Introduction
Epithelial cell adhesion molecule (EpCAM) is a
40-kDa glycosylated transmembrane cell surface protein that plays
an important role in Ca2+ independent hemophilic
cell-to-cell adhesion, cell signaling, migration, proliferation and
differentiation (1). EpCAM was
reported to be exclusively present in epithelial tissues and is
highly expressed across a large number of epithelial cancers
(2–11). Particular interest has been focused
on EpCAM expression as a poor prognostic biomarker across a large
number of carcinomas (2, 4–6,
8–10, 12–15).
EpCAM is expressed on carcinosarcomas, which are
rare biphenotypic tumors that display characteristics of epithelial
as well as sarcomatous elements (16, 17). The expression of EpCAM is
considered to be absent on purely non-epithelial tumors, such as
sarcomas and hematopoietic cancers. However, a recent comprehensive
analysis of EpCAM expression across human tumors clearly
demonstrated that EpCAM is expressed at weak to intense levels in
several soft tissue sarcomas, including angiosarcoma (25%),
epitheloid sarcoma (50%), fibrosarcoma (22%) and synovial sarcoma
(100%) (18). An abstract
presented at the European Society for Medical Oncology indicated
that EpCAM-positive circulating cells were detectable in just under
half of all the soft tissue sarcoma patients investigated, although
the authors were unable to determine whether the EpCAM-positive
cells were of epithelial origin due to direct tumor invasion of
blood vessels from the surrounding tissue or tumor cells at the
moment of mesenchymal-to-epithelial transition (19). Moreover, induced multidrug
resistance in osteosarcomas has been shown to increase the
expression of cell adhesion markers, including EpCAM (20).
Despite the substantial evidence described above,
whether EpCAM is actually expressed on sarcomas remains debatable.
Furthermore, it has not yet been determined whether the previously
reported expression of EpCAM on sarcomas is of prognostic
significance, as has been reported for multiple carcinomas. In this
study, we aimed to determine whether EpCAM is indeed expressed at
detectable levels in a subset of sarcomas and provide evidence
supporting that the steady-state protein expression level of EpCAM
is statistically correlated with the degree of cytological atypia
in leiomyosarcomas.
Materials and methods
Meta-analysis of EpCAM expression
The normalized intensity values of EpCAM mRNA
were evaluated using the Cancer Cell Line Encyclopedia (CCLE,
www.broadinstitute.org/ccle/home). This dataset
compares the mRNA expression, chromosomal copy number variation and
massively parallel sequencing data from 947 diverse human cancer
lines (21).
Case material
Tissue arrays of formalin-fixed and
paraffin-embedded human angiosarcomas (cat no. SO8010),
osteosarcomas (cat no. OS804a) and leiomyosarcomas (cat no. SO804)
were obtained from US BioMax, Inc. (Rockville, MD, USA) These
clinically characterized tumor samples consisted of 2-mm cores with
a section thickness of 4 microns and totaled 6 angiosarcoma, 40
osteosarcoma and 80 leiomyosarcoma cases. The cases were reviewed
by a pathologist and the diagnoses were confirmed by
histomorphology per established morphological criteria.
Immunohistochemistry
The sections were deparaffinized, rehydrated and
treated for antigen retrieval using Trilogy solution (Cell Marque,
Rocklin, CA, USA; cat no. 920P-10). To block non-specific binding,
the sections were incubated in background block solution (Cell
Marque; cat no. 927B-05) at room temperature for 10 min prior to
application of the anti-EpCAM primary antibody diluted 1:100 as per
the manufacturer's suggestions (Abcam, Cambridge, UK; cat no.
ab71916). The sections were then washed in phosphate-buffered
saline with Tween-20 (Cell Marque; cat no. 934B-09) three times for
5 min per wash and incubated with the CytoScan HRP Detection System
(Cell Marque; cat no. 951D-20). Immunostaining was performed using
the DAB Substrate kit (Cell Marque; cat no. 957D-20) and
counterstained with hematoxylin.
Quantitation of
immunohistochemistry
EpCAM immunopositivity was scored semiquantitatively
for the percentage of tumor cells stained and staining intensity
(0, negative; +, weak; ++, moderate; and +++, strong). For
statistical analysis, scoring was converted to numerical values (0,
0; +, 1; ++, 2; and +++, 3) and the mean values ± standard error of
the mean for leiomyosarcomas exhibiting mild, moderate and severe
cytological atypia were calculated. Two tailed t-tests were
performed to determine statistical significance, which was set at
P≤0.05.
Results
Expression of EpCAM across a diverse
array of cancer cell lines
CCLE is a publicly accessible cancer genomic
database jointly developed by Novartis and the Broad Institute to
systematically interpret mRNA expression, chromosomal copy number
variation and massively parallel sequencing data from 947 human
cancer lines (21). While these
groups primarily utilized this database for predictive modeling of
anticancer drug sensitivity, a plethora of genomic data awaits
meta-analysis to generate and test potential hypotheses that are
formulated by bioinformaticians. Utilizing the data housed in the
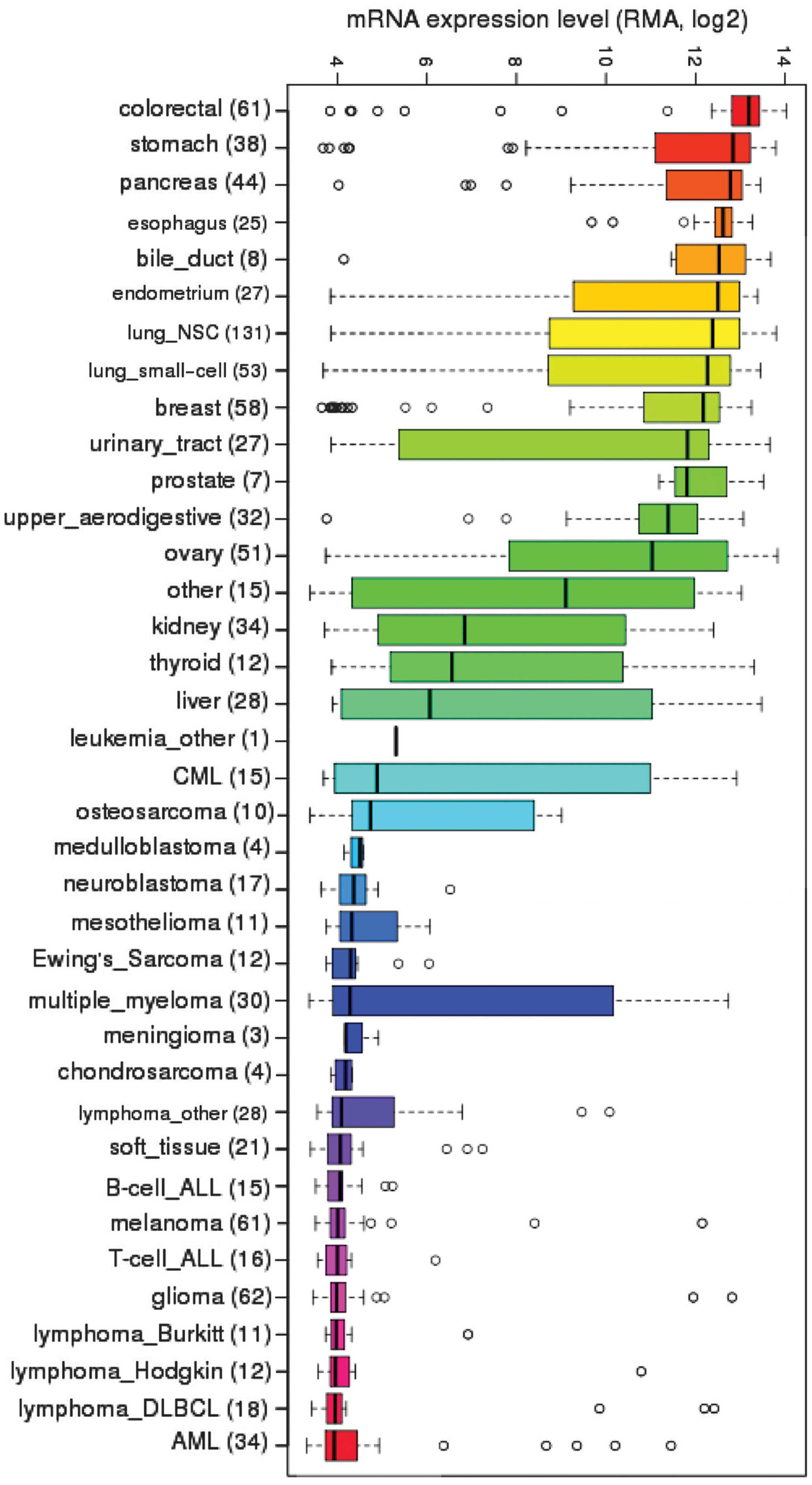
CCLE, we investigated the steady-state mRNA expression of
EpCAM across the diverse array of cancer cell lines
(Fig. 1). As expected,
EpCAM transcripts were highly expressed in a large number of
carcinomas and least expressed in hematopoietic cancers, such as
lymphomas. Surprisingly, sarcomas exhibited variable degrees of
expression; osteosarcomas displayed moderate levels, while Ewing's
sarcoma, chondrosarcoma and mixed soft tissue sarcomas exhibited
low levels of EpCAM mRNA expression.
EpCAM mRNA and protein expression in
sarcomas
Given the unexpected levels of EpCAM mRNA
expression in osteosarcomas based on our genomic meta-analysis, we
sought to verify these findings at the protein level in three
sarcoma types that our laboratory is currently investigating,
namely osteosarcomas, leiomyosarcomas and angiosarcomas. We
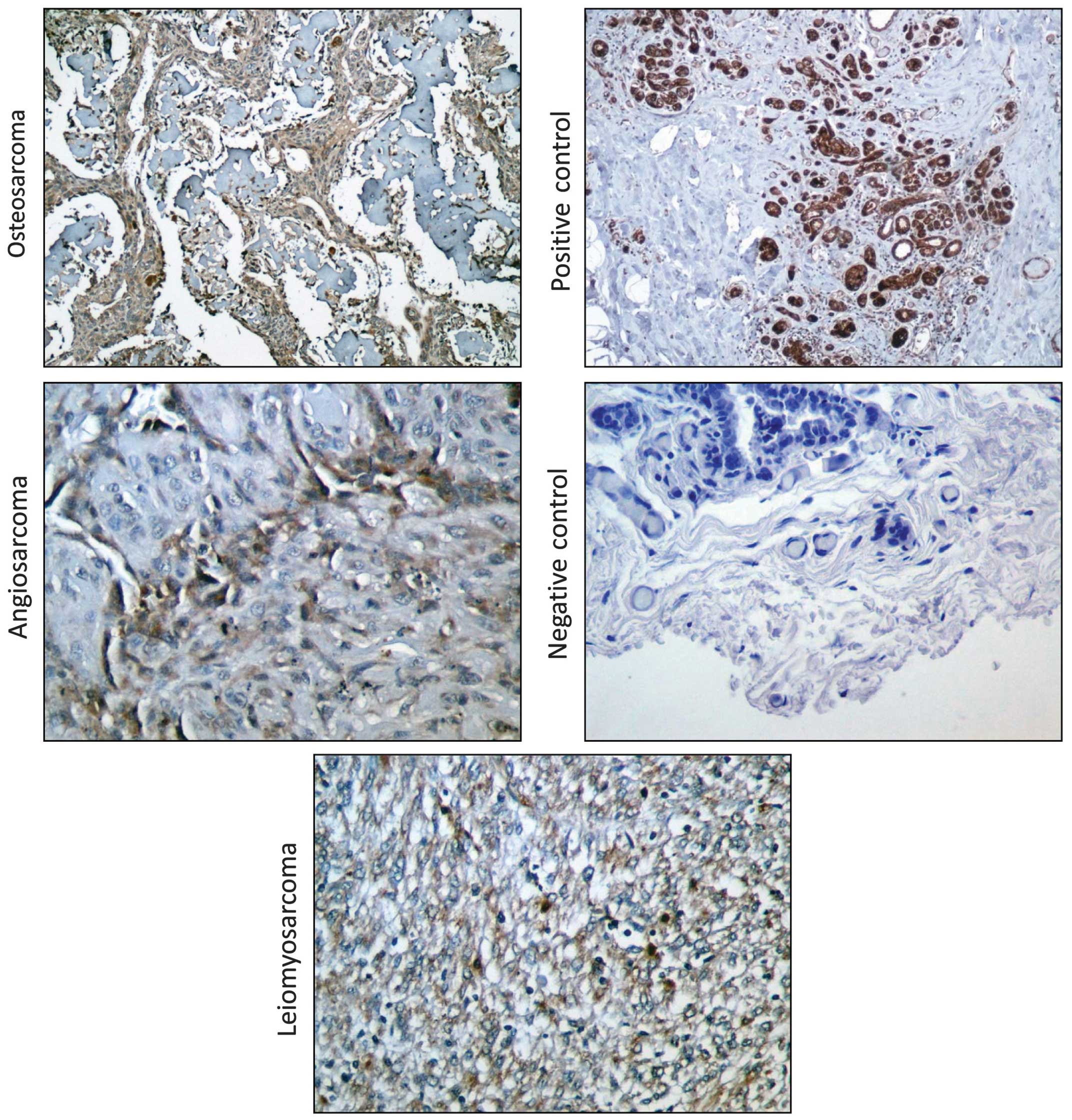
utilized immunohistochemistry to detect EpCAM protein levels in
clinically evaluated human tumor sections from 6 angiosarcomas, 40
osteosarcomas and 80 leiomyosarcomas. EpCAM protein was detected at
low levels in half of the angiosarcomas (Table I, Fig.
2) and it was detectable in all the investigated osteosarcomas,
with 10% of the tumors exhibiting weak expression, 87.5% exhibiting
moderate expression and only 1 tumor sample exhibiting strong EpCAM
expression (Table II, Fig. 2). We observed mild to moderate
staining for EpCAM on 62.5% of the leiomyosarcoma sections that
were examined in our analysis, with 51% of the tumors exhibiting
weak and 11% moderate EpCAM staining (Table III, Fig. 2). Collectively, these data indicate
that, contrary to the established scientific belief that EpCAM
expression is solely limited to tissues and tumors of epithelial
origin, this ‘epithelial-specific’ protein is indeed expressed in a
subset of sarcomas of mesenchymal origin.
 | Table IEpCAM protein expression in
angiosarcomas. |
Table I
EpCAM protein expression in
angiosarcomas.
| Gender | Age (years) | Organ | Score |
|---|
| M | 16 | Fibrous tissue | 0 |
| F | 65 | Fallopian tube | 0 |
| M | 42 | Spleen | 0 |
| M | 47 | Heart | + |
| M | 80 | Liver | + |
| F | 65 | Blood vessel | + |
 | Table IIEpCAM protein expression in
osteosarcomas. |
Table II
EpCAM protein expression in
osteosarcomas.
| Gender | Age (years) | Stage | TNM | Score |
|---|
| F | 38 | IA | T1N0M0 | ++ |
| M | 43 | IA | T1N0M0 | ++ |
| F | 17 | IB | T2N0M0 | +++ |
| M | 41 | IB | T2N0M0 | ++ |
| M | 19 | IB | T2N0M0 | ++ |
| F | 16 | IIA | T2N0M0 | ++ |
| M | 41 | IIA | T1N0M0 | ++ |
| F | 15 | IIA | T2N0M0 | + |
| F | 12 | IIA | T1N0M0 | ++ |
| M | 13 | IIA | T1N0M0 | ++ |
| F | 44 | IIA | T1N0M0 | ++ |
| M | 37 | IIA | T1N0M0 | ++ |
| M | 29 | IIA | T1N0M0 | ++ |
| M | 32 | IIA | T1N0M0 | ++ |
| M | 47 | IIB | T2N0M0 | + |
| M | 38 | IIB | T2N0M0 | ++ |
| M | 32 | IIB | T2N0M0 | + |
| M | 42 | IIB | T2N0M0 | ++ |
| M | 11 | IIB | T2N0M0 | ++ |
| M | 38 | IIB | T2N0M0 | ++ |
| F | 32 | IIB | T2N0M0 | + |
| M | 51 | IIB | T2N0M0 | ++ |
| F | 14 | IIB | T2N0M0 | ++ |
| F | 47 | IIB | T2N0M0 | ++ |
| F | 14 | IIB | T2N0M0 | ++ |
| F | 32 | IIB | T2N0M0 | ++ |
| F | 14 | IIB | T2N0M0 | ++ |
| M | 16 | IIB | T2N0M0 | ++ |
| M | 18 | IIB | T2N0M0 | ++ |
| M | 23 | IIB | T2N0M0 | ++ |
| M | 60 | IIB | T2N0M0 | ++ |
| M | 31 | IIB | T2N0M0 | ++ |
| M | 30 | IIB | T2N0M0 | ++ |
| F | 32 | IIB | T2N0M0 | ++ |
| M | 64 | IIB | T2N0M0 | ++ |
| M | 35 | IIB | T2N0M0 | ++ |
| M | 21 | IIB | T2N0M0 | ++ |
| M | 51 | IIB | T2N0M0 | ++ |
| M | 44 | IIB | T2N0M0 | ++ |
| M | 17 | IIB | T2N0M0 | ++ |
 | Table IIIEpCAM protein expression in
leiomyosarcomas. |
Table III
EpCAM protein expression in
leiomyosarcomas.
| Gender | Age (years) | Organ | Cytological
atypia | Score |
|---|
| F | 50 | Gallbladder | Mild | 0 |
| M | 42 | Abdominal
cavity | Mild | 0 |
| F | 75 | Esophagus | Mild | 0 |
| M | 50 | Mesentery | Mild | 0 |
| M | 65 | Abdominal
cavity | Mild | 0 |
| F | 63 | Mesentery | Mild | 0 |
| M | 37 | Mediastinum | Mild | 0 |
| F | 62 | Pelvic cavity | Mild | 0 |
| F | 56 |
Retroperitoneum | Mild | 0 |
| F | 48 | Colon | Mild | 0 |
| F | 39 |
Retroperitoneum | Mild | 0 |
| M | 72 | Gallbladder | Mild | 0 |
| M | 74 | Liver | Mild | 0 |
| F | 35 |
Retroperitoneum | Mild | 0 |
| F | 45 |
Retroperitoneum | Mild | 0 |
| F | 51 | Abdominal
cavity | Mild | + |
| F | 38 | Fibrous tissue | Mild | + |
| M | 46 | Colon | Mild | + |
| M | 61 | Esophagus | Mild | + |
| F | 38 |
Retroperitoneum | Mild | + |
| F | 76 | Mesentery | Mild | + |
| F | 43 | Colon | Mild | ++ |
| F | 41 |
Retroperitoneum | Mild | ++ |
| F | 46 |
Retroperitoneum | Mild | ++ |
| F | 24 | Pelvic cavity | Moderate | 0 |
| M | 36 | Liver | Moderate | 0 |
| F | 38 | Pelvic cavity | Moderate | 0 |
| F | 48 | Pelvic cavity | Moderate | 0 |
| M | 64 |
Retroperitoneum | Moderate | 0 |
| M | 80 | Epiploon | Moderate | 0 |
| F | 51 | Abdominal
cavity | Moderate | 0 |
| M | 57 | Fibrous tissue | Moderate | 0 |
| F | 54 |
Retroperitoneum | Moderate | 0 |
| F | 41 |
Retroperitoneum | Moderate | 0 |
| M | 78 | Esophagus | Moderate | 0 |
| M | 52 | Abdominal
cavity | Moderate | 0 |
| M | 65 | Esophagus | Moderate | 0 |
| M | 45 | Abdominal
cavity | Moderate | + |
| F | 39 |
Retroperitoneum | Moderate | + |
| F | 53 |
Retroperitoneum | Moderate | + |
| M | 61 |
Retroperitoneum | Moderate | + |
| F | 47 |
Retroperitoneum | Moderate | + |
| F | 60 | Abdominal
cavity | Moderate | + |
| F | 63 |
Retroperitoneum | Moderate | + |
| F | 57 |
Retroperitoneum | Moderate | + |
| M | 72 |
Retroperitoneum | Moderate | + |
| F | 50 |
Retroperitoneum | Moderate | + |
| M | 57 |
Retroperitoneum | Moderate | + |
| M | 45 | Tongue | Moderate | + |
| M | 60 | Abdominal
cavity | Moderate | + |
| F | 78 |
Retroperitoneum | Moderate | + |
| F | 42 |
Retroperitoneum | Moderate | + |
| F | 59 | Abdominal
cavity | Moderate | + |
| F | 41 |
Retroperitoneum | Moderate | + |
| F | 44 |
Retroperitoneum | Moderate | + |
| F | 60 | Tongue | Moderate | + |
| M | 49 | Nose | Moderate | + |
| F | 43 |
Retroperitoneum | Moderate | + |
| F | 40 |
Retroperitoneum | Moderate | + |
| M | 42 | Abdominal
cavity | Moderate | + |
| F | 45 |
Retroperitoneum | Moderate | + |
| M | 67 | Lung | Moderate | + |
| M | 84 | Skin | Moderate | + |
| F | 42 |
Retroperitoneum | Moderate | + |
| F | 66 | Skin | Moderate | + |
| M | 38 | Fibrous tissue | Moderate | + |
| F | 61 |
Retroperitoneum | Moderate | ++ |
| F | 46 |
Retroperitoneum | Moderate | ++ |
| F | 35 |
Retroperitoneum | Moderate | ++ |
| F | 82 | Adrenal gland | Moderate | ++ |
| M | 69 | Abdominal
cavity | Moderate | ++ |
| F | 57 | Breast | Severe | 0 |
| M | 56 | Epiploon | Severe | 0 |
| F | 73 |
Retroperitoneum | Severe | + |
| F | 52 |
Retroperitoneum | Severe | + |
| F | 52 |
Retroperitoneum | Severe | + |
| F | 58 | Abdominal
cavity | Severe | + |
| F | 46 | Epiploon | Severe | + |
| F | 35 | Breast | Severe | + |
| F | 52 | Abdominal
cavity | Severe | ++ |
Correlation of EpCAM expression with
cytological atypia
In addition to the pathological classification, we
obtained clinical data on the osteosarcomas and leiomyosarcomas
that were utilized in our analysis of EpCAM protein expression.
These data included patient gender and age for both tumor types,
tumor staging and TNM classification of the osteosarcoma samples
and the degree of cytological atypia for the leiomyosarcomas. Given
the established prognostic significance of EpCAM expression in
human carcinomas, we sought to elucidate whether this extended into
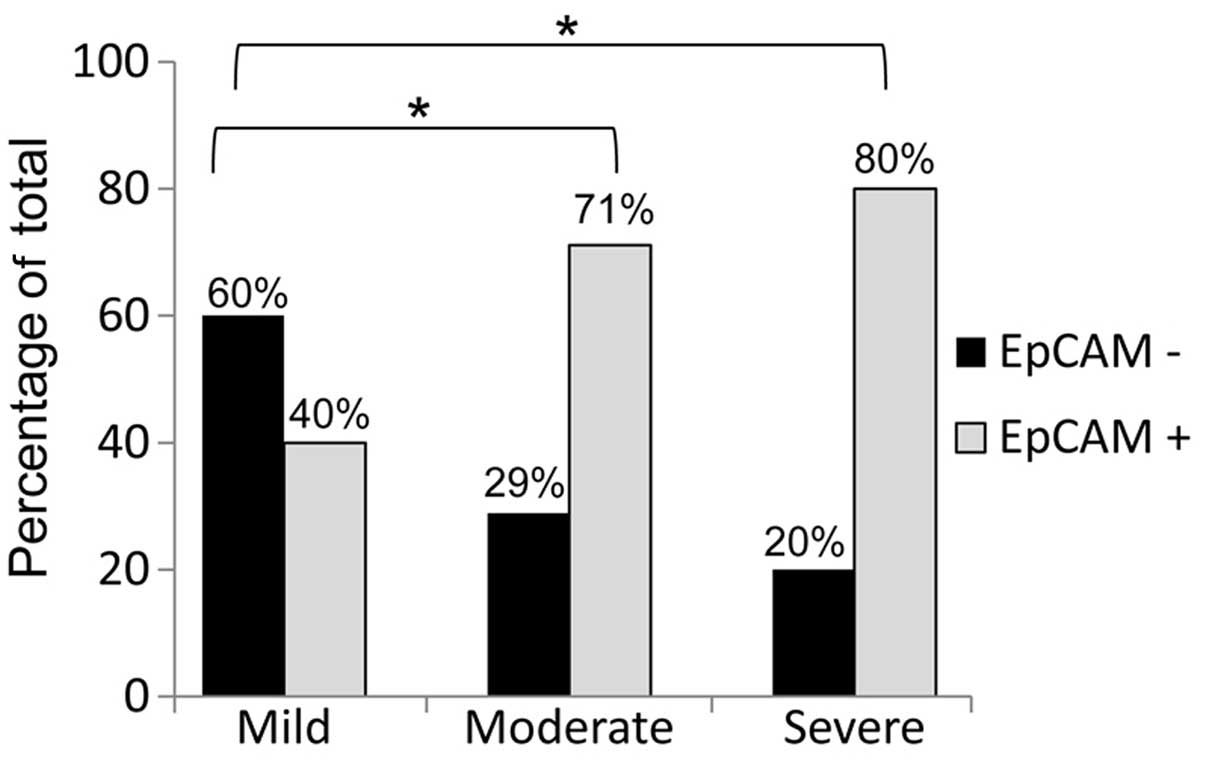
sarcomas that expressed this protein. We observed that
leiomyosarcomas with moderate and severe cytological atypia
exhibited a significantly higher percentage of EpCAM-positive
staining compared to tumors with mild cytological atypia (Fig. 3). While 60% of leiomyosarcomas
displaying mild cytological atypia were rated as EpCAM-negative,
only 29 and 20% of EpCAM-negative leiomyosarcomas displayed
moderate and severe cytological atypia, respectively. We utilized
statistical analysis as described in the Materials and methods
section to correlate EpCAM expression to each known clinical
characteristic and demonstrated that EpCAM expression was
significantly correlated with the degree of cytological atypia in
leiomyosarcomas (EpCAM expression was calculated as follows: mild,
0.50±0.14; moderate, 0.83±0.09; and severe, 0.89±0.20; P≤0.05 for
all comparisons). These data indicate that the increase in EpCAM
expression is directly correlated with the increase in the degree
of cytological atypia.
Discussion
EpCAM has historically been shown to be expressed
across epithelial tissues, with a few exceptions (22). Additionally, EpCAM is highly
overexpressed across a wide range of carcinomas and was originally
described as the dominant antigen in patients with colon carcinoma
(23, 24). The expression of this protein has
been associated with poor clinical prognosis in patients with a
number of carcinomas by functioning as an oncogene and suppressing
CD4+ T-cell-dependent immune responses (2, 5,
25, 26). Moreover, use of the EpCAM-specific
monoclonal antibody edrecolomab and the tri-functional antibody
catumaxomab in patients with metastatic cancers has demonstrated
positive antitumor effects (27–30),
suggesting that targeting EpCAM may prove to be efficacious against
certain carcinomas.
By mining the gene expression data of various cancer
cell lines deposited in the CCLE portal, we quickly noticed that,
despite the common scientific belief that EpCAM is strictly
expressed in tissues of epithelial origin, tumors of mesenchymal
origin, such as osteosarcomas, displayed moderate expression of
this gene. This prompted us to delve further into EpCAM expression
to demonstrate that, of the mesenchymal tumors investigated in our
analysis, all osteosarcomas and over half of the leiomyosarcomas
and angiosarcomas expressed EpCAM protein at detectable levels, as
determined by immunohistochemistry. Our data confirmed the previous
findings by Went et al (18), suggesting that 22–100% of sarcomas
express weak to intense levels of the EpCAM protein. Given that
EpCAM has been extensively reported to be a prognostic biomarker
for carcinomas, we sought to determine whether the prognostic
significance of this protein extends to sarcomas. Using established
clinical data from our panel of osteosarcoma and leiomyosarcoma
samples, we identified a direct statistical correlation between
EpCAM expression and the degree of cytological atypia in
leiomyosarcomas. Cytological atypia is one of the most significant
prognostic factors for leiomyosarcoma (31); thus, similar to the results
obtained for carcinomas, EpCAM expression is a negative prognostic
factor for leiomyosarcomas.
This study, contrary to the current clinical
perception that EpCAM is solely expressed in epithelial tissues,
suggests that EpCAM mRNA and protein expression may be used as a
prognostic biomarker for leiomyosarcoma severity and will
potentially expand the number of cancers exhibiting susceptibility
to immunotherapy against EpCAM.
References
|
1
|
Litvinov SV, Bakker HA, Gourevitch MM,
Velders MP and Warnaar SO: Evidence for a role of the epithelial
glycoprotein 40 (Ep-CAM) in epithelial cell-cell adhesion. Cell
Adhes Commun. 2:417–428. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gastl G, Spizzo G, Obrist P, Dünser M and
Mikuz G: Ep-CAM overexpression in breast cancer as a predictor of
survival. Lancet. 356:1981–1982. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Heinzelmann-Schwarz VA, Gardiner-Garden M,
Henshall SM, et al: Overexpression of the cell adhesion molecules
DDR1, Claudin 3, and Ep-CAM in metaplastic ovarian epithelium and
ovarian cancer. Clin Cancer Res. 10:4427–4436. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Songun I, Litvinov SV, van de Velde CJ,
Pals ST, Hermans J and van Krieken JH: Loss of Ep-CAM (CO17-1A)
expression predicts survival in patients with gastric cancer. Br J
Cancer. 92:1767–1772. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Spizzo G, Went P, Dirnhofer S, et al:
Overexpression of epithelial cell adhesion molecule (Ep-CAM) is an
independent prognostic marker for reduced survival of patients with
epithelial ovarian cancer. Gynecol Oncol. 103:483–488. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kimura H, Kato H, Faried A, et al:
Prognostic significance of EpCAM expression in human esophageal
cancer. Int J Oncol. 30:171–179. 2007.PubMed/NCBI
|
|
7
|
Shim HS, Yoon BS and Cho NH: Prognostic
significance of paired epithelial cell adhesion molecule and
E-cadherin in ovarian serous carcinoma. Hum Pathol. 40:693–698.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schmidt M, Hasenclever D, Schaeffer M, et
al: Prognostic effect of epithelial cell adhesion molecule
overexpression in untreated node-negative breast cancer. Clin
Cancer Res. 14:5849–5855. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schmidt M, Petry IB, Böhm D, et al: Ep-CAM
RNA expression predicts metastasis-free survival in three cohorts
of untreated node-negative breast cancer. Breast Cancer Res Treat.
125:637–646. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Akita H, Nagano H, Takeda Y, et al: Ep-CAM
is a significant prognostic factor in pancreatic cancer patients by
suppressing cell activity. Oncogene. 30:3468–3476. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pietzner K, Woopen H, Richter R, et al:
Expression of epithelial cell adhesion molecule in paired tumor
samples of patients with primary and recurrent serous ovarian
cancer. Int J Gynecol Cancer. 23:797–802. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Battista MJ, Cotarelo C, Jakobi S, et al:
Overexpression of epithelial cell adhesion molecule protein is
associated with favorable prognosis in an unselected cohort of
ovarian cancer patients. J Cancer Res Clin Oncol. 140:1097–1102.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Seligson DB, Pantuck AJ, Liu X, et al:
Epithelial cell adhesion molecule (KSA) expression: pathobiology
and its role as an independent predictor of survival in renal cell
carcinoma. Clin Cancer Res. 10:2659–2669. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Varga M, Obrist P, Schneeberger S, et al:
Overexpression of epithelial cell adhesion molecule antigen in
gallbladder carcinoma is an independent marker for poor survival.
Clin Cancer Res. 10:3131–3136. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stoecklein NH, Siegmund A, Scheunemann P,
et al: Ep-CAM expression in squamous cell carcinoma of the
esophagus: a potential therapeutic target and prognostic marker.
BMC Cancer. 6(165)2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Choijamts B, Jimi S, Kondo T, et al:
CD133+ cancer stem cell-like cells derived from uterine
carcinosarcoma (malignant mixed Müllerian tumor). Stem Cells.
29:1485–1495. 2011.
|
|
17
|
Paniz Mondolfi AE, Jour G, Johnson M, et
al: Primary cutaneous carcinosarcoma: insights into its clonal
origin and mutational pattern expression analysis through
next-generation sequencing. Hum Pathol. 44:2853–2860. 2013.
|
|
18
|
Went PT, Lugli A, Meier S, et al: Frequent
EpCam protein expression in human carcinomas. Hum Pathol.
35:122–128. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vincenzi B, Rossie E, Zoccoli A, et al:
Circulating tumor cells in soft tissue sarcomas. In: Poster
presented at 37th EMSO Congress; 2012; http://www.oxfordjournals.org/our_journals/annonc/downloads/annonc23s9-v2.pdfAccessed.
June. 2014
|
|
20
|
Sanchez-Carbayo M, Belbin TJ, Scotlandi K,
et al: Expression profiling of osteosarcoma cells transfected with
MDR1 and NEO genes: regulation of cell adhesion, apoptosis, and
tumor suppression-related genes. Lab Invest. 83:507–517. 2003.
View Article : Google Scholar
|
|
21
|
Barretina J, Caponigro G, Stransky N, et
al: The Cancer Cell Line Encyclopedia enables predictive modelling
of anticancer drug sensitivity. Nature. 483:603–607. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schmelzer E and Reid LM: EpCAM expression
in normal, non-pathological tissues. Front Biosci. 13:3096–3100.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Herlyn M, Steplewski Z, Herlyn D and
Koprowski H: Colorectal carcinoma-specific antigen: detection by
means of monoclonal antibodies. Proc Natl Acad Sci USA.
76:1438–1442. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Patriarca C, Macchi RM, Marschner AK and
Mellstedt H: Epithelial cell adhesion molecule expression (CD326)
in cancer: a short review. Cancer Treat Rev. 38:68–75. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gutzmer R, Li W, Sutterwala S, et al: A
tumor-associated glycoprotein that blocks MHC class II-dependent
antigen presentation by dendritic cells. J Immunol. 173:1023–1032.
2004. View Article : Google Scholar
|
|
26
|
Münz M, Kieu C, Mack B, Schmitt B, Zeidler
R and Gires O: The carcinoma-associated antigen EpCAM upregulates
c-Myc and induces cell proliferation. Oncogene. 23:5748–5758.
2004.PubMed/NCBI
|
|
27
|
Fagerberg J, Hjelm AL, Ragnhammar P,
Frödin JE, Wigzell H and Mellstedt H: Tumor regression in
monoclonal antibody-treated patients correlates with the presence
of anti-idiotype-reactive T lymphocytes. Cancer Res. 55:1824–1827.
1995.
|
|
28
|
Chelius D, Ruf P, Gruber P, et al:
Structural and functional characterization of the trifunctional
antibody catumaxomab. MAbs. 2:309–319. 2010. View Article : Google Scholar
|
|
29
|
Seimetz D, Lindhofer H and Bokemeyer C:
Development and approval of the trifunctional antibody catumaxomab
(anti-EpCAM × anti-CD3) as a targeted cancer immunotherapy. Cancer
Treat Rev. 36:458–467. 2010.
|
|
30
|
Ströhlein MA and Heiss MM: The
trifunctional antibody catumaxomab in treatment of malignant
ascites and peritoneal carcinomatosis. Future Oncol. 6:1387–1394.
2010.PubMed/NCBI
|
|
31
|
Bell SW, Kempson RL and Hendrickson MR:
Problematic uterine smooth muscle neoplasms. A clinicopathologic
study of 213 cases. Am J Surg Pathol. 18:535–558. 1994. View Article : Google Scholar : PubMed/NCBI
|