Introduction
Flap endonuclease 1 (FEN1) is a versatile, structure
specific and multifunctional nuclease involved in DNA replication
and repair (1,2). Human FEN1, which is the archetypal
member of the Rad2 nuclease family (3,4), is
located on chromosome 11q12 and consists of two exons and one
intron. FEN1 efficiently removed the 5′-flaps generated by Polδ/ε
during repair synthesis of long-patch base-excision repair (LP-BER)
and removed primers during lagging-strand DNA synthesis and Okazaki
fragment processing (3,5,6).
Furthermore, FEN1 can be stimulated to promote apoptotic DNA
fragmentation following apoptotic stimuli, acting as a 5′
exonuclease (1) and a gap-dependent
endonuclease (7,8), as reported via its ability to
participate in multiple protein-protein interactions. Thus far,
>30 FEN1-interacting proteins have been identified (2). Of these FEN1 interaction partners,
proliferating cell nuclear antigen (PCNA), which was initially
identified as a replication accessory protein, accompanies FEN1 in
all FEN1-involved DNA metabolic pathways except for the apoptotic
DNA fragmentation pathway, suggesting a critical role of the
FEN1/PCNA interaction in regulating LP-BER (9). A tumor suppressor function for FEN1 has
been shown in preclinical models (10–14).
Therefore, FEN1 has been considered as a key factor during
maintenance of genomic stability and protecting against
carcinogenesis.
However, being a multifunctional factor, mutation of
FEN1 has been suggested to cause genomic instability and
predisposition to cancer. The functional impairment of yeast RAD27
(the homolog of mammalian FEN1) leads to a marked increase
in the rate of spontaneous mutations (8,15,16). A recent study showed that groups of
FEN1 mutations in cancer specimens that abrogated two of the
three nuclease activities lead to cancer initiation and progression
(17). Yang et al (18) identified two single-nucleotide
polymorphisms (SNP), −69G>A (rs174538, in the gene promoter
region) and 4150G>T (rs4246215, in gene 3′-untranslated region),
following thorough re-sequencing of the FEN1 locus in 30
Chinese Han healthy volunteers. The study identified that the
−69G>A change leads to elevated promoter activity, which is most
likely due to a higher binding affinity of the G allele with
certain unknown transcriptional inhibitors. The −69G>A and
4150G>T SNPs influenced gene expression in vivo
subsequent to examining FEN1 mRNA in 38 lung normal tissues,
15 esophagus normal tissues, 12 stomach normal tissues and 13
normal tissues through quantitative analyses (18). Abnormal expression and/or function of
FEN1 resulting from SNPs may possibly contribute to
different cancer susceptibility. On the basis of the previous
findings mentioned, we hypothesized that the functional genetic
variants in the FEN1 gene may affect cancer risk.
Meta-analysis is a statistical technique for combining results from
different studies to produce a single estimate of the major effect
with enhanced precision (19).
Therefore, a meta-analysis of the published studies was conducted
to derive a more precise estimation of the association between
FEN1 polymorphisms and cancer risk.
Materials and methods
Identification and eligibility of
relevant studies
Computer searches were carried out by two
investigators independently in Embase, Pubmed, ISI Web of Knowledge
and Chinese National Knowledge Infrastructure databases (until
March 31, 2014) to collect case-control studies of the FEN1
SNPs (rs174538 and rs4246215) association with cancer risk. The
keywords were as follows: Cancer/carcinoma, Flap
endonuclease-1/FEN1, −69G>A/rs174538 and
4150G>T/rs4246215 and polymorphism/genotype/SNP. In addition,
reference lists of the main studies and reviews were also assessed
by a manual search to identify additional relevant publications.
The following criteria were used to select studies for further
meta-analysis: i) Case-control studies; ii) studies that evaluated
the association of FEN1 SNPs (rs174538 and rs4246215) on
cancer risk; iii) studies that contained at least two comparison
groups (cancer vs. control group); and iv) studies that included
detailed genotyping data.
The following exclusion criteria were used
accordingly: i) The design of the experiments were not case-control
studies; ii) the source of cases and controls, and other essential
information were not provided; iii) the genotype distribution of
the control population was departure from Hardy-Weinberg
Equilibrium; and iv) reviews and duplicated publications.
Data extraction
Evaluations of studies were performed independently
by two investigators and data with discrepancies in identification
were discussed by all investigators. For each included study, the
following information was collected: First author, year of
publication, country of origin, ethnicity, source of control,
number of cases and controls, genotyping methods for rs174538 and
rs4246215, total number of cases and controls, as well as number of
cases and controls with A/A, A/G, G/G and T/T, T/G, G/G genotypes.
All the case and control groups were well-controlled.
Statistical analysis
For the control group of each study, the allelic
frequency was calculated. The strength of associations between
FEN1 SNPs (rs174538 and rs4246215) and cancer risk were
measured by odds ratio (OR) with 95% confidence interval (CI). For
rs174538, the AA genotype was used as the reference genotype in all
analyses. The risks of the GG and GA genotypes for cancers were
estimated, compared to the AA homozygote, and subsequently the
risks of GA+GG for cancer were evaluated, respectively.
Accordingly, for rs4246215, the TT genotype was used as the
reference genotype in all the analyses. The risks of the GT and GG
genotypes for cancer were estimated, compared to the TT homozygote,
and subsequently the risks of GT+GG for cancers were evaluated. The
significance of the pooled OR was determined by the Z test.
Statistical heterogeneity among studies was assessed with the Q and
I2 statistics. The Q test and I2 were claimed
to test the variation, which was due to heterogeneity or by random
error. When the P-value of heterogeneity tests was P≤0.1, the
random effects model was used. When the P-value of heterogeneity
test was P≥0.1, the fixed effects model was used. Sensitivity
analysis was also tested by removing one study at a time to
calculate the overall homogeneity and effect size. Publication bias
was evaluated by the funnel plot and further assessed by the method
of Egger's linear regression test. All the statistical analyses
were carried out with the Review Manager version 5.1 software
(Revman; The Cochrane Collaboration, Oxford, United Kingdom). All
P-values in the meta-analysis were two-sided, and P<0.05 were
considered to indicate a statistically significant difference.
Results
Characteristics of studies
A total of 19 sub-studies from 8 studies that
fulfilled our search criteria were preliminarily identified for
further detailed evaluation (Fig. 1).
One study was excluded as the designs of the experiments were not
case-control studies. Two studies did not focused on FEN1
SNPs (rs174538 and rs4246215) and cancer risk. Two studies were
excluded as there was no detailed genotyping data. Four studies
were review comments. Finally, 13 sub-studies from 4 studies on
rs174538 and rs4246215 genotypes and cancer risk were identified
(18,20–22),
including a total of 5,108 cancer cases and 6,382 case-free
controls. The characteristics of the included studies are listed in
Table I. The included studies were
all based on Chinese populations. All were case-control studies,
including lung cancer, breast cancer, glioma, hepatocellular
carcinoma, esophageal cancer, gastric cancer and colorectal cancer.
All the types of cancer were confirmed by histology or pathology.
Additionally, the controls were mainly matched on age, of which all
the studies were hospital-based.
 | Table I.Characteristics of the studies
included in the meta-analysis. |
Table I.
Characteristics of the studies
included in the meta-analysis.
| First author, year
(Ref.) | Country | Ethnicity | Cancer type | Genotyping
method | Source of
control | Total sample size
(case/control) |
|---|
| Lv, 2014 (20) |
|
|
|
|
|
|
| 1 | China | Asian | Breast cancer | PCR-RFLP | Hospital | 800/800 |
| 2 | China | Asian | Breast cancer | PCR-RFLP | Hospital | 300/600 |
| Chen, 2013 (21) | China | Asian | Glioma | PCR-RFLP | Hospital | 317/802 |
| Liu, 2012 (22) |
|
|
|
|
|
|
| 1 | China | Asian | Hepatocellular
carcinoma | PCR-RFLP | Hospital | 411/423 |
| 2 | China | Asian | Esophageal
cancer | PCR-RFLP | Hospital | 266/386 |
| 3 | China | Asian | Gastric cancer | PCR-RFLP | Hospital | 220/250 |
| 4 | China | Asian | Colorectal
cancer | PCR-RFLP | Hospital | 126/162 |
| 5 | China | Asian | Hepatocellular
carcinoma | PCR-RFLP | Hospital | 237/315 |
| 6 | China | Asian | Esophageal
cancer | PCR-RFLP | Hospital | 289/337 |
| 7 | China | Asian | Gastric cancer | PCR-RFLP | Hospital | 192/204 |
| 8 | China | Asian | Colorectal
cancer | PCR-RFLP | Hospital | 110/145 |
| Yang, 2009
(18) |
|
|
|
|
|
|
| 1 | China | Asian | Lung cancer | PCR-RFLP | Hospital | 1,013/1,131 |
| 2 | China | Asian | Lung cancer | PCR-RFLP | Hospital | 827/827 |
Quantitative synthesis
The frequency of the A allele varied widely across
the 13 studies, ranging from 23 to 46% in rs174538 among 6,381
healthy controls (Table II). The
frequency of the T allele ranged from 35 to 46% in rs4246215 among
6,381 healthy controls (Table III).
The average frequencies of the A and T allele in the two
polymorphisms (rs174538 and rs4246215) were 39 and 41%,
respectively.
 | Table II.rs174538 polymorphism genotype
distribution and allele frequency in cases and controls. |
Table II.
rs174538 polymorphism genotype
distribution and allele frequency in cases and controls.
|
| Genotype, n | Allele frequency, n
(%) |
|---|
|
|
|
|
|---|
|
| Case | Control | Case | Control |
|---|
|
|
|
|
|
|
|---|
| First author, year
(Ref.) | Total | GG | GA | AA | Total | GG | GA | AA | G | A | G | A |
|---|
| Chen, 2013
(21) |
317 | 160 | 122 | 35 |
802 | 316 | 356 | 130 | 437
(69) | 197 (31) | 988
(62) | 616 (38) |
| Liu, 2012 (22) |
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
410 | 203 | 173 | 34 |
423 | 174 | 185 | 64 | 579
(71) | 241 (29) | 533
(63) | 313 (37) |
| 2 |
266 | 137 | 105 | 24 |
386 | 163 | 168 | 55 | 379
(71) | 153 (29) | 494
(64) | 278 (36) |
| 3 |
220 | 118 | 86 | 16 |
250 | 108 | 108 | 34 | 322
(73) | 118 (27) | 324
(65) | 176 (35) |
| 4 |
126 | 64 | 51 | 11 |
162 | 65 | 73 | 24 | 178
(71) | 73
(29) | 203
(63) | 121 (37) |
| 5 |
238 | 87 | 117 | 34 |
315 | 96 | 149 | 70 | 281
(59) | 195 (41) | 341
(54) | 289 (46) |
| 6 |
289 | 107 | 144 | 38 |
336 | 100 | 163 | 73 | 358
(62) | 220 (38) | 393
(54) | 309 (46) |
| 7 |
192 | 71 | 96 | 25 |
204 | 61 | 101 | 42 | 138
(49) | 146 (51) | 223
(55) | 185 (45) |
| 8 |
110 | 40 | 53 | 17 |
145 | 44 | 71 | 30 | 133
(60) | 87
(40) | 159
(55) | 131 (45) |
| Lv, 2014 (20) |
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
800 | 401 | 317 | 82 |
800 | 315 | 355 | 130 | 1,119 (70) | 481 (30) | 985
(62) | 615 (38) |
| 2 |
300 | 146 | 120 | 34 |
600 | 200 | 284 | 116 | 412
(69) | 188 (31) | 784
(65) | 416 (35) |
| Yang, 2009
(18) |
|
|
|
|
|
|
|
|
|
|
|
|
| 1 | 1,013 | 505 | 402 | 106 | 1,131 | 467 | 496 | 168 | 1,402 (70) | 614 (30) | 1,430 (63) | 832 (37) |
| 2 |
827 | 286 | 394 | 147 |
827 | 257 | 384 | 186 | 966
(58) | 688 (42) | 898
(54) | 756 (46) |
 | Table III.rs4246215 polymorphism genotype
distribution and allele frequency in cases and controls. |
Table III.
rs4246215 polymorphism genotype
distribution and allele frequency in cases and controls.
|
| Genotype, n | Allele frequency, n
(%) |
|---|
|
|
|
|
|---|
|
| Case | Control | Case | Control |
|---|
|
|
|
|
|
|
|---|
| First author, year
(Ref.) | Total | GG | GT | TT | Total | GG | GT | TT | G | T | G | T |
|---|
| Chen 2013 (21) |
314 | 160 | 120 | 34 |
802 | 309 | 363 | 130 | 440
(70) | 188 (30) | 981
(61) | 623 (39) |
| Liu, 2012 (22) |
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
411 | 195 | 177 | 39 |
423 | 176 | 187 | 60 | 567
(69) | 255 (31) | 539
(64) | 307 (36) |
| 2 |
249 | 115 | 114 | 20 |
386 | 161 | 172 | 53 | 344
(69) | 154 (31) | 494
(64) | 278 (36) |
| 3 |
210 | 111 | 82 | 17 |
250 | 107 | 110 | 33 | 304
(72) | 116 (28) | 324
(65) | 176 (35) |
| 4 |
119 | 61 | 47 | 11 |
161 | 65 | 74 | 22 | 169
(71) | 69
(29) | 204
(63) | 118 (37) |
| 5 |
237 | 85 | 118 | 34 |
315 | 98 | 148 | 69 | 288
(61) | 186 (39) | 344
(55) | 286 (45) |
| 6 |
289 | 110 | 141 | 38 |
337 | 101 | 164 | 72 | 361
(62) | 217 (38) | 366
(54) | 308 (46) |
| 7 |
192 | 72 | 95 | 25 |
204 | 59 | 102 | 43 | 239
(62) | 145 (38) | 220
(54) | 188 (46) |
| 8 |
110 | 39 | 55 | 16 |
145 | 44 | 71 | 30 | 133
(60) | 87
(40) | 159
(55) | 131 (45) |
| Lv, 2014 (20) |
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
800 | 365 | 335 | 100 |
800 | 308 | 362 | 130 | 1,065 (67) | 535 (33) | 978
(61) | 622 (39) |
| 2 |
300 | 152 | 114 | 34 |
600 | 195 | 289 | 116 | 418
(70) | 182 (30) | 679
(57) | 521 (43) |
| Yang, 2009
(18) |
|
|
|
|
|
|
|
|
|
|
|
|
| 1 | 1,013 | 468 | 421 | 124 | 1,131 | 460 | 500 | 171 | 1,357 (67) | 669 (33) | 1,420 (63) | 842 (37) |
| 2 |
827 | 286 | 394 | 147 |
827 | 257 | 383 | 187 | 966
(58) | 688 (42) | 897
(54) | 757 (46) |
The main results of the meta-analysis are listed in
Tables IV and V. Overall, there was evidence of an
association between the variant genotypes and cancer risk in
different genetic models when all the studies were pooled into the
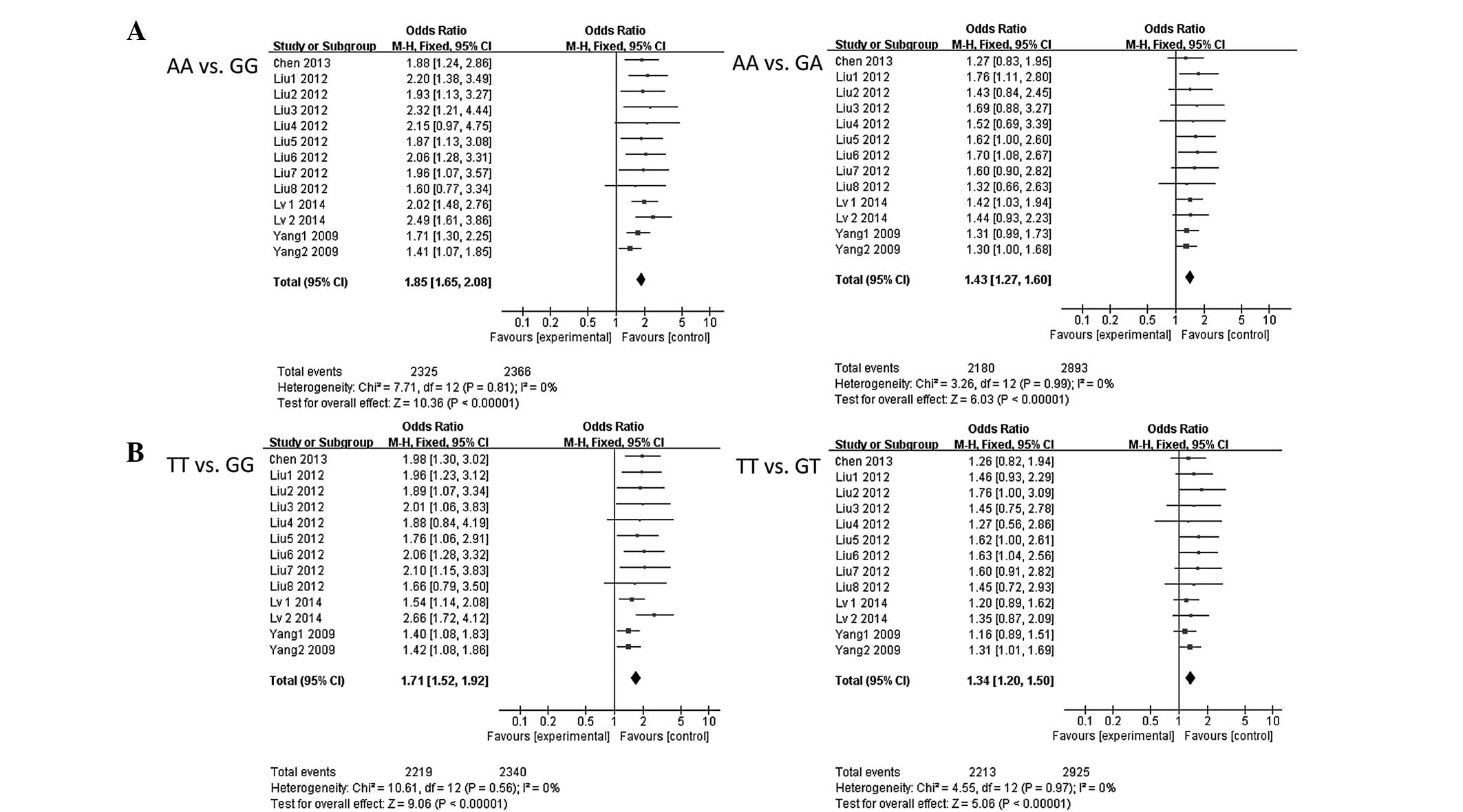
meta-analysis. As shown in Table IV,
carriers of the FEN1 −69GG genotype showed a significantly
elevated risk of cancer compared to −69AA carriers (OR, 1.85; 95%
CI, 1.65–2.08; P<0.00001). Logistic regression analyses also
revealed that individuals with FEN1 −69GA genotypes were
significantly associated with increased cancer risk compared to
−69AA genotypes (OR, 1.43; 95% CI, 1.27–1.60; P<0.00001). In
addition, the variant GG+GA genotypes were associated with an
increased cancer risk when compared to the −69AA genotypes (OR,
1.28; 95% CI, 1.16–1.42; P<0.00001). Similar results were
observed for the 4150G>T polymorphism. As shown in Table V, the FEN1 4150GG genotype
showed a significantly elevated risk of cancer compared to 4150TT
carriers (OR, 1.71; 95% CI, 1.52–1.92; P<0.00001). Logistic
regression analyses also revealed that individuals with the
FEN1 4150GT genotypes were significantly associated with an
increased cancer risk compared to the 4150TT genotypes (OR, 1.34;
95% CI, 1.20–1.50; P<0.00001). The variant GG+GT genotypes were
associated with an increased cancer risk when compared to the
4150TT genotypes (OR, 1.50; 95% CI, 1.35–1.67; P<0.00001).
 | Table IV.Risk of cancer associated with the
genotypes of FEN1 −69G>A (rs174538). |
Table IV.
Risk of cancer associated with the
genotypes of FEN1 −69G>A (rs174538).
|
|
|
|
| Heterogeneity |
|
|---|
|
|
|
|
|
|
|
|---|
| Genotype | OR | 95% CI | P-value | I2,
% | P-value | Effects model |
|---|
| AA | 1 (Reference) |
|
|
|
|
|
| GA | 1.43 | 1.27–1.60 | <0.00001 | 0 | 0.99 | F |
| GG | 1.85 | 1.65–2.08 | <0.00001 | 0 | 0.05 | R |
| GA+GG | 1.28 | 1.16–1.42 | <0.00001 | 91 | <0.00001 | R |
 | Table V.Risk of cancer associated with the
genotypes of FEN1 4150G>T (rs4246215). |
Table V.
Risk of cancer associated with the
genotypes of FEN1 4150G>T (rs4246215).
|
|
|
|
| Heterogeneity |
|
|---|
|
|
|
|
|
|
|
|---|
| Genotype | OR | 95% CI | P-value | I2,
% | P-value | Effects model |
|---|
| TT | 1 (Reference) |
|
|
|
|
|
| GT | 1.34 | 1.20–1.50 | <0.00001 | 0 | 0.97 | F |
| GG | 1.71 | 1.52–1.92 | <0.00001 | 0 | 0.56 | F |
| GT+GG | 1.50 | 1.35–1.67 | <0.00001 | 91 | 0.91 | F |
Tests of heterogeneity
Statistically significant heterogeneity was observed
between trials of the following analyses using the Q statistic. As
shown in Fig. 2, for AA vs. GG:
Pheterogeneity=0.05, I2=0, and
therefore, a random effect model was performed; for AA vs. GA:
Pheterogeneity=0.99, I2=0, a
fixed-effect model was performed; for TT vs. GG:
Pheterogeneity=0.56, I2=0, a
fixed-effect model was performed; and for TT vs. GT:
Pheterogeneity=0.91, I2=0, a
fixed-effect model was performed.
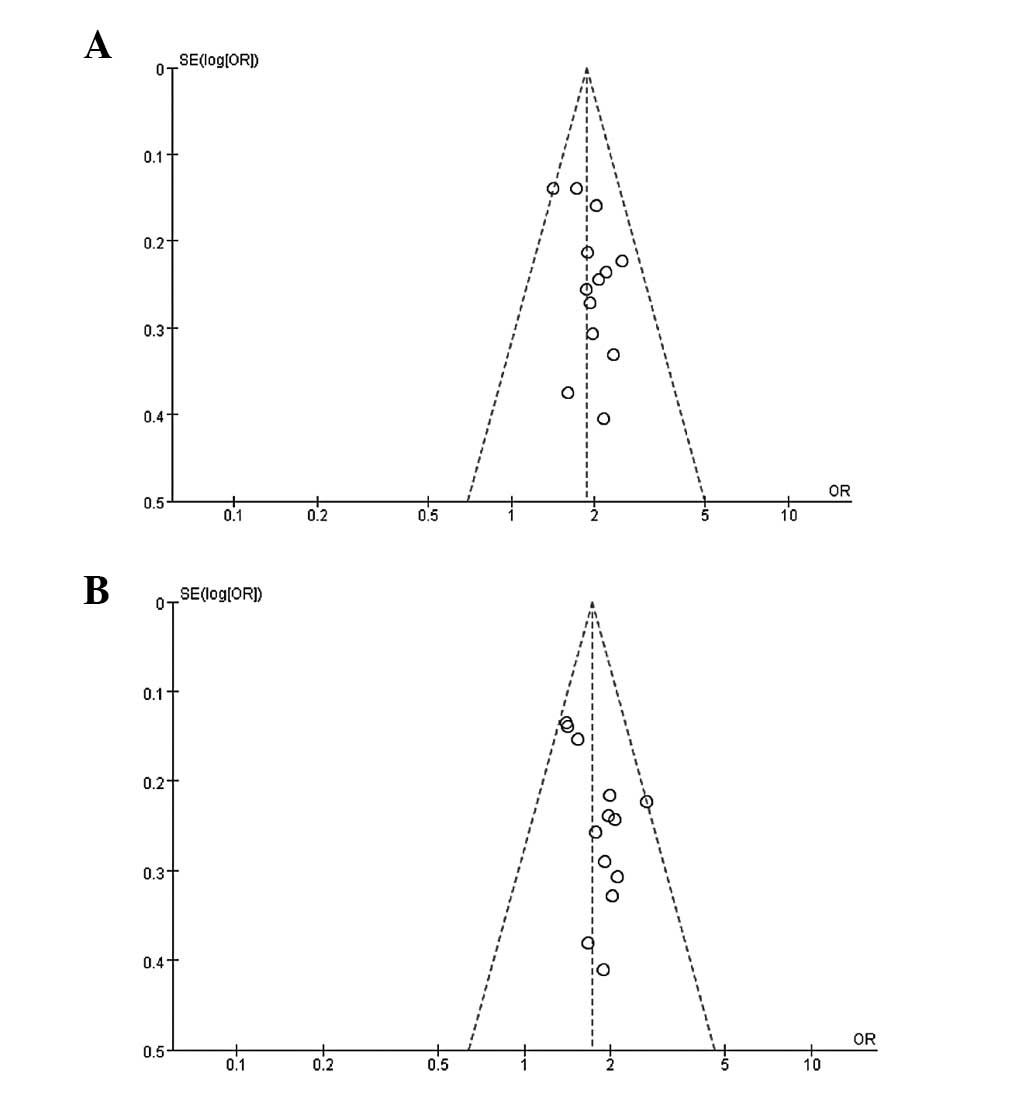
Publication bias
Begg's funnel plot and Egger's test were performed
to assess the publication bias. The funnel plots did not reveal any
clear asymmetry in all the genotypes (Fig. 3), and the results of Egger's test
revealed no publication bias (P>0.05).
Discussion
FEN1 exhibits a prominent role in maintaining
genomic stability and protecting against malignant transformation
through its involvement in DNA repair and other multiple DNA
metabolic pathways (8). Therefore,
the structure or functional deficiency of FEN1 may destroy the
genomic stability to increase the risk of cancer. As previously
mentioned, FEN1 mutations reduced nuclease activity to lead
to cancer initiation and development (15). FEN1 −69 G and 4150 G alleles,
which were correlated to significantly decreased FEN1 mRNA
expression in normal gastrointestinal tissues, were associated with
increased gastrointestinal cancer risks compared to −69A and 4150T
alleles in two independent case-control cohorts (21). The FEN1 polymorphisms, rs174538
and rs4246215, may be common cancer risk factors.
In the present meta-analysis, all 13 case-control
studies were pooled in a Chinese population to estimate the overall
cancer risk of the SNPs. In the present study, the AA and TT
genotypes were used as a reference genotype in all the analyses.
For rs174538, it was found that individuals exhibiting the GG and
GG/GA genotypes were significantly associated with an increased
risk of cancer compared to the −69AA genotype. In the combined
meta-analyses, the −69GG genotype had a 1.85-fold increased risk
for cancer. Similar results were identified for rs4246215, all the
genotypes were significantly associated with an increased cancer
risk compared to the TT genotype (P<0.01). The present study
indicated that functional rs174538 and rs4246215 were significantly
associated with an increased risk of cancer. These results are
consistent to the findings in breast cancer, lung cancer,
hepatocellular carcinoma, esophageal cancer, gastric cancer,
colorectal cancer and glioma from different medical centers of
China (18,20–22), which
confirmed our speculation that the functional genetic variants in
the FEN1 gene may affect cancer risk as common factors.
When interpreting the results of the present study,
certain limitations of the meta-analysis must be considered.
Firstly, all the cancer cases and controls were hospital-based, and
inherent selection bias may exist. Thus, it is important to
validate these findings in a population-based prospective study.
Secondly, the meta-analysis was based on pooled data and no
individual data was available; thus, the risk of cancer could not
be assessed according to stratification of gene-environment and
other risk factors of cancer. Thirdly, all the subjects were
Chinese, and all the genotyping methods in the included studies
were polymerase chain reaction-restriction fragment length
polymorphism. Further large-scale multicenter studies with more
detailed individual data, with different environmental backgrounds
are warranted to further validate the gene-gene and
gene-environment interactions on SNPs and cancer risk.
In conclusion, the present meta-analysis provides
evidence of the effects of FEN1 SNPs (rs174538 and
rs4246215) on the cancer risk. The study indicated that functional
rs174538 and rs4246215 were significantly associated with an
increased risk of cancer in the Chinese population. Further studies
based on different ethnicity are warranted to verify these
findings.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81471670); the
International Cooperative Project of Shaanxi province, China (grant
no. 2013KW-32-01); the Fundamental Research Funds for the Central
Universities, China; and the Specialized Research Fund of the
Second Affiliated Hospital of Xi'an Jiaotong University, China
[grant no. RC (GG) 201203].
References
|
1
|
Liu Y, Kao HI and Bambara RA: Flap
endonuclease 1: a central component of DNA metabolism. Annu Rev
Biochem. 73:589–615. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zheng L, Jia J, David Finger LD, Guo Z, et
al: Functional regulation of FEN1 nuclease and its link to cancer.
Nucleic Acid Res. 39:781–794. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lieber MR: The FEN-1 family of
structure-specific nucleases in eukaryotic DNA replication,
recombination and repair. Bioessays. 19:233–240. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tomlinson CG, Atack JM, Chapados B, et al:
Substrate recognition and catalysis by flap endonucleases and
related enzymes. Biochem Soc Trans. 38:433–437. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Harrington JJ and Lieber MR: The
characterization of a mammalian DNA structure-specific
endonuclease. EMBO J. 13:1235–1246. 1994.PubMed/NCBI
|
|
6
|
Shen B, Singh P, Liu R, et al: Multiple
but dissectible functions of FEN-1 nucleases in nucleic acid
processing, genome stability and diseases. Bioessays. 27:717–729.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reagan MS, Pittenger C, Siede W and
Friedberg EC: Characterization of a mutant strain of Saccharomyces
cerevisiae with a deletion of the RAD27 gene, a structural homolog
of the RAD2 nucleotide excision repair gene. J Bacteriol.
177:364–371. 1995.PubMed/NCBI
|
|
8
|
Zheng L, Zhou M, Chai Q, et al: Novel
function of the flap endonuclease 1 complex in processing stalled
DNA replication forks. EMBO Rep. 6:83–89. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zheng L, Dai H, Qiu J, et al: Disruption
of the FEN-1/PCNA interaction results in DNA replication defects,
pulmonary hypoplasia, pancytopenia and newborn lethality in mice.
Mol Cell Biol. 27:3176–3186. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Henneke G, FriedrichHeineken E and
Hubscher U: Flap endonuclease 1: a novel tumour suppresser protein.
Trends Biochem Sci. 28:384–390. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Henneke G, Koundrioukoff S and Hubscher U:
Phosphorylation of human Fen1 by cyclin-dependent kinase modulates
its role in replication fork regulation. Oncogene. 22:4301–4313.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kucherlapati M, Yang K, Kuraguchi M, et
al: Haploinsufficiency of flap endonuclease (Fen1) leads to rapid
tumor progression. Proc Natl Acad Sci. 99:9924–9929. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu Z, Lin Y, Xu H, et al: High risk of
benzo [alpha] pyrene-induced lung cancer in E160D FEN1 mutant mice.
Mutat Res. 731:85–91. 2001. View Article : Google Scholar
|
|
14
|
Xu H, Zheng L, Dai H, et al:
Chemical-induced cancer incidence and underlying mechanisms in Fen1
mutant mice. Oncogene. 30:1072–1081. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tishkoff DX, Filosi N, Gaida GM and
Kolodner RD: A novel mutation avoidance mechanism dependent on S.
cerevisiae RAD27 is distinct from DNA mismatch repair. Cell.
88:253–263. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Parrish JZ, Yang CL, Shen BH and Xue D:
CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates
with CPS-6/EndoG to promote apoptotic DNA degradation. EMBO J.
22:3451–3460. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng L, Dai H, Zhou M, et al: Fen1
mutations result in autoimmunity, chronic inflammation and cancers.
Nat Med. 13:812–819. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang M, Guo H, Wu C, et al: Functional
FEN1 polymorphisms are associated with DNA damage levels and lung
cancer risk. Hum Mutat. 30:1320–1328. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dai ZJ, Wang XJ, Kang AJ, et al:
Association between APE1 single nucleotide polymorphism (rs1760944)
and cancer risk: a meta-analysis based on 6,419 cancer cases and
6,781 case-free controls. J Cancer. 5:253–259. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lv Z, Liu W, Li D, et al: Association of
functional FEN1 genetic variants and haplotypes and breast cancer
risk. Gene. 538:42–45. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen YD, Zhang X, Qiu XG, et al:
Functional FEN1 genetic variants and haplotypes are associated with
glioma risk. J Neurooncol. 111:145–151. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu L, Zhou C, Zhou L, et al: Functional
FEN1 genetic variants contribute to risk of hepatocellular
carcinoma, esophageal cancer, gastric cancer and colorectal cancer.
Carcinogenesis. 33:119–123. 2012. View Article : Google Scholar : PubMed/NCBI
|