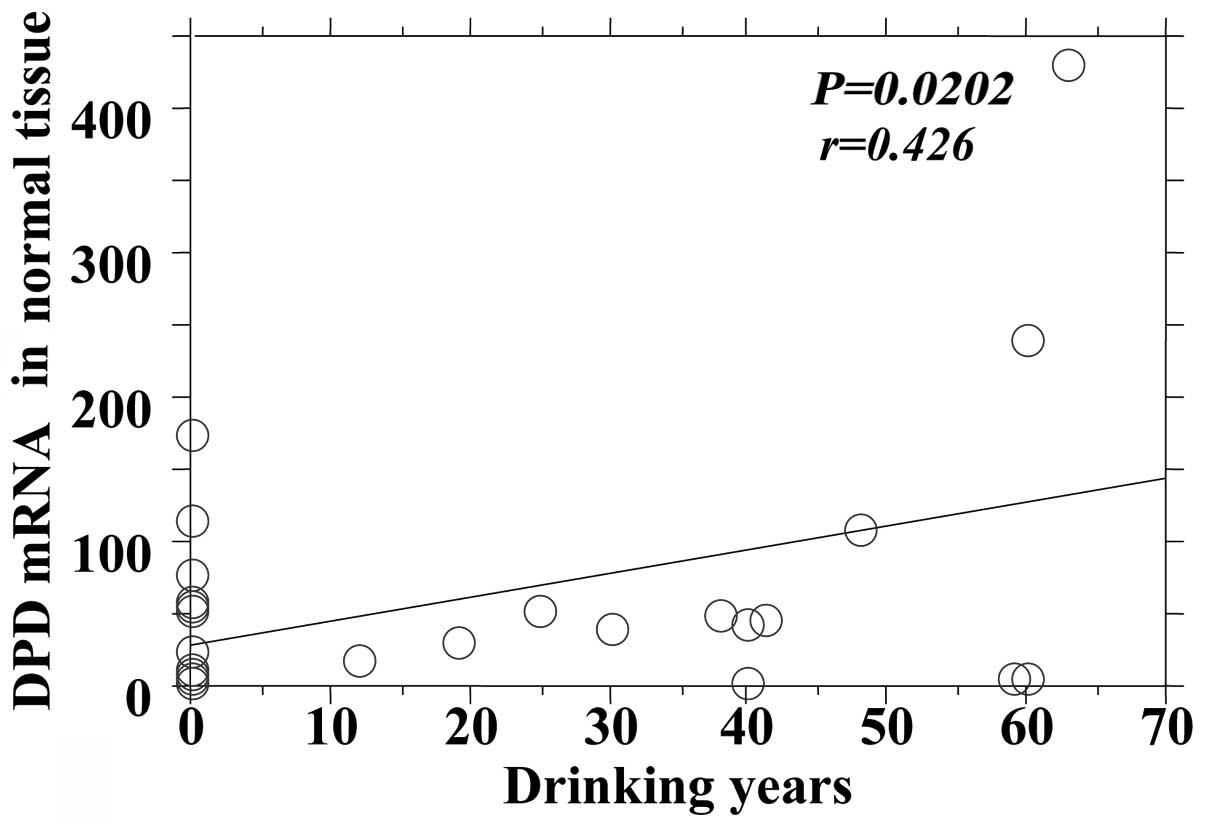
the DPD mRNA expression in tumors and adjacent normal tissues
(Figs. 4 and 5).
 | Table IV.P-values and the correlation
coefficients between the drinking years and the mRNA expression of
5-FU-related metabolic enzymes. |
Table IV.
P-values and the correlation
coefficients between the drinking years and the mRNA expression of
5-FU-related metabolic enzymes.
| Enzymes | P-value | Correlation
coefficient |
|---|
| TS | | |
| Tumor | 0.6687 | 0.084 |
| Normal | 0.3528 | 0.818 |
| DPD | | |
| Tumor | 0.0444 | 0.375 |
| Normal | 0.0202 | 0.426 |
| OPRT | | |
| Tumor | 0.2879 | 0.205 |
| Normal | 0.3902 | 0.167 |
| TP | | |
| Tumor | 0.1286 | 0.290 |
| Normal | 0.9183 | 0.020 |
Discussion
5-FU is metabolized into two different active forms,
5-fluorodeoxyuridine monophosphate (FdUMP) and 5-fluorouridine
triphosphate, which inhibit DNA synthesis and RNA function,
respectively. FdUMP and its coenzyme, 5,10-methylene
terahydrofolate, form a covalent ternary complex with the
DNA-synthesizing enzyme TS (1).
This complex inhibits the conversion of deoxyuridine monophosphate
to deoxythymidine monophosphate, which is an essential step of DNA
synthesis, thus exerting anticancer effects (2). Consequently, the over-expression of
TS possibly decreases the inhibitory effect of 5-FU on DNA
synthesis; several studies suggested that high amounts of TS in
tumor cells may decrease the efficiency of 5-FU and lead to the
development of drug resistance (3–7).
DPD is the initial and rate-limiting enzyme in the
catabolic pathway of 5-FU. Therefore, DPD may reduce the anticancer
effects of 5-FU in tumor cells and several studies indicated that
the level of DPD expression is significantly correlated with
resistance to 5-FU (8–10).
OPRT is the first key enzyme in the phosphorylation
of 5-FU, converting 5-FU to 5-fluorouridine monophosphate. OPRT is
considered to predominantly inhibit RNA synthesis. Thus, it was
reported that high amounts of intratumoral OPRT are correlated with
sensitivity to 5-FU (11–13).
TP catalyzes the reversible conversion of 5-FU to
5-deoxy-5-fluorouridine and high expression of this enzyme in
tumors is correlated with a high response rate to
5-deoxy-5-fluorouridine (14,15)
(Fig. 1). Thus, the anticancer
effects of 5-FU are closely correlated with the activity of its
metabolic enzymes, which have been extensively investigated in oral
cancers (3,4,8,9,11,16).
A previous clinical study demonstrated that
neoadjuvant chemotherapy may affect the 5-FU-related metabolic
enzyme status, although the effects of lifestyle habits, including
smoking and alcohol consumption, on 5-FU-related metabolic enzymes
were not investigated (22). The
results of the present study on mRNA analysis indicated that mRNA
expression of TS, TP and OPRT in tumor and adjacent normal tissues
was not affected by smoking and alcohol consumption. However, the
mRNA expression of DPD was dependently enhanced in tumor and
adjacent normal tissues by both the Brinkman index and alcohol
drinking years. DPD is the initial and rate-limiting enzyme that
catabolizes 5-FU in the liver. Therefore, DPD in tumor cells may
reduce the anticancer effects of 5-FU and several studies reported
that the levels of DPD expression are significantly correlated with
resistance to 5-FU (8–10,17).
Lifestyle habits, including cigarette smoking and
alcohol consumption, are known to affect drug treatment through
pharmacokinetic and pharmacodynamic mechanisms (19–21,23).
Cigarette smoking contains thousands of different compounds, some
of which are known carcinogens. In addition, exposure to these
compounds may facilitate the development of detoxification enzyme
systems and cigarette smoking is a known enzyme inducer (19). Smoking may reduce the intake and
serum concentrations of nutrients, such as β-carotene and vitamin
A, which are known to protect against the development of epithelial
cancers. Smoking is also associated with an increased blood
carboxyhemoglobin concentration, which causes a leftward shift in
the hemoglobin-oxygen dissociation curve, leading to relative
tissue hypoxia and alterations in the enzyme status (23).
Alcohol is metabolized through several processes or
pathways in the liver. The alcohol dehydrogenase pathway, the
microsomal ethanol oxidizing system and the catalase pathway, play
significant roles in alcohol metabolism. The alcohol molecule is
metabolized to aldehyde and, subsequently, to acetate, which is
broken down into water and carbon dioxide and easily eliminated
from the body. As regards the effect of alcohol on these enzymes,
it was previously reported that alcohol interacts with certain
membrane-associated signal transduction systems, resulting in
positive and/or negative regulation of adenylate cyclase,
phospholipase C and phospholipase A2; furthermore, it was reported
that alcohol may repress TS and thymidine kinase activity following
hepatectomy (20,21).
In the present study, the DPD mRNA expression in
tumor and adjacent normal tissues was found to be correlatively
enhanced by the Brinkman index and drinking years. It is known that
the DPD status corresponds to the alternations in the 5-FU
pharmacokinetic profile. For example, DPD deficiency may lead to
severe 5-FU-associated toxicities and, in theory, a DPD excess may
reduce the therapeutic effects of 5-FU. It is suggested that the
diversities in the DPD status originate in individual DPD genomic
polymorphism and may be modulated by smoking and alcohol
consumption, which may affect DPD functionality in the liver.
The exact mechanism underlying the modulation of the
5-FU metabolic enzymes by lifestyle habits has not been fully
elucidated and further investigation is required, including a
clinical evaluation of the 5-FU efficiency and accompanying side
effects. However, the results of the present study suggest that
smoking and alcohol consumption may reduce the anticancer activity
of 5-FU, possibly through inducing DPD activity. Cigarette smoking
and alcohol consumption have long been known as risk factors for
head and neck cancers, including oral cancers. It is suggested that
a significant proportion of oral cancer patients follow this
lifestyle, thus enhancing DPD activity, which subsequently leads to
a reduction in 5-FU efficiency. S-1 is a novel, orally administered
anticancer agent that is a combination of tegafur, gimeracil and
oterasil potassium. Gimeracil is a competitive inhibitor of DPD and
maintains efficacious 5-FU concentrations in the plasma and tumor
tissues. Therefore, anticancer drugs including DPD inhibitors, such
as S-1, may be recomended for cancer patients exhibiting smoking
and/or alcohol consumption habits.
Abbreviations:
|
OSCC
|
oral squamous cell carcinoma;
|
|
TS
|
thymidylate synthase;
|
|
DPD
|
dihydropyrimidine dehydrogenase;
|
|
OPRT
|
orotate phosphoribosyl
transferase;
|
|
TP
|
thymidine phosphorylase;
|
|
5-FU
|
5-fluorouracil;
|
|
UFT
|
tegafur/uracil
|
Acknowledgements
This study was supported in part by
Grants-in-Aid (2059.2324) for Scientific Research from the Ministry
of Education, Culture, Sports, Science and Technology of Japan.
References
|
1.
|
Danenberg PV: Thymidylate synthase - a
target enzyme in cancer chemotherapy. Biochem Biophys Acta.
473:73–92. 1977.
|
|
2.
|
Santi DV, MacHenry CS and Sommer H:
Mechanism of interaction of thymidylate synthetase with
5-fluorodeoxyuridylate. Biochemistry. 13:471–481. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Harada K, Kawashima Y, Yoshida H and Sato
M: Thymidylate synthase expression in oral squamous cell carcinoma
predicts response to S-1. Oncol Rep. 15:1417–1423. 2006.PubMed/NCBI
|
|
4.
|
Kawano K, Goto H, Kanda T and Yanagisawa
S: Predictive value of immunohistochemical thymidylate synthase
expression for histological response to tegafur/uracil (UFT) in
oral squamous cell carcinoma. Int J Oral Maxillofac Surg.
32:633–637. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Leichman CG, Lenz HJ, Leichman L, et al:
Quantitation of intratumoral thymidylate synthase expression
predicts for disseminated colorectal cancer response and resistance
to protracted-infusion fluorouracil and weekly leucovorin. J Clin
Oncol. 15:3223–3229. 1997.
|
|
6.
|
Beck A, Etienne MC, Chéradame S, et al: A
role for dihydropyrimidine dehydrogenase and thymidylate synthase
in tumor sensitivity to fluorouracil. Eur J Cancer. 30:1517–1522.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Lenz HJ, Leichman CG, Danenberg KD, et al:
Thymidylate synthase mRNA level in adenocarcinoma of the stomach: a
predictor for primary tumor response and overall survival. J Clin
Oncol. 14:176–182. 1996.PubMed/NCBI
|
|
8.
|
Hoque M, Kawamata H, Nakashiro K, et al:
Dihydropyrimidine dehydrogenase mRNA level correlates with the
response to 5-fluorouracil-based chemo-immuno-radiation therapy in
human oral squamous cell cancer. Int J Oncol. 19:953–958. 2001.
|
|
9.
|
Kobayashi H, Koike T, Nakatsuka A, et al:
Dihydropyrimidine dehydrogenase expression predicts survival
outcome and chemo-sensitivity to 5-fluorouracil in patients with
oral squamous cell carcinoma. Oral Oncol. 41:38–47. 2005.
View Article : Google Scholar
|
|
10.
|
Etienne MC, Chéradame S, Fischel JL, et
al: Response to fluorouracil therapy in cancer patients: the role
of tumoral dihydropyrimidine dehydrogenase activity. J Clin Oncol.
13:1663–1670. 1995.PubMed/NCBI
|
|
11.
|
Yoshitomi I, Kawasaki G, Yanamoto S and
Mizuno A: Orotate phosphoribosyl transferase mRNA expression in
oral squamous cell carcinoma and its relationship with the
dihydropyrimidine dehydrogenase expression and the clinical effect
of 5-fluorouracil. Oral Oncol. 42:880–887. 2006. View Article : Google Scholar
|
|
12.
|
Peters GJ, Laurensse E, Leyva A, Lankelma
J and Pinedo HM: Sensitivity of human, murine, and rat cells to
5-fluorouracil and 5’-deoxy-5-fluorouridine in relation to
drug-metabolizing enzymes. Cancer Res. 46:20–28. 1986.
|
|
13.
|
Peters GJ, Braakhuis BJ, de Brujin EA,
Laurensse EJ, van Walsum M and Pinedo HM: Enhanced therapeutic
efficacy of 5’-deoxy-5-fluorouridine in 5-fluorouracil-resistant
head and neck tumors in relation to 5-fluorouracil metabolizing
enzymes. Br J Cancer. 56:327–334. 1989.
|
|
14.
|
Koizumi W, Saigenji K, Nakamaru N, Okayasu
I and Kurihara M: Prediction of response to 5’-deoxy-5fluorouridine
(5’-DFUR) in patients with inoperable advanced gastric cancer by
immunostaining of thymidine phosphorylase/platelet-derived
endothelial cell growth factor. Oncology. 56:215–222. 1999.
|
|
15.
|
Sawada N, Ishikawa T, Fukase Y, Nishida M,
Yoshikubo T and Ishitsuka H: Induction of thymidine phosphorylase
activity and enhancement of capecitabine efficacy by taxol/taxotere
in human cancer xenografts. Clin Cancer Res. 4:1013–1019.
1998.PubMed/NCBI
|
|
16.
|
Kawasaki G, Yoshitomi I, Yamamoto S and
Mizuno A: Thymidylate synthase and dihydropyrimidine dehydrogenase
expression in oral squamous cell carcinoma: An immunohistochemical
and clinicopathologic study. Oral Surg Oral Med Oral Pathol Radiol
Endod. 94:717–723. 2002. View Article : Google Scholar
|
|
17.
|
Yang CG, Ciccolini J, Blesius A, et al:
DPD-based adaptive dosing of 5-FU in patients with head and neck
cancer: impact on treatment efficacy and toxicity. Cancer Chemother
Pharmacol. 67:49–56. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Freedman ND, Schatzkin A, Leitzmann MF,
Hollenbeck AR and Abent CC: Alcohol and head and neck cancer risk
in a prospective study. Br J Cancer. 96:1469–1474. 2007.PubMed/NCBI
|
|
19.
|
Browman GP, Wong G, Hodson I, et al:
Influence of cigarette smoking on the efficacy of radiation therapy
in head and neck cancer. N Engl J Med. 328:159–163. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Yoshida Y, Komatsu M, Ozeki A, Nango R and
Tsukamoto I: Ethanol represses thymidylate synthase and thymidine
kinase at mRNA level in regenerating rat liver after partial
hepatectomy. Biochim Biophys Acta. 1336:180–186. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Lieber CS: Ethanol metabolism, cirrhosis
and alcoholism. Clin Chim Acta. 257:59–84. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Yamashita T, Toida M, Kato K, et al: The
effect of neoadjuvant therapy on the 5-fluorouracil metabolic and
relative enzymes of oral squamous cell carcinoma. Oncol Rep.
22:501–507. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Zevin S and Benowitz NL: Drug interactions
with tobacco smoking. An update. Clin Pharmacokinet. 36:425–438.
1999. View Article : Google Scholar : PubMed/NCBI
|