Introduction
Asthma is a major global health concern, primarily
resulting from interactions between genetic and environmental
factors. A significant association was previously demonstrated
between single-nucleotide polymorphisms (SNPs) and haplotypes of
the a disintegrin and metalloprotease 33 (ADAM33) gene and asthma
in ethnically diverse populations (1). An association with ADAM33 variants in
adult-onset asthma has been reported in several populations
(2–4). However, the majority of the currently
available data on ADAM33 and asthma have been obtained from
Caucasians, whereas the availability of data on ADAM33 SNP
association with asthma in Asian populations is limited. Certain
studies reported no significant association between SNPs and asthma
susceptibility in the Chinese and other populations (5,6). The
precise biological role of ADAM33 gene polymorphisms in asthma
remains to be elucidated. Sharma et al (6) recently reviewed the association of
all investigated ADAM33 gene polymorphisms with asthma worldwide
and concluded that causal polymorphisms have yet to be identified.
Data from different populations are available, demonstrating the
association between ADAM33 gene polymorphisms and asthma; however,
no study has yet demonstrated an association of SNPs T2 and ST+5
with asthma in the Chinese Uyghur population.
Xinjiang is located at the middle of the Silk Road
that once extended from Rome to China. Several ethnicities, such as
the Uyghur (48%), Han (38%) and Kazakh people (7%), reside in this
area (7). The Uyghurs are a
Mongoloid-Caucasoid mixed population living in Northwest China who
are mainly herdsmen or peasants in pastures. The Uyghur religion,
lifestyle and customs differ from those of other Chinese ethnic
groups; they live under relatively fixed conditions over a long
period of time, which favors the development of a specific
population and the stability of certain genes. It was previously
demonstrated that the Uyghur allele distribution differs distinctly
from that of other populations in Northwest China, such as Han, Hui
and Mongolian.
The present study was conducted to investigate the
role of ADAM33 SNPs T2 and ST+5 in asthma in the Chinese Uyghur
population of Xinjiang.
Materials and methods
Subjects
This study included 183 unrelated adults with a
clinical diagnosis of asthma (mean age, 42.93±13.48 years; range,
18–80 years) and 155 healthy control subjects (mean age,
41.14±14.07 years; range, 18–71 years). The subjects were recruited
between February, 2012 and May, 2013 at the Department of
Respiratory Medicine, Xinjiang Medical University, Xinjiang,
Urumuqi, China. There were no statistically significant differences
in age and gender between the two groups (P>0.05). The diagnosis
of suspected asthma was confirmed and the severity of asthma
attacks was assessed according to the 2008 revision of the
diagnostic criteria of the Chinese Medical Association for
Respiratory Diseases (Asthma Study Group ) (8).
All the subjects met the following criteria: i) no
family history of diseases involving the candidate genes, such as
immune system diseases; ii) no other severe diseases, such as
severe infections, new-onset myocardial infarction or stroke, or
history of atopy; and iii) long-term residence in the Xinjiang area
(third generation or more).
The study protocol was reviewed and approved by the
Ethics Committee of Xinjiang Medical Hospital and written informed
consent was obtained from all the participants.
Reagents and instruments
The Genomic DNA Isolation kit (catalog no., SK8224)
was purchased from Sangon Biotech Co., Ltd. (Shanghai, China). The
PCR reaction contained TaqDNA polymerase, 10X polymerase chain
reaction (PCR) buffer without Mg2+ [100 mM Tris-HCl, pH
8.8 at 25°C; 500 mM KCl; 0.8% (v/v) Nonidet], 25 mM magnesium
chloride (MgCl2), and 10 mM deoxyribonucleoside
triphosphates (dNTPs), all purchased from Sangon Biotech Co. The
DNA marker was purchased from BBI Canada Inc. (Markham, ON,
Canada), and the 6X DNA Loading dye from Sangon Biotech Shanghai,
China). The PCR products were purified on with the DNA extraction
kit (catalog SK1141; Sangon Biotech), and the restriction
endonuclease for the digestion of DNA was purchased from MBI
Fermentas (Beijing, China). The PCR amplification reaction was
performed using a PCR thermocycler (BBI Canada Inc.) on a SW-CJ-1D
clean bench (Jiang Su Sujie purification plant). A DK-8D-type
electric heated tank (Hysen letter Experimental Instrument Co.) was
used to digest the sample. Additional instruments used in this
study were: a DYY-8 regulator steady flow electrophoretic to
maintain a stable electric volume (Hai Qite Analytical Instruments
Co., Ltd., Dalian, China); a YXJ-2 centrifuge machine (Gordon Ltd.,
Shanghai, China). a H6-1 micro-electrophoresis tank (Shanghai
Xingyi Plexiglas Co., Ltd., Shanghai, China); a Gene Genius gel
imaging system by Alibaba Co. (Beijing, China); a U-3010 UV
spectrophotometer (Hitachi Co., Tokyo, Japan); and pipettes (range,
100–1,000, 20–200, and 0.5–10 μl) by BBI Canada Inc.
Genomic DNA extraction
Peripheral venous whole-blood samples (2 ml) from
all the subjects were collected in EDTA anticoagulant tubes and
stored at −80°C. The Genomic DNA Isolation kit (SK8224; Sangon
Biotech, Co., Ltd.) was used for genomic DNA extraction according
to the manufacturer’s protocol.
ADAM33 genotyping by polymerase chain
reacytion fragment length polymorphism
The preliminary determination of genetic samples
were performedby Bioengineering Co., Ltd. (Shanghai, China). The
primers used were as follows: T2 (rs2280090) forward (F),
5-AGGCTTTGAATCCAGGTCC-3, and reverse (R), 5-GGCAATAACCCACTCAGGAT-3;
ST+5 (rs597980) F, 5-CTCTGTCTCACCAGTTTTCGG-3, and R,
5-TTATGTGACTCCCCACTCCG-3. The PCR reaction for the amplification of
T2 and ST+5 was performed on atotal volume of 15 μl, containing 0.5
μl of DNA, 1.5 μl of 10X TaqDNA polymerase buffer, 0.3 μl of dNTPs,
0.1 μl of TaqDNA polymerase, 1.2 μl of MgCl2, 0.4 μl of
primers, TaqDNA polymerase 0.1 μl, and 11 μl of double distilled
water. The cycling conditions were: initial denaturation at 95°C
for 5 min; 40 cycles of denaturation at 95°C for 30 sec; annealing
at 68/67.5°C for 45 sec; and extension at 72°C for 60 sec; final
extension at 72°C for 6 min. The digestion reaction was performed
in a total volume of 15 μl, containing contain 10 μl of the PCR
products, 1.5 μl of the 10X buffer R (Received data buffer), 0.1 μl
of endonuclease (both from Sangon Biotech Co.), and 3.4 μl of
double distilled water. The samples were digested overnight at
37°C.
Electrophoresis
The digestion products were run on a 3% agarose gel
containing ethidium bromide at a voltage of 150 V for 30 min. Band
detection was then performed on a gel imaging system. Based on the
acquired images, genotyping data were then entered into a NCBI
database.
ADAM33 gene (T2 and ST+5) and its
association with lung function
Of the 183 patients carrying the ADAM33 gene, 149
were asthmatic. The percentage of predicted forced expiratory
volume in 1 sec (FEV1%) and percentage of predicted forced vital
capacity (FVC%) were compared between the two groups,
respectively.
Statistical analyses
Statistical analysis was performed using SPSS
software, version 17.0 (SPSS Inc., Chicago, IL, USA). The
Hardy-Weinberg equilibrium (HWE) was calculated using the
χ2 test, with expected frequencies derived from the
actual frequencies of each SNP in both groups. Measurement data
results are expressed as means ± standard deviation. Statistical
significance was set at P=0.05. The two groups of genotypes were
compared using analysis of variance, applying the LSD t-test.
Results
Subject characteristics
As shown in Table
I, there were no significant differences in gender and age
between the two groups. As regards lung function tests, there was a
significant difference in FEV1% between asthmatic patients and
healthy controls (70.96±17.44 vs. 104.21±8.20, P<0.05).
Conventional risk factors, including a history of allergy and a
family history of asthma, were higher among asthma patients.
 | Table IDescription of study population. |
Table I
Description of study population.
| Characteristics | Cases | Control |
|---|
| No. of patients | 183 | 155 |
| Age, years (mean ±
SD) | 42.93±13.48 | 41.14±14.07 |
| Gender, no. (%) |
| Male | 73 (39.89) | 66 (42.58) |
| Female | 110 (60.11) | 89 (57.42) |
| FEV1% (mean ±
SD) | 70.96±17.44 | 104.21±8.20 |
Product and restriction fragment length
polymorphism analysis of ADAM33 gene amplified by PCR
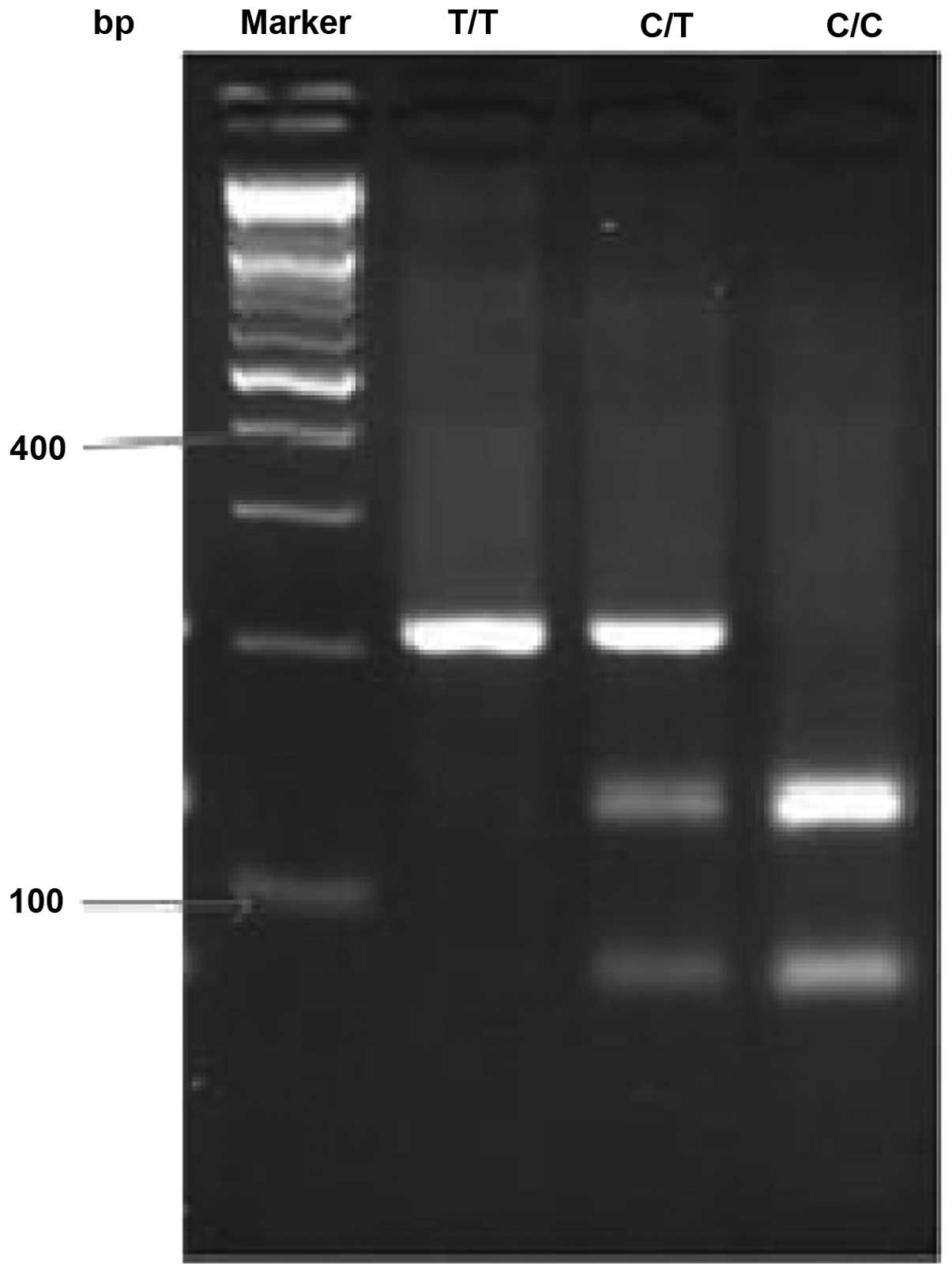
The ST+5 polymorphism genotype is for the C/T,
following the PCR, the product was 206-bp in length. The digestion
of the PCR product yielded 206-bp bands in TT homozygotes, 128- and
78-bp bands in CC homozygotes and all three bands in heterozygotes
(Fig. 1). The T2 polymorphism
genotype is for the A/G, following the PCR, the product was 425-bp
in length. The digestion of the product yielded 402- and 23-bp
bands in GG homozygotes, 289-, 113- and 23-bp bands in AA
homozygotes and all four bands in heterozygotes (Fig. 2).
Distribution of ADAM33 gene
polymorphisms
As regards the T2 and ST+5 polymorphisms, the
asthmatic patients (χ2=1.338 and 3.663, respectively;
P>0.05) and healthy controls (χ2=3.601 and 3.353,
respectively; P>0.05) were in HWE. There were no significant
differences in the distribution of the genotypes and alleles of
ADAM33 T2 (P=0.291) and ST+5 (P=0.501) between asthmatic patients
and healthy controls (Table
II).
 | Table IIGenotype and allele distribution of
ADAM33 gene polymorphisms. |
Table II
Genotype and allele distribution of
ADAM33 gene polymorphisms.
| Genotype | Patient no.
(n=183) | Control no.
(n=155) | χ2 | P-value | Allele | Patient no.
(n=366) | Control no.
(n=310) | χ2 | P-value |
|---|
| T2 |
| AA | 4 | 6 | 0.830 | 0.362 | A | 42 | 44 | 1.117 | 0.291 |
| AG | 34 | 32 | 0.588 | 0.443 | G | 324 | 266 | | |
| GG | 145 | 117 | 0.667 | 0.410 | | | | | |
| ST+5 |
| TT | 37 | 35 | 0.276 | 0.599 | C | 216 | 175 | 0.453 | 0.501 |
| CT | 76 | 65 | 0.006 | 0.940 | T | 150 | 135 | | |
| CC | 70 | 55 | 0.279 | 0.597 | | | | | |
Haplotype frequencies of the ADAM33
gene
The haplotype structure was analyzed using two SNPs
(T2 and ST+5) and nine haplotypes were identified in the two groups
(Table III). There was no
significant difference in the distribution of haplotypes between
asthmatic patients and healthy controls.
 | Table IIIFrequencies of SNP haplotypes. |
Table III
Frequencies of SNP haplotypes.
| SNP position | Haplotype
frequency | | |
|---|
|
|
| | |
|---|
| Haplotype | T2 | ST+5 | Patient no.
(n=183) | Control no.
(n=155) | χ2 | P-value |
|---|
| 1 | GG | TT | 37 | 34 | | |
| 2 | AG | TT | 0 | 1 | 1.072 | 0.300 |
| 3 | AA | TT | 0 | 0 | - | - |
| 4 | GG | CC | 48 | 33 | 0.784 | 0.376 |
| 5 | AG | CC | 18 | 15 | 0.054 | 0.817 |
| 6 | AA | CC | 4 | 5 | 0.188 | 0.665 |
| 7 | GG | CT | 59 | 48 | 0.157 | 0.692 |
| 8 | AG | CT | 17 | 18 | 0.118 | 0.732 |
| 9 | AA | CT | 0 | 1 | 1.072 | 0.300 |
Association of the ADAM33 phenotypes with
FEV1% and FVC%
As regards the T2 and ST+5 polymorphisms (Table IV), there was no significant
difference in FEV1% and FVC% among the different genotypes.
 | Table IVGenotype distribution of ADAM33 gene
polymorphisms among asthmatic patients.a |
Table IV
Genotype distribution of ADAM33 gene
polymorphisms among asthmatic patients.a
| Genotype | Patient no.
(n=149) | FEV1% | FVC% |
|---|
| T2 |
| AA | 4 | 74±31 | 86±25 |
| AG | 29 | 74±15 | 85±13 |
| GG | 116 | 70±17 | 86±15 |
| F-value | | 0.763 | 1.164 |
| P-value | | 0.468 | 0.315 |
| ST+5 |
| TT | 65 | 72±17 | 84±14 |
| CT | 57 | 70±17 | 78±15 |
| CC | 27 | 70±20 | 81±18 |
| F-value | | 0.222 | 2.233 |
| P-value | | 0.802 | 0.111 |
Discussion
In this study, we analysed the T2 and ST+5
polymorphisms of the ADAM33 gene to investigate their association
with asthma. This association was previously investigated in Asian
populations with inconclusive results (9,10).
Over the last few years, a number of studies demonstrated that
ADAM33 SNPs are significantly associated with asthma in different
populations, including the Han Chinese; however, no single SNP of
the ADAM33 gene (T2 and ST+5) was found to be associated with
asthma across all the studies published thus far in the Uyghur
population. We did not identify any association of individual SNPs
with the presence of asthma or its severity.
The ADAM33 gene is located on chromosome 20p13 and
37 SNPs were initially identified (11). The identification of ADAM33 as an
asthma susceptibility gene marks the entry of asthma research into
the genomic area and provides a new insight to the understanding of
the molecular basis underlying the pathogenesis of asthma. The
heterogeneity of this disease may be attributed to different
mutations within the gene and a combined effect of gene-gene and
gene-environment interactions (12,13).
Another possible reason may be that the linkage disequilibrium
patterns that exist between the identified SNPs in the gene may
differ among different populations (9). Four SNPs (F+1, S1, ST+5 and V4) were
previously found to be significantly associated with specific
airway resistance at the age of 5 years (P<0.04) when children
aged 3 and 5 years were investigated, but no study reported a
positive association with T2 and ST+5 in older populations
(9). The polymorphisms were found
to be associated with reduced lung function at the ages of 3 and 5
years, with a more significant association with the age of 5 years,
which may reflect the increased sample size. ADAM33 polymorphisms
were originally shown to be associated with bronchial
hyperresponsiveness. In previous studies, T1, T2, S2 and V-3 sites
were reported among Japanese adult populations (14,15),
whereas they were negative in this study. Minor alleles of the
ADAM33 SNPs S+1, ST+4 and T2 were overtransmitted to the
asthma-affected offspring, unlike any particular haplotype. In a
study on Chinese subjects, adult asthma was associated with ADAM33
SNPs V4, T2, T1 and Q-1, but not with T+1 and S1, or with V4 and
T+1, whereas the association with T1 sites was found to be
significant (16), which proves
that there is a difference in genetic association among different
population in China, particularly between the Han and Uyghur
Chinese people. The potential mechanism underlying the association
of the T1 locus SNP with asthma may be that the T1 locus SNP leads
to decreased FEV1 and increased susceptibility to asthma. There was
no significant difference in the FVC and the FEV1 between patients
with ADAM33 gene genotypes T2 and ST+5 in this study, as shown in
Table IV.
A possible role for the ADAM33 gene in childhood
asthma was excluded in a Mexican population and in a Caucasian
population of North America (17,18).
A particular subset of SNPs may represent a risk factor for asthma
in Caucasians, whereas a different subset may increase the risk in
Asian populations. However, no SNP has been consistently associated
with asthma across ethnically diverse groups, similar to this
study. Therefore, different SNPs of the ADAM33 gene may contribute
to asthma susceptibility in specific ethnic populations. Additional
studies with a larger sample size and long-term follow-up may lead
to a more thorough understanding of the role of the T2 and ST+5
ADAM33 gene polymorphisms in the Uyghur population. Considering
that the limited sample size may yield relative risk estimates
lacking adequate precision, extended analyses with a larger sample
size should be performed on patients of different ethnic origins to
further verify this association.
In conclusion, our results revealed no association
between the T2 and ST+5 ADAM33 gene polymorphisms and asthma in the
Chinese Uyghur population of Xinjiang. Further investiagtion of the
SNPs in genes surrounding ADAM33 is required and should
specifically address the molecular mechanisms that may confer
asthma susceptibility. The replication of consistent allelic
associations in additional populations may help resolve this
issue.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81160004).
References
|
1
|
Howard TD, Postma DS, Jongepier H, et al:
Association of a disintegrin and metalloprotease 33 (ADAM33) gene
with asthma in ethnically diverse populations. J Allergy Clin
Immunol. 4:717–722. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thongngarm T, Jameekornrak A, Limwongse C,
et al: Association between ADAM33 polymorphisms and asthma in a
Thai population. Asian Pac J Allergy Immunol. 26:205–211.
2008.PubMed/NCBI
|
|
3
|
Bijanzadeh M, Ramachandra NB, Mahesh PA,
et al: Association of IL-4 and ADAM33 gene polymorphisms with
asthma in an Indian population. Lung. 188:415–422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Awasthi S, Tripathi P, Ganesh S and Husain
N: Association of ADAM33 gene polymorphisms with asthma in Indian
children. J Hum Genet. 56:188–195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang P, Liu QJ, Li JS, et al: Lack of
association between ADAM33 gene and asthma in a Chinese population.
Int J Immunogenet. 33:303–306. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sharma N, Tripathi P and Awasthi S: Role
of ADAM33 gene and associated single nucleotide polymorphisms in
asthma. Allergy Rhinol (Providence). 2:63–70. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mayinu and Chen X: Evaluation of LOXL1
polymorphisms in exfoliation syndrome in the Uygur population. Mol
Vis. 17:734–744. 2011.
|
|
8
|
Branch of Chinese Medical Association for
Respiratory Diseases (BCMARD). Bronchial asthma therapeutic
guidelines (definition, diagnosis, treatment of bronchial asthma).
Chin J Tuberc Respir Dis. 31:177–185. 2008.(In Chinese).
|
|
9
|
Vercelli D: Genetics, epigenetic, and the
environment: switching, buffering, releasing. J Allergy Clin
Immunol. 113:381–386. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Diemen CC, Postma DS, Vonk JM,
Bruinenberg M, Schouten JP and Boezen HM: A disintegrin and
metalloprotease 33 polymorphisms and lung function decline in the
general population. Am J Respir Crit Care Med. 172:329–333.
2005.PubMed/NCBI
|
|
11
|
Van Eerdewegh P, Little RD, Dupuis J, et
al: Association of the ADAM33 gene with asthma and bronchial
hyperresponsiveness. Nature. 418:426–430. 2002.PubMed/NCBI
|
|
12
|
Postma DS and Howard T: ADAM33 gene:
confirming a gene without linkage. Clin Exp Allergy. 34:1–3. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Simpson A, Maniatis N, Jury F, et al:
Polymorphisms in a disintegrin and metalloprotease 33 (ADAM33)
predict impaired early-life lung function. Am J Respir Crit Care
Med. 172:55–60. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Noguchi E, Ohtsuki Y, Tokunaga K, et al:
ADAM33 polymorphisms are associated with asthma susceptibility in a
Japanese population. Clin Exp Allergy. 36:602–608. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sakagami T, Jinnai N, Nakajima T, et al:
ADAM33 polymorphisms are associated with aspirin-intolerant asthma
in the Japanese population. J Hum Genet. 52:66–72. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Su D, Zhang X, Sui H, et al: Association
of ADAM33 gene polymorphisms with adult allergic asthma and
rhinitis in a Chinese Han population. BMC Med Genet. 9:822008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lind DL, Choudhry S, Ung N, et al: ADAM33
is not associated with asthma in Puerto Rican or Mexican
populations. Am J Respir Crit Care Med. 168:1312–1316. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Raby BA, Silverman EK, Kwiatkowski DJ, et
al: ADAM33 polymorphisms and phenotype associations in childhood
asthma. J Allergy Clin Immunol. 113:1071–1078. 2004. View Article : Google Scholar : PubMed/NCBI
|