Introduction
Thrombosis is one of the most frequent complications
in cancer and the second leading cause of death among patients with
malignant diseases (1). Despite the
strong association between thrombosis and cancer, thrombosis
remains underdiagnosed and undertreated in such patients.
Predicting the advent of thrombosis may be difficult due to lack of
reliable markers.
The protein C (PC) pathway plays a major role in
regulating coagulation. PC is activated when thrombin binds to the
endothelial cell surface receptor thrombomodulin. Activated PC
(APC), in the presence of its cofactor, protein S, inactivates
factors Va and VIIIa, thus limiting the progression of the
coagulation cascade. The interaction between endothelial PC
receptor (EPCR) and its ligand increases the affinity of PC for the
thrombin-thrombomodulin complex. EPCR, which binds PC or APC with
the same affinity, may be released in the plasma as a free soluble
form (sEPCR) from membrane-associated EPCR. Circulating sEPCR is
formed through the action of a metalloprotease, which is activated
by thrombin and by certain inflammatory mediators (2–4), or as a
result of proteolytic cleavage (a disintegrin and metalloprotease
17/tumor necrosis factor α-converting enzyme) (5). sEPCR circulates in the plasma and
inhibits PC activation and APC anticoagulant function (6). sEPCR, by binding free PC, reduces the
availability of the latter for the EPCR present on endothelial
cells, leading to an increase in the incidence of thrombosis
(7). EPCR is a 46-kDa type 1
transmembrane glycoprotein. The plasma variation of sEPCR is under
genetic control. The EPCR gene consists of 4 exons and is located
on chromosome 20q-11.2. Several studies have reported certain EPCR
gene polymorphisms as candidate risk factors for thrombosis
(8).
The 6936A/G single-nucleotide polymorphism (SNP)
results in a serine-to-glycine substitution at residue 219 in the
transmembrane domain and is associated with increased levels of
sEPCR and increased thrombotic risk (9). In a previous study, we reported that
malignant cells express and secrete EPCR and that sEPCR is
associated with hypercoagulability in human hematological
malignancies (10,11). We also reported that the EPCR gene
sequence of HL-60 myeloblastic leukemia cells is quite identical to
that of endothelial cells, in that it harbors the different SNPs
(11) as previously reported for the
endothelial cell gene (12). However,
in the HL-60 cell gene, a thymidine insertion (locus 260) and an
adenosine deletion (locus 840) were also identified within the 5′
untranslated region. Moreover, a thymidine repetition was detected
within the untranslated region (locus 7650) of the exon IV
(10). The HL-60 cells are malignant
leukemic cells and, therefore, finding additional modifications
inherent in the EPCR gene structure was not surprising. In this
study, we demonstrated that, while the risk of thrombosis is high
in cancer patients, the presence of 6936A/G SNP further increases
this risk, underlining the synergetic role of the EPCR gene
polymorphisms in coagulation.
Patients and methods
Patients
In order to determine the role of the EPCR 6936A/G
SNP as a possible indicator of the risk of thrombosis in leukemia,
we conducted a retrospective study on 205 patients with diagnosed
hematological malignancies. DNA from patients with leukemia, who
were admitted at the Hematology Departments of Hôtel-Dieu and
Saint-Antoine hospitals (Paris, France) between 1995 and 2012, were
obtained from the Leukemia Tumor Bank (Table IA). Blood samples from healthy donors
were obtained from the Hôtel-Dieu blood bank and used as controls
(Table IA). The patients (117 men and
88 women) were divided into 3 groups on the basis of their
pathology as follows: Chronic lymphocytic leukemia (CLL, n=76),
acute myeloid leukemia (AML, n=72) and acute lymphoblastic leukemia
(ALL, n=33). In addition to these 3 groups, a fourth group included
other hematological pathologies, namely lymphoma (n=13), chronic
myelogenous leukemia (n=8), chronic myelomonocytic leukemia (n=1),
B-cell prolymphocytic leukemia (n=1) and myeloproliferative
neoplasm (n=1).
 | Table I.Subject data. |
Table I.
Subject data.
| A, Disease-wise split
of patients/healthy donors by gender and age |
|---|
|
|---|
| Subjects | No. | M/F | Age, years mean ± SD
(range) |
|---|
| Healthy donors | 63 | 23/40 | 35±15
(22–57) |
| Patients | | | |
|
Total | 205 | 117/88 | 59±19
(15–91) |
| CLL | 76 | 39/37 | 68±10
(90–44) |
| AML | 72 | 43/29 | 59±17
(25–91) |
| ALL | 33 | 18/15 | 38±17
(15–80) |
|
Othersa | 24 | 17/7 | 61±16
(34–83) |
|
| B, Patients with
available and complete clinical data |
|
| Patients | No. | M/F | Age, years mean ± SD
(range) |
|
| Total | 110 | 56/46 | 59±19
(15–91) |
| CLL | 25 | 9/16 | 71±9 (88–50) |
| AML | 55 | 29/26 | 60±18
(25–91) |
| ALL | 16 | 6/10 | 37±18
(15–76) |
| Others | 14 | 10/4 | 59±16
(34–79) |
|
| C, Distribution of
types of thrombosis |
|
|
| Thrombosis type |
|
|
|
| Total no. of
patients | DIC | DVT | PE | IS | AT | SVT | C | S | NS |
|
| 23 | 5 | 9 | 2 | 2 | 1 | 1 | 1 | 1 | 1 |
Only 110 medical records were available for
profiling analysis. We were therefore obliged to disregard the
remaining 95 cases (Table IB). The
diagnosed types of thrombosis are presented in Table IC.
Ethical approval
All patients were informed and consent was obtained
in accordance with the Declaration of Helsinki. Authorization for
conservation and preparation of elements of the human body for
scientific purposes no. AC-2013–1992.
Blood mononuclear cells and DNA
samples
Following patient informed consent, venous blood
samples were collected in 5-ml tubes containing heparin or EDTA.
The blood was centrifuged and the plasma discarded. Mononuclear
cells were isolated by Ficoll density gradient centrifugation using
lymphocyte separation medium (GE Healthcare Ltd.,
Velizy-Villacoublay, France), according to the manufacturer's
recommendations and the resulting pellets were frozen at −80°C and
conserved in the leukemia cell bank until use. DNA from
5×106 cells was extracted and purified using Qiagen spin
columns (Qiagen, Courtaboeuf, France). The purity and concentration
of the DNA were determined by optical density measured at 260 nm
using a NanoDrop™ 2000c spectrophotometer (Thermo Fisher
Scientific, Villebon-sur-Yvette, France). The DNA samples were
frozen at −20°C until use.
Genotyping of SNP
SNP genotyping is the measurement of genetic
variations of SNPs between members of a species. The EPCR 6936 SNP
was determined using 10 ng/ml DNA in the patented SNP genotyping
system (KBioscience UK Ltd., Hoddesdon, UK) based on fluorescent
resonance energy transfer; this is a homogenous fluorescent
genotyping system using a unique form of competitive
allele-specific polymerase chain reaction (PCR) (the Kompetitive
Allele Specific PCR genotyping system; KASPar system). Genotyping
was performed using GenoScreen (Lille, France). The use of two
competitive allele-specific tailed forward primers (primer for the
6936A allele, AGCCACACCAGCAATGATGAAACT; and primer for the 6936G
allele, GCCACACCAGCAATGATGAAACC) and one reverse primer
(GGAGCCAAACAAGCCGCTCCTA) provided increased locus-specific
discrimination.
Statistical analysis
The Fisher's exact test was employed in order to
verify whether the distribution of 6936A/G SNP was identical
between healthy donors and leukemia patients. We then investigated
the distribution of 6936A/G SNP as a function of gender and type of
hematological malignancy. Finally, we also investigated the
incidence of thrombosis as a function of the type of pathology and
the 6936A/G SNP by resorting to the logistic regression model. All
the tests were in bilateral comparison with a significance level of
0.05. The results were obtained using R version 3.0.0 and version
3.0.1 software.
Results
Allele distribution
The distribution of the 6936A and 6936G alleles was
similar in all subjects (healthy donors and patients; n=268)
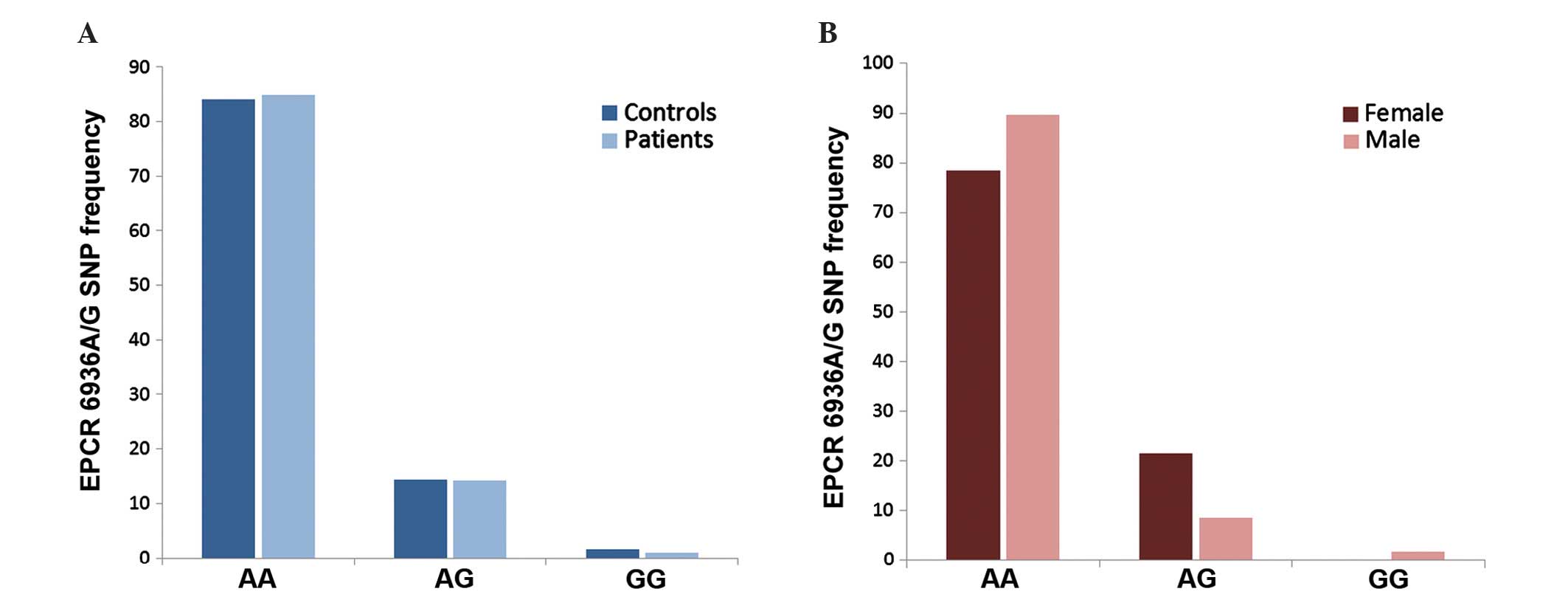
(P=0.859) (Fig. 1A). In the cohort of
patients (n=205), this distribution was found to change according
to gender (Fig. 1B), with the
frequency of the 6936AG genotype being 2-fold lower in men (9%)
compared with that in women (22%) (P=0.009). The male (n=43) and
female (n=29) AML patients presented 11.6 and 31% of the 6936AG
genotype, respectively (data not shown).
Genotype distribution
The genotype distribution according to pathology is
presented in Table II. No
significant difference was observed between the different types of
malignancies (P=0.684).
 | Table II.EPCR 6936A/G SNP as a function of
pathology. |
Table II.
EPCR 6936A/G SNP as a function of
pathology.
|
| Genotype, no.
(%) |
|---|
|
|
|
|---|
| Pathology | AA | AG | GG | Total | P-value |
|---|
| CLL | 66
(87) | 10 (13) | 0 (0) | 76
(100) |
|
| AML | 58
(80) | 12 (17) | 2 (3) | 72
(100) |
|
| ALL | 30
(91) | 3 (9) | 0 (0) | 33
(100) |
|
| Othersa | 20
(83) | 4
(17) | 0 (0) | 24
(100) |
|
| Total | 174 (85) | 29 (14) | 2 (1) | 205 (100) | 0.684 |
Of the 205 patients, case files mentioning whether
they had a previous thrombotic event were available for only 110.
The number of patients, grouped according to each covariate, is
presented in Table III.
 | Table III.Characteristics of patients included
in the regression model (n=110). |
Table III.
Characteristics of patients included
in the regression model (n=110).
| Characteristics | Patient no. |
|---|
| Gender |
|
| Male | 56 |
|
Female | 54 |
| EPCR 6936A/G SNP |
|
| AA | 87 |
| AG | 21 |
| GG | 2 |
| Pathology |
|
| CLL | 25 |
| AML | 55 |
| ALL | 16 |
|
Othersa | 14 |
Incidence of thrombosis
As there were only 2 homozygous patients with the
6936GG genotype in our database, estimates of the effect of the
6936GG genotype on thrombosis would be extremely unreliable;
therefore, they were excluded from the present study, thus leaving
a total of 108 cases for further analysis. In addition, 6 patients
received medication for thrombosis prevention. This medication may
lower the risk of thrombosis, which, in the present analysis, would
introduce a confusion bias. However, given the low number of
patients treated, a stratification or adjustment based on treatment
would result in erroneous estimation. To ensure that the inclusion
of the patients treated for thrombosis prevention did not alter our
findings, we set up the computation with only the 102 non-treated
patients and assessed whether the results were similar to those
obtained for the 108 patients. Of the 108 patients tested for
thrombotic events, 21.3% developed at least one thrombotic episode.
The thrombosis rate in all pathologies for male and female patients
is presented in Table IV.
 | Table IV.Incidence of thrombosis as a function
of EPCR 6936A/G SNP and pathology. |
Table IV.
Incidence of thrombosis as a function
of EPCR 6936A/G SNP and pathology.
| Variables | Patient no.
(M/F) | Thrombosis, %
(patient no., M/F) |
|---|
| All patients | 108
(52/46) | 21.3 (23,
11/12) |
| AA |
87 (47/40) | 17.2 (15, 8/7) |
| AG |
21 (15/16) | 38.1 (8, 3/5) |
| CLL | 25
(9/16) | 12.0 (3, 2/1) |
| AA | 20
(8/12) | 10.0 (2, 2/0) |
| AG | 5
(1/4) | 20.0 (1, 0/1) |
| AML |
53 (27/26) | 28.3 (15, 6/9) |
| AA |
41 (24/17) | 22.0 (9, 3/6) |
| AG | 12 (3/9) | 50.0 (6, 3/3) |
| ALL | 16
(6/10) | 12.5 (2, 1/1) |
| AA | 15 (6/9) | 13.3 (2, 1/1) |
| AG | 1 (0/1) |
0 |
| Othersa | 14
(10/4) | 21.4 (3, 2/1) |
| AA | 11 (9/2) | 18.2 (2, 2/0) |
| AG | 3
(1/2) | 33.3 (1, 0/1) |
The data analysis revealed that thrombosis occurred
in 12% of patients with CLL, 28.3% of patients with AML, 12.5% of
patients with ALL and 21.4% of patients with other hematological
malignancies (lymphoma, 13; chronic myelogenous leukemia, 8; and
chronic myelomonocytic leukemia, B-cell prolymphocytic leukemia and
myeloproliferative neoplasm, 1 each).
On analysis of the data regarding EPCR 6936A/G SNP,
we found that the incidence of thrombotic events was higher when
the 6936AG genotype (38.1%) was present, compared with the 6936AA
genotype (17.2%); in patients with the 6936AG genotype, thrombotic
event(s) occurred in 50% of AML, 20% of CLL and 33.3% of the other
patients, whereas in patients with the 6936AA genotype, thrombosis
developed in 22% of AML, 10% of CLL and 18.2% of other
hematological pathologies.
Of note, no thrombotic disorder was reported in ALL
patients with the 6936AG genotype. Therefore, thrombotic disorders
were prevalent only in ALL patients harboring the 6936AA genotype,
occurring in 13.3% of the cases. This may be due to the fact that
patients with ALL were younger (mean age, 37 years) compared with
those with other malignant hematological diseases.
Logistic regression model
In the univariate logistic model for thrombosis and
6936A/G SNP, it was demonstrated that patients with the 6936AG
genotype (n=21) were more susceptible to thrombotic episodes
compared with those with the 6936AA genotype (n=87). The odds ratio
was estimated at 2.95, which is significantly higher than 1
(P=0.047) (Table V). The data from
the logistic regression model show a significant difference in the
incidence of thrombosis between AML patients with the 6936AA (n=41)
and 6936AG (n=12) genotypes. The odds ratio was estimated at 4.65
(P=0.018). The risk of developing thrombosis is therefore higher
for patients with AML harboring the 6936AG genotype.
 | Table V.Logistic regression model. |
Table V.
Logistic regression model.
| Explanatory
variable | Category | Reference | OR | 95% CI |
P-valuec |
|---|
|
Genotypea | AG | AA | 2.95 | 1.04–8.37 | 0.047 |
| AMLb | AML*AG | AML*AA | 4.65 | 1.34–16.17 | 0.018 |
Discussion
One of the EPCR polymorphisms, 6936A/G SNP, results
in the substitution of the serine at residue 219 with glycine in
the transmembrane domain. This mutation is associated with
increased plasma levels of sEPCR and is a candidate risk factor for
thrombosis (5). The presence of
6936A/G SNP (EPCR Gly 219) was also found at a higher frequency in
coronary heart disease and has been associated with increased
thrombosis risk in type 2 diabetic patients (13).
We previously reported that EPCR is expressed in
human malignant blood cells, often resulting in higher plasma sEPCR
levels (11). Using a retrospective
clinical study (n=110), we observed that, when the plasma sEPCR
level rises above the 200 ng/ml threshold, the risk of thrombosis
also rises considerably (40%) in hematological pathologies
(11). In this study, we demonstrated
that the thrombotic event incidence increases in leukemic patients
when plasma sEPCR is high, due to the presence of the 6936AG
genotype. This synergism increased the incidence of thrombotic
events in AML patients with high plasma sEPCR, from 41.7% reported
previously (11) to 50% for AML
heterozygous patients with the 6936AG genotype.
Several thrombotic events are under genetic control.
EPCR gene polymorphism is one among several genetic traits that
affect venous thrombosis. There are several other genes that are
involved in thrombotic events, including Serpin C-1, ABO locus, PC,
protein S1, factor V Leiden (resistant to APC), prothrombin,
fibrinogen γ chain, factor XI, glycoprotein VI, HIVEPI (protein
that participates in the transcriptional regulation of inflammatory
target genes) and kininogen 1 (gene encoding high-molecular-weight
kininogen) genes (14). However, the
mechanism by which this large panel of genes intervene in cancer
thrombophilia remains obscure. In brief, we observed that leukemic
cells not only harbor EPCR gene polymorphisms but also secrete
large amounts of the EPCR protein (11). In a previous retrospective study, the
association between EPCR SNPs and the incidence of thrombosis has
been investigated in several myeloma patients (15).
The data presented in this study indicate that, in
patients with malignant hemopathies, the presence of the EPCR
6936A/G SNP results in an increase in the incidence of thrombosis.
Despite the difference in the incidence rates, there was no
statistically significant difference in the 6936A/G SNP
distribution between the 4 patient groups (CLL, AML, ALL and other
hematological malignancies) and the healthy donors group. The EPCR
6936A/G SNP distribution is under genetic control, is independent
of the occurrence of any pathology and remains identical for any
population.
Among patients with malignant hemopathies, those
with AML harboring the 6936AG genotype were found to be the most
prone to thrombosis. Thrombosis affected ~50% of individuals with
the 6936AG genotype, whereas this rate was lower for CLL, ALL and
other malignant hemopathies. The difference in the thrombosis rate
between the 6936AA and 6936AG genotypes in AML was found to be
statistically significant (P<0.01).
Our cohort of patients was rather small (n=110) and
the frequency of the 6936GG genotype in any population is low.
Therefore, a definitive statistical statement on the 6936GG
genotype cannot be made with a limited number of cases.
In conclusion, this pilot study was the first to
demonstrate a significant association of the 6936AG genotype
(6936A/G SNP) of EPCR with thrombotic events in AML. The presence
of the 6936AG genotype in patients with malignant hematological
diseases may be a risk factor for thrombosis and its determination
in patients, particularly in those with AML, may be crucial for
thrombosis prevention and management. To the best of our knowledge,
this is the first report on the association of the 6936AG genotype
with the risk of thrombosis in leukemia.
Acknowledgements
We would like to thank Professor Amu Therwath (UMR,
Paris Diderot, Paris 7 University, Lariboisière Hospital, INSERM
U965, Paris, France) for his valuable assistance.
References
|
1
|
Khorana AA: Venous thromboembolism and
prognosis in cancer. Thromb Res. 125:490–493. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rao Mohan LV, Esmon CT and Pendurthi UR:
Endothelial cell protein C receptor: A multiliganded and
multifunctional receptor. Blood. 124:1553–1562. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gu JM, Katsuura Y, Ferrell GL, Grammas P
and Esmon CT: Endotoxin and thrombin elevate rodent endothelial
cell protein C receptor mRNA levels and increase receptor shedding
in vivo. Blood. 95:1687–1693. 2000.PubMed/NCBI
|
|
4
|
Xu J, Qu D, Esmon NL and Esmon CT:
Metalloproteolytic release of endothelial cell protein C receptor.
J Biol Chem. 275:6038–6044. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qu D, Wang Y, Song Y, Esmon NL and Esmon
CT: The Ser219→Gly dimorphism of the endothelial protein C receptor
contributes to the higher soluble protein levels observed in
individuals with the A3 haplotype. J Thromb Haemost. 4:229–235.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
de Willige Uitte S, Van Marion V,
Rosendaal FR, Vos HL, de Visser MC and Bertina RM: Haplotypes of
the EPCR gene, plasma sEPCR levels and the risk of deep venous
thrombosis. J Thromb Haemost. 2:1305–1310. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liaw PC, Neuenschwander PF, Smirnov MD and
Esmon CT: Mechanisms by which soluble endothelial cell protein C
receptor modulates protein C and activated protein C function. J
Biol Chem. 275:5447–5452. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dennis J, Johnson CY, Adediran AS, de
Andrade M, Heit JA, Morange PE, Trégouët DA and Gagnon F: The
endothelial protein C receptor (PROCR) Ser219Gly variant and risk
of common thrombotic disorders: A HuGE review and meta-analysis of
evidence from observational studies. Blood. 119:2392–2400. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saposnik B, Reny JL, Gaussem P, Emmerich
J, Aiach M and Gandrille S: A haplotype of the EPCR gene is
associated with increased plasma levels of sEPCR and is a candidate
risk factor for thrombosis. Blood. 103:1311–1318. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ducros E, Mirshahi S, Azzazene D, et al:
Endothelial protein C receptor expressed by ovarian cancer cells as
a possible biomarker of cancer onset. Int J Oncol. 41:433–440.
2012.PubMed/NCBI
|
|
11
|
Ducros E, Mirshahi SS, Faussat AM, et al:
Soluble endothelial protein C receptor (sEPCR) is likely a
biomarker of cancer-associated hypercoagulability in human
hematologic malignancies. Cancer Med. 1:261–267. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fukudome K and Esmon CT: Identification,
cloning and regulation of a novel endothelial cell protein
C/activated protein C receptor. J Biol Chem. 269:26486–26491.
1994.PubMed/NCBI
|
|
13
|
Ireland H, Konstantoulas CJ, Cooper JA,
Hawe E, Humphries SE, Mather H, Goodall AH, Hogwood J, Juhan-Vague
I and Yudkin JS: EPCR Ser219Gly: Elevated sEPCR, prothrombin F1+2,
risk for coronary heart disease, and increased sEPCR shedding in
vitro. Atherosclerosis. 183:283–292. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Morange PE and Trégouët DA: Current
knowledge on the genetics of incident venous thrombosis. J Thromb
Haemost. 11(Suppl 1): 111–121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dri AP, Politou M, Gialeraki A, Bagratuni
T, Kanellias N and Terpos E: Decreased incidence of EPCR 4678G/C
SNP in multiple myeloma patients with thrombosis. Thromb Res.
132:400–401. 2013. View Article : Google Scholar : PubMed/NCBI
|