Introduction
Breast cancer is the most frequently diagnosed
cancer and the leading cause of cancer-related mortality among
women worldwide, accounting for 1.38 million new cancer cases and
458,400 deaths in 2008 (1). Over
the last few years, the incidence rates of breast cancer have
increased in most countries. Several studies have found that the
development of breast cancer is possibly associated with tobacco,
alcohol consumption and other environmental factors (2,3).
Furthermore, interindividual differences, including
single-nucleotide polymorphisms, may affect protein activity and
alter the susceptibility to developing breast cancer. The
nucleotide excision repair (NER) system plays an important role in
DNA repair; this system recognizes the DNA damage and incises the
DNA strand on both sides of the lesion, removes the oligonucleotide
containing the damage and reconstructs the corrected fragment.
The xeroderma pigmentosum complementation group G
(XPG) gene is an important component of the NER system and is also
referred to as excision repair cross-complementation group 5
(ERCC5). The fundamental structure of the human ERCC5 protein
contains N- and I-nuclease domains that are highly conserved and
collectively form the nuclease core. The N- and I-nuclease domains
are separated by 600 amino acids that constitute a critical region
for protein-protein interactions, including with transcription
factor IIH (TFIIH) and replication protein A (RPA) and combine
ERCC5 with the sites of NER (4).
The mutation of nucleotides may alter gene function
and affect protein construction, which in turn alters the
mechanical interactions and the function of the NER system during
cellular DNA repair. The Asp1104His (G>C) polymorphism (rs17655)
results in an aspartic acid to histidine transition at position
1104 in exon 15, which may affect protein activity and interaction
with TFIIH, affect the NER system and alter genetic susceptibility
to cancer (5,6).
It was previously demonstrated that the variant
genotype may affect susceptibility to different diseases, such as
lung cancer (7) and bladder cancer
(8), in different ethnicities and
increase the risk of progression from HIV infection to AIDS
(9). In 2003, Kumar et al
(10) reported the first study on
the association between the XPG Asp1104His polymorphism and breast
cancer risk. To date, several studies on the XPG Asp1104His
polymorphism and breast cancer have been conducted. However, the
results of those studies have been inconsistent or even
contradictory. Therefore, we performed the meta-analysis to assess
the association between the XPG Asp1104His polymorphism and breast
cancer risk based on the currently available published studies.
Materials and methods
Search strategy
The US National Library of Medicine’s PubMed
database was searched using the terms ‘breast cancer’, ‘XPG’,
‘ERCC5’, ‘polymorphism’ and their combinations for all genetic
studies on the association between Asp1104His polymorphism and
breast cancer risk during the time period from 2003, when the first
study was reported by Kumar et al (10), to May, 2014. The ‘Related Articles’
application was used to identify additional studies on the same
subject. All the studies were selected using the following three
criteria: i) case-control study of the XPG Asp1104His polymorphism
and breast cancer; ii) sufficient published data for estimating
odds ratios (ORs) with 95% confidence intervals (CIs); and iii)
when multiple publications reported similar or overlapping data, we
selected the largest or most recent publication, as recommended by
Little et al (11) The
characteristics of the studies are summarized in Table I.
 | Table ICharacteristics of case-control
studies on XPG Asp1104His polymorphism and breast cancer risk
included in the meta-analysis. |
Table I
Characteristics of case-control
studies on XPG Asp1104His polymorphism and breast cancer risk
included in the meta-analysis.
| | | | | | | Genotype
distribution | | |
|---|
| | | | | | |
| | |
|---|
| | | | | | | Cases | Controls | | |
|---|
| | | | | | |
|
| | |
|---|
| First author | Year | Country | Racial descent | Source of
controls | Cases | Controls | Asp/Asp | Asp/His | His/His | Asp/Asp | Asp/His | His/His | P for HWEa | (Refs.) |
|---|
| Kumar | 2003 | Finland | Caucasian | PB | 220 | 308 | 108 | 96 | 16 | 182 | 107 | 19 | 0.54 | (10) |
| Mechanic | 2006 | America | Caucasian | PB | 1,249 | 1,133 | 771 | 409 | 69 | 661 | 412 | 60 | 0.69 | (16) |
| Mechanic | 2006 | America | African | PB | 757 | 674 | 231 | 387 | 139 | 231 | 320 | 123 | 0.51 | (16) |
| Shen | 2006 | America | Caucasian | Sisters | 154 | 151 | 83 | 63 | 8 | 82 | 62 | 7 | 0.27 | (17) |
| Crew | 2007 | America | Caucasian | PB | 999 | 1,051 | 562 | 371 | 66 | 571 | 409 | 71 | 0.85 | (18) |
| Jorgensen | 2007 | America | Caucasian | PB | 264 | 275 | 159 | 93 | 12 | 165 | 95 | 15 | 0.78 | (19) |
| Rajaraman | 2008 | America | Mixed | PB | 819 | 1,079 | 482 | 288 | 49 | 674 | 352 | 53 | 0.42 | (20) |
| Smith | 2008 | America | Caucasian | HB | 320 | 408 | 195 | 113 | 12 | 256 | 124 | 28 | 0.02 | (21) |
| Smith | 2008 | America | African | HB | 52 | 75 | 13 | 32 | 7 | 18 | 37 | 20 | 0.91 | (21) |
| Ming-Shiean | 2010 | China | Asian | HB | 401 | 531 | 134 | 191 | 76 | 159 | 243 | 129 | 0.06 | (22) |
Data extraction
Two investigators (Xu and Xie) independently
extracted the following data from the 8 publications: first
author’s name, publication year, country of origin, source of
controls, racial descent of the study population (Asian, African,
European and mixed), number of different genotypes and
Hardy-Weinberg equilibrium (HWE) in controls.
Statistical analysis
Crude ORs with 95% CIs were computed to assess the
strength of the association between the XPG Asp1104His polymorphism
and breast cancer risk for the allele contrast (His vs. Asp),
codominant model (His/His vs. Asp/Asp; Asp/His vs. Asp/Asp),
dominant model (His/His+Asp/His vs. Asp/Asp) and recessive model
(His/His vs. Asp/Asp+Asp/His). Subgroup statistical analysis was
only conducted in Europeans, owing to the small sample of African
and Asian subjects. Heterogeneity was assessed with the
Chi-square-based Q test (12) and
the pooled OR estimation of each study was calculated with the
random-effects model (DerSimonian and Laird method) when P<0.10
(13); otherwise, the
fixed-effects model (Mantel-Haenszel method) was used (14). Publication bias was evaluated with
the funnel plot and the linear regression asymmetry test by Egger
et al (15). P<0.05 was
considered to reflect significant publication bias. Statistical
analysis was performed with STATA software, version 11.0
(StataCorp, College Station, TX, USA), using two-sided
P-values.
Results
Study characteristics
A total of 8 eligible articles (10 case-control
studies) including 5,235 patients with breast cancer and 5,685
healthy control subjects, were included in this meta-analysis
(10,16–22).
Of the 10 studies, 6 were conducted in European, 2 in African, 1 in
Asian and 1 in mixed populations. The genotyping methods comprised
polymerase chain reaction-restriction fragment length polymorphism,
TaqMan and sequence detection system. The distribution of genotypes
in the controls was in agreement with HWE, as in the study by Smith
et al (21) in European
subjects.
Meta-analysis
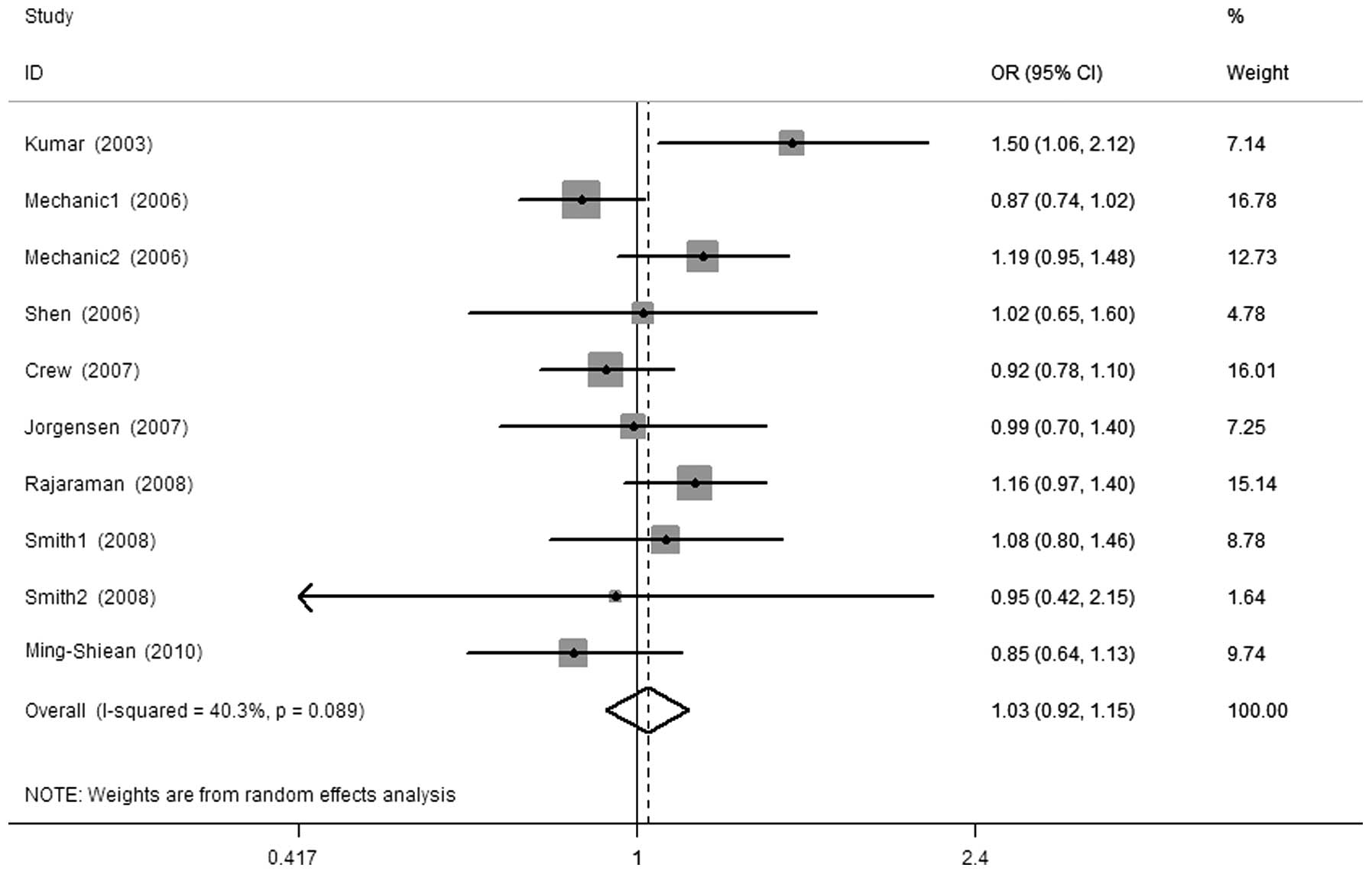
The results of this meta-analysis and heterogeneity
assessment are presented in Table
II. Overall, there were no significant associations between the
XPG Asp1104His polymorphism and breast cancer risk (His vs. Asp,
OR=1.00, 95% CI: 0.91–1.08, Ph=0.082; His/His vs.
Asp/Asp, OR=0.96, 95% CI: 0.83–1.11, Ph=0.265; Asp/His
vs. Asp/Asp, OR=1.02, 95% CI: 0.94–1.11, Ph=0.098;
His/His+Asp/His vs. Asp/Asp, OR=1.03, 95% CI: 0.92–1.15,
Ph=0.089, Fig. 1; and
His/His vs. Asp/Asp+Asp/His, OR=0.93, 95% CI: 0.81–1.06,
Ph=0.286). In the subgroup analysis by ethnicity, we
also did not identify any significant associations between the XPG
Asp1104His polymorphism and breast cancer risk in European
subjects. Further analysis was performed only with studies that
fulfilled HWE and no significant associations were observed.
 | Table IISummary ORs and 95% CIs of XPG
Asp1104His polymorphism and breast cancer risk. |
Table II
Summary ORs and 95% CIs of XPG
Asp1104His polymorphism and breast cancer risk.
| | His vs. Asp | His/His vs.
Asp/Asp | Asp/His vs.
Asp/Asp | His/His+Asp/His vs.
Asp/Asp | His/His vs.
Asp/Asp+Asp/His |
|---|
| |
|
|
|
|
|
|---|
| Variables | No.a | OR | 95% CI | P |
Phb | OR | 95% CI | P |
Phb | OR | 95% CI | P |
Phb | OR | 95% CI | P |
Phb | OR | 95% CI | P |
Phb |
|---|
| Total | 10 | 1.00 | 0.91–1.08 | 0.92 | 0.082 | 0.96 | 0.83–1.11 | 0.62 | 0.265 | 1.02 | 0.94–1.11 | 0.57 | 0.098 | 1.03 | 0.92–1.15 | 0.62 | 0.089 | 0.93 | 0.81–1.06 | 0.29 | 0.286 |
| HWE | 9 | 1.00 | 0.91–1.10 | 0.98 | 0.054 | 0.99 | 0.85–1.15 | 0.88 | 0.362 | 1.04 | 0.92–1.17 | 0.57 | 0.088 | 1.02 | 0.91–1.16 | - | 0.061 | 0.95 | 0.83–1.09 | 0.48 | 0.420 |
| Ethnicity |
| Caucasian | 7 | 1.00 | 0.94–1.08 | 0.90 | 0.148 | 1.01 | 0.84–1.22 | 0.94 | 0.491 | 1.01 | 0.92–1.10 | 0.90 | 0.058 | 1.03 | 0.91–1.18 | 0.63 | 0.072 | 1.01 | 0.84–1.21 | 0.95 | 0.556 |
| African | 2 | 1.05 | 0.91–1.21 | 0.50 | 0.173 | 1.06 | 0.79–1.42 | 0.68 | 0.152 | 1.21 | 0.96–1.52 | 0.10 | 0.983 | 1.17 | 0.94–1.45 | 0.15 | 0.603 | 0.74 | 0.33–1.66 | 0.47 | 0.088 |
| Design |
|
Population-based | 6 | 1.04 | 0.94–1.15 | 0.49 | 0.074 | 1.08 | 0.91–1.27 | 0.38 | 0.756 | 1.02 | 0.93–1.11 | 0.68 | 0.021 | 1.06 | 0.91–1.23 | 0.45 | 0.021 | 1.04 | 0.89–1.22 | 0.62 | 0.928 |
|
Hospital-based | 3 | 0.87 | 0.15–1.00 | 0.05 | 0.573 | 0.65c | 0.48–0.89c | 0.006c | 0.748c | 1.06 | 0.86–1.30 | 0.61 | 0.509 | 0.95 | 0.78–1.16 | 0.61 | 0.526 | 0.66c | 0.50–0.87c | 0.003c | 0.456c |
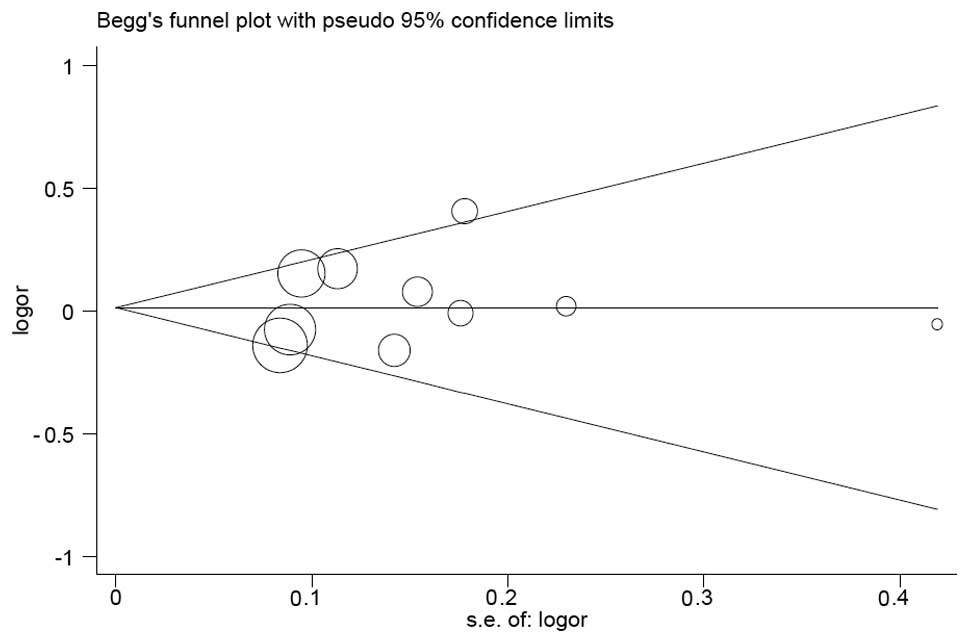
Publication bias
Funnel plots were drawn and Egger’s tests were
performed to access publication bias. The shape of the funnel plots
revealed symmetricity (Fig. 2,
His/His+Asp/His vs. Asp/Asp model). These results were further
supported by analysis via Egger’s tests, which suggested that all
models without significant publication bias (P=0.986 for His vs.
Asp; P=0.456 for His/His vs. Asp/Asp; P=0.217 for Asp/His vs.
Asp/Asp; P=0.484 for His/His+Asp/His vs. Asp/Asp; and P=0.440 for
His/His vs. Asp/Asp+Asp/His).
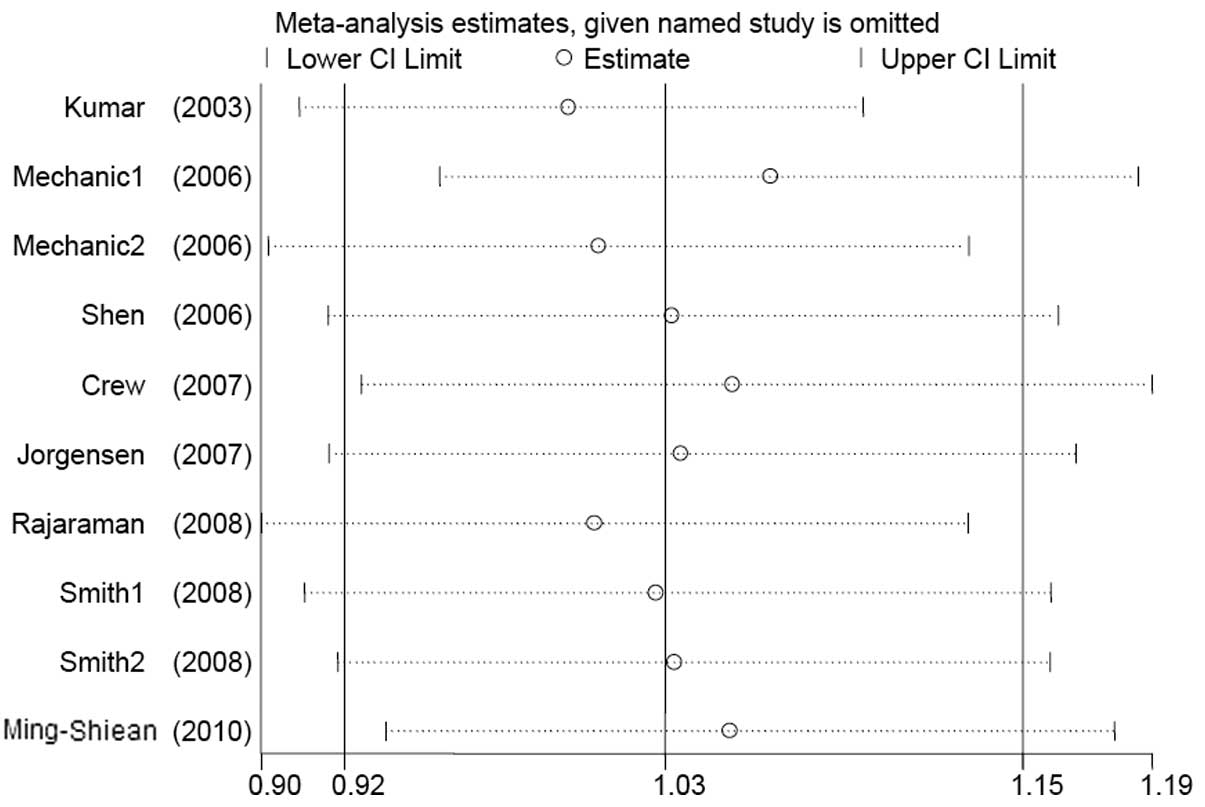
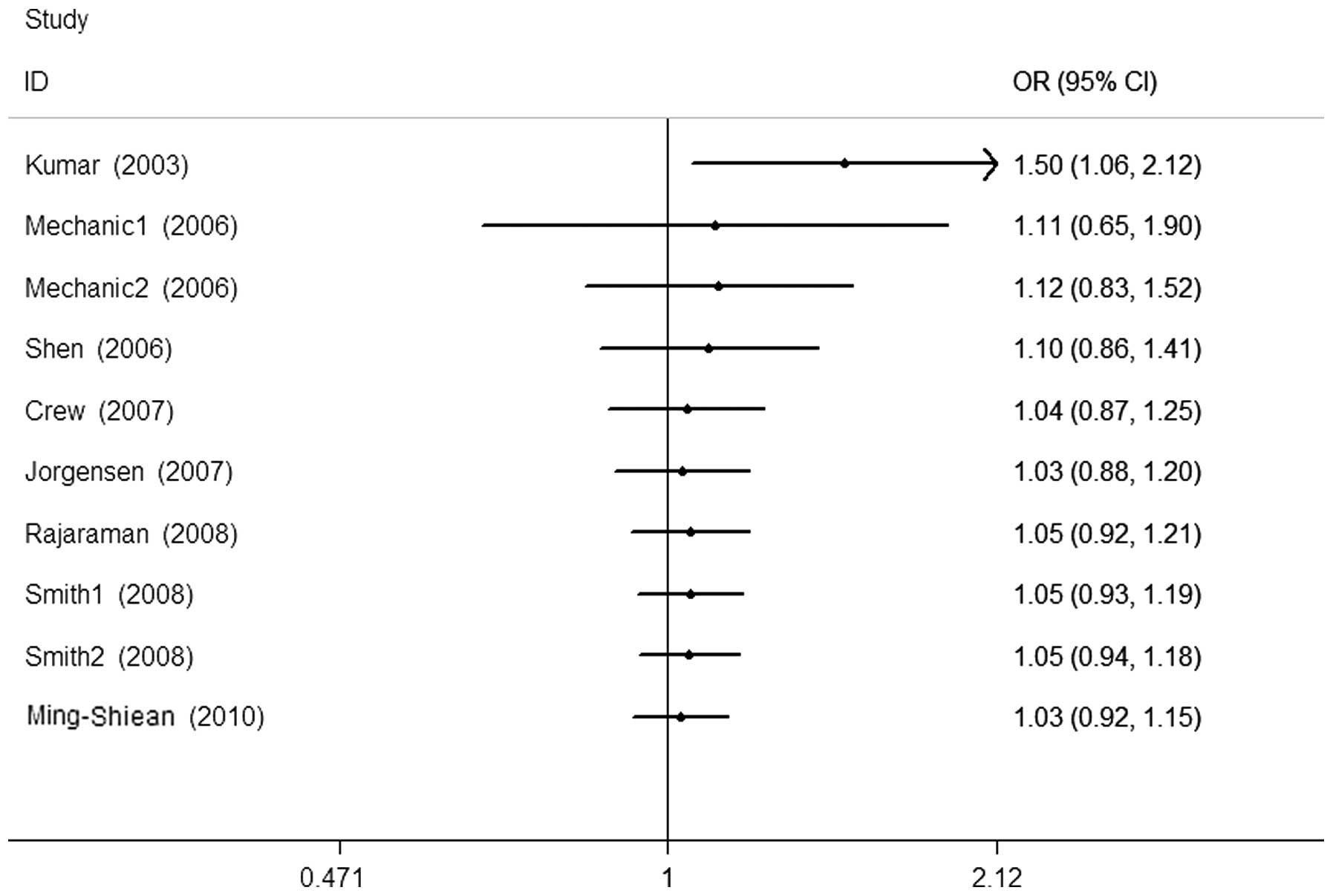
Cumulative and sensitivity analysis
Studies were sequentially deleted to determine the
effect of the individual dataset on the pooled ORs (Fig. 3, His/His+Asp/His vs. Asp/Asp
model). The results were consistent in all the genetic models,
indicating that our results are statistically robust. In the
cumulative meta-analysis, results that became negative from the
second study were accumulated (Fig.
4, His/His+Asp/His vs. Asp/Asp model).
Discussion
The XPG gene is located on chromosome 13q33 and
encodes a 1,186-amino acid structure-specific endonuclease, which
is a member of the flap endonuclease family and plays an important
role in the NER system (23). This
enzyme may combine actions with XPB helicase and ERCC2/XPD helicase
at the DNA damage site (7) and
make 3′-incisions in human NER through incising DNA at a junction
of single- to double-stranded DNA, such as bubbles and loop
structures (24). A dual incision
may be performed, with ERCC1-XPF making the 5′-incision.
Additionally, XPG may involved in the stabilization of a
pre-incision complex on the damaged DNA and stimulate the binding
of human endonuclease III to thymine and glycol-containing DNA
(25). A previous molecular study
reported that the deficiency of XPG may result in certain
epithelial diseases, such as XP (26). Furthermore, several studies also
indicated that mutations in the XPG gene are associated with the
development of diseases such as lung cancer and osteosarcoma
(27,28).
Previous reports on the association between the XPG
Asp1104His polymorphism and breast cancer were discrepant or even
contradictory. Kumar et al (10) found that the genotype with the C
allele (His) was associated with a ~1.5-fold increased risk for
breast cancer in European subjects (OR=1.5, 95% CI: 1.04–2.16) in
2003. By contrast, Ming-Shiean et al (22) considered the G allele variant (Asp)
to be significantly associated with breast cancer in Asian subjects
(OR=1.42, 95% CI: 1.08–1.97). However, other studies reported no
association between the XPG Asp1104His polymorphism and breast
cancer risk.
This meta-analysis included 10 case-control studies,
involving 5,235 patients with breast cancer and 5,685 healthy
controls. No significant association was found between the XPG
Asp1104His polymorphism and breast cancer risk, not even in the
subgroup analysis of European subjects. According to the results,
certain limitations of this meta-analysis need to be addressed.
First, the sample of breast cancer patients and controls was
inadequate to reach a definitive conclusion. Second, we were unable
to obtain more original data and the results were based on
unadjusted estimates, lacking the evaluation of the covariates of
age, menopausal status, smoking and alcohol consumption and other
environmental factors, which limited the evaluation of the
interaction effect of genes and environmental or other factors.
Third, there was some heterogeneity in different models, but it was
successfully removed or alleviated in the subgroup analysis.
Despite these limitations, the statistical assessment of
publication bias, cumulative and sensitivity analyses all indicated
that our results are credible.
In conclusion, our meta-analysis indicated that the
XPG Asp1104His polymorphism is not associated with breast cancer
risk. However, further, large-scale epidemiological studies are
required to validate these conclusions.
Acknowledgements
We gratefully acknowledge the support of the
subjects who participated in this study. This study was partly
supported by grants from the Foundation of the Ministry of
Education of Hubei Province (no. D20142102), the Foundation of
Hubei University of Medicine (no. 2013GPY07) and the Taihe Hospital
(nos. EBM2013006 and EBM2013031).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Conlon MS, Johnson KC, Bewick MA, Lafrenie
RM and Donner A: Smoking (active and passive), N-acetyltransferase
2, and risk of breast cancer. Cancer Epidemiol. 34:142–149. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Beasley JM, Coronado GD, Livaudais J, et
al: Alcohol and risk of breast cancer in Mexican women. Cancer
Causes Control. 21:863–870. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thorel F, Constantinou A, Dunand-Sauthier
I, et al: Definition of a short region of XPG necessary for TFIIH
interaction and stable recruitment to sites of UV damage. Mol Cell
Biol. 24:10670–10680. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Collins A and Harrington V: Repair of
oxidative DNA damage: assessing its contribution to cancer
prevention. Mutagenesis. 17:489–493. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Berwick M and Vineis P: Markers of DNA
repair and susceptibility to cancer in humans: an epidemiologic
review. J Natl Cancer Inst. 92:874–897. 2000. View Article : Google Scholar
|
|
7
|
Kiyohara C and Yoshimasu K: Genetic
polymorphisms in the nucleotide excision repair pathway and lung
cancer risk: a meta-analysis. Int J Med Sci. 4:59–71. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sanyal S, Festa F, Sakano S, et al:
Polymorphisms in DNA repair and metabolic genes in bladder cancer.
Carcinogenesis. 25:729–734. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sobti RC, Berhane N, Mehedi SA, et al:
Association and impact of XPG Asp 1104 His gene polymorphism in HIV
1 disease progression to AIDS among north Indian HIV seropositive
individuals. Mol Biol Rep. 37:317–324. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kumar R, Hoglund L, Zhao C, Forsti A,
Snellman E and Hemminki K: Single nucleotide polymorphisms in the
XPG gene: determination of role in DNA repair and breast cancer
risk. Int J Cancer. 103:671–675. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Little J, Bradley L, Bray MS, et al:
Reporting, appraising, and integrating data on genotype prevalence
and gene-disease associations. Am J Epidemiol. 156:300–310. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lau J, Ioannidis JP and Schmid CH:
Quantitative synthesis in systematic reviews. Ann Intern Med.
127:820–826. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mantel N and Haenszel W: Statistical
aspects of the analysis of data from retrospective studies of
disease. J Natl Cancer Inst. 22:719–748. 1959.PubMed/NCBI
|
|
15
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mechanic LE, Millikan RC, Player J, et al:
Polymorphisms in nucleotide excision repair genes, smoking and
breast cancer in African Americans and whites: a population-based
case-control study. Carcinogenesis. 27:1377–1385. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen J, Desai M, Agrawal M, et al:
Polymorphisms in nucleotide excision repair genes and DNA repair
capacity phenotype in sisters discordant for breast cancer. Cancer
Epidemiol Biomarkers Prev. 15:1614–1619. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Crew KD, Gammon MD, Terry MB, et al:
Polymorphisms in nucleotide excision repair genes, polycyclic
aromatic hydrocarbon-DNA adducts, and breast cancer risk. Cancer
Epidemiol Biomarkers Prev. 16:2033–2041. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jorgensen TJ, Visvanathan K, Ruczinski I,
Thuita L, Hoffman S and Helzlsouer KJ: Breast cancer risk is not
associated with polymorphic forms of xeroderma pigmentosum genes in
a cohort of women from Washington County, Maryland. Breast Cancer
Res Treat. 101:65–71. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rajaraman P, Bhatti P, Doody MM, et al:
Nucleotide excision repair polymorphisms may modify ionizing
radiation-related breast cancer risk in US radiologic
technologists. Int J Cancer. 123:2713–2716. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smith TR, Levine EA, Freimanis RI, et al:
Polygenic model of DNA repair genetic polymorphisms in human breast
cancer risk. Carcinogenesis. 29:2132–2138. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ming-Shiean H, Yu JC, Wang HW, et al:
Synergistic effects of polymorphisms in DNA repair genes and
endogenous estrogen exposure on female breast cancer risk. Ann Surg
Oncol. 17:760–771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Harrington JJ and Lieber MR: Functional
domains within FEN-1 and RAD2 define a family of structure-specific
endonucleases: implications for nucleotide excision repair. Genes
Dev. 8:1344–1355. 1994. View Article : Google Scholar
|
|
24
|
Matsunaga T, Park CH, Bessho T, Mu D and
Sancar A: Replication protein A confers structure-specific
endonuclease activities to the XPF-ERCC1 and XPG subunits of human
DNA repair excision nuclease. J Biol Chem. 271:11047–11050. 1996.
View Article : Google Scholar
|
|
25
|
Bessho T: Nucleotide excision repair 3′
endonuclease XPG stimulates the activity of base excision repair
enzyme thymine glycol DNA glycosylase. Nucleic Acids Res.
27:979–983. 1999.
|
|
26
|
Fagbemi AF, Orelli B and Scharer OD:
Regulation of endonuclease activity in human nucleotide excision
repair. DNA Repair (Amst). 10:722–729. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chang JS, Wrensch MR, Hansen HM, et al:
Nucleotide excision repair genes and risk of lung cancer among San
Francisco Bay Area Latinos and African Americans. Int J Cancer.
123:2095–2104. 2008. View Article : Google Scholar
|
|
28
|
Biason P, Hattinger CM, Innocenti F, et
al: Nucleotide excision repair gene variants and association with
survival in osteosarcoma patients treated with neoadjuvant
chemotherapy. Pharmacogenomics J. 12:476–483. 2012. View Article : Google Scholar : PubMed/NCBI
|