Introduction
Lung cancer is the most common malignancy affecting
both genders and remains the main cause of cancer-related mortality
worldwide (1). Despite advances in
diagnosis and treatment of lung cancer, it remains a disease with
high morbidity and mortality. Lung cancers are classified according
to histological type and this classification has important
implications for the clinical management and prognosis of this
disease. The two main histological groups are non-small-cell lung
cancer (NSCLC) and small-cell lung cancer (SCLC). Approximately 85%
of lung tumors are NSCLCs. NSCLC includes three major histological
subtypes: Adenocarcinoma, squamous cell carcinoma and large-cell
carcinoma (2). Despite the new
therapeutic approaches, the overall survival of patients with lung
cancer remains low. The 5-year survival for SCLC is lower compared
with that of NSCLC (6 vs. 18%, respectively). Therefore, the
identification of highly sensitive and specific biomarkers that
highlight pathological changes early during the course of the
disease, in order to allow timely clinical intervention, is
crucial. A better understanding of biomarkers associated with lung
cancer may be of clinical value in improving treatment selection
and prognostication, and may even set the base for the development
of future novel targeted therapies.
Krüppel-like factors (KLFs) are a family of
evolutionarily conserved mammalian zinc finger transcription
factors, named after their homology with Krüppel, which is a
Drosophila melanogaster protein (3). KLFs are involved in a number of
important cellular processes, such as growth, development,
differentiation, proliferation and apoptosis (4–6). KLF4
(also referred to as gut-enriched KLF or GKLF) is one of the first
KLF family members identified (7,8).
KLF4 is a transcription factor expressed in a wide
variety of human tissues, which is important for a number of
different physiological processes, including development,
differentiation and maintenance of normal tissue homeostasis. KLF4
is a bifunctional transcription factor able to either activate or
repress transcription using different mechanisms, depending on the
target gene. Thus, depending on the cell type or cell context, KLF4
may act either as a tumor suppressor gene or as an oncogene.
KLF4 is implicated as a tumor suppressor gene in the
gastrointestinal tract epithelium, as its expression is decreased
in human colon and gastric cancers (9–11). The
loss of KLF4 is associated with poor survival (10) and it was found to be downregulated in
gastric cancer, with evidence of hypermethylation of the
5′-untranslated region and loss of heterozygosity of the KLF4 locus
or point mutations in the coding region (12–14). A
similar tumor suppressor role is also observed in colorectal cancer
(9), esophageal cancer (15), lung cancer (16), bladder cancer (17), medulloblastoma (12) and T-cell leukemia (13).
Conversely, KLF4 may function as a transforming
oncogene. KLF4-transformed rat kidney epithelial cells exhibit
morphological transformation and an increased tumorigenicity in
athymic mice (14). Increased KLF4
expression has been reported in human head and neck squamous cell
carcinoma and breast cancer (14,18).
Moreover, KLF4 expression has been demonstrated to be a poor
prognostic factor for early breast cancer and skin cancer (19,20),
corroborating its oncogenic role. In the skin, overexpression of
KLF4 results in hyperplasia and dysplasia (21), eventually leading to the development
of squamous cell carcinoma (22).
Whether KLF4 acts as a tumor suppressor or an oncogene is likely
determined by differences in cell context, expression patterns of
other genes and the chromatin environment of individual cells.
However, the mechanism underlying these differences remains
unknown.
A recent study demonstrated that KLF4 may function
as a tumor suppressor gene in lung cancer. The expression of KLF4
was downregulated in 21 of 25 primary lung cancers and ectopic
expression of KLF4 suppressed lung cancer cell proliferation and
clonogenic formation in vitro. Moreover, transfection of
lung cancer cells with the KLF4 gene also suppressed tumor growth
in vivo (16). However, the
molecular mechanism underlying the tumor-suppressive function of
KLF4 in lung cancer remains to be determined, as only few studies
have investigated the role and differences in expression of KLF4
among different histological groups of lung cancer. In this study,
the KLF4 protein expression level was investigated in lung tumors
(31 adenocarcinomas and 16 SCLCs) and normal tissues and the
clinical significance of KLF4 expression for diagnosis and
treatment decision-making was evaluated using immunohistochemical
analysis.
Materials and methods
Sample collection and clinical
data
A total of 47 formalin-fixed paraffin-embedded lung
cancer samples (31 adenocarcinomas and 16 SCLCs) and normal tissue
samples from healthy donors (n=13) were collected between January,
2014 and July, 2015 from the Department of Pathological Anatomy of
the Notre Dame de Secours University Hospital (Byblos, Lebanon) and
the National Institute of Pathology (Baabda, Lebanon). All the
tumor and normal tissue samples were obtained from surgical
specimens of patients with lung cancer. This study was approved by
the Institutional Review Board of the Notre Dame de Secours
University Hospital.
Immediately following surgical removal, all the
tissue samples were fixed in formalin and embedded in paraffin
prior to sectioning for histological and immunohistochemical
analyses. All the cancer tissue samples were graded by a
pathologist and histologically classified. Epidemiological and
clinical information were collected from patient records and
registries (Table I).
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
| Characteristics | No. (%) |
|---|
| Total subjects | 60 |
|
Healthy | 13 (21.66) |
|
NSCLC | 31 (51.66) |
| SCLC | 16 (26.66) |
| Gender |
|
| Male | 40 (66.66) |
| Female | 20 (33.33) |
| Age in years, median
(range) | 63 |
|
Healthy | 57 (20–87) |
|
NSCLC | 66 (21–84) |
| SCLC | 65 (23–83) |
| Stage (NSCLC) |
|
| I | 8 |
| II | 9 |
| III | 7 |
| IV | 7 |
Immunohistochemistry and scoring
Paraffin-embedded tissue sections (4 µm) were
subjected to immunostaining using the Ventana automated stainer
(BenchMark XT; Roche Diagnostics GmbH, Mannheim, Germany) at the
National Institute of Pathology (Baabda, Lebanon). The tissue
sections were hydrated through xylene and graded ethanols and
equilibrated in phosphate-buffered saline prior to undergoing
antigen retrieval. Endogenous peroxidase activity was quenched with
0.3% hydrogen peroxide for 5 min and the tissue sections were
incubated with a mouse monoclonal anti-KLF4 antibody (cat. no.
SAB5300069; clone 1E6; Sigma-Aldrich, St. Louis, MO, USA) at a
dilution of 1:200 for 1 h at room temperature. The appropriate
secondary antibody was horseradish peroxidase (HRP)-conjugated
rabbit anti-mouse IgG (cat. no. A9044; Sigma-Aldrich) at a dilution
of 1:200 for 1 h at room temperature. HRP detection was achieved
using 3,3′-diaminobenzidine substrate (Sigma-Aldrich) and the
slides were counterstained with hematoxylin.
Immunostaining was blindly evaluated by two
investigators (G.A. and E.H.) in an effort to achieve a consensus
on staining patterns by light microscopy. A quantitative score was
estimated by adding the score of the staining area and that of
staining intensity for each case to assess the expression levels of
the protein. The quantitative score was estimated by calculating
the percentage of immunopositive cells as follows: 0, no staining
of cells in any microscopic fields; 1+, <30% of cells stained
positive; 2+, 30–60% stained positive; and 3+, >60% stained
positive. The intensity was scored by evaluating the average
staining intensity of the positive cells as follows: 0, no
staining; 1+, mild staining; 2+, moderate staining; and 3+, intense
staining.
Data analysis
All statistical analyses were performed using SPSS
software for Windows, version 18.0 (SPSS Inc., Chicago, IL, USA). A
paired t-test was used to compare the KLF4 expression level between
tumors and matched normal tissues and among different histological
tumor types.
The patients were classified into two groups, namely
NSCLC and SCLC. The χ2 test was applied to determine the
correlation between the KLF4 level and clinicopathological
parameters. P<0.05 was considered to indicate statistically
significant differences.
Results
Patient characteristics
The characteristics of the patients included in this
study are summarized in Table I. Lung
cancer tissues were obtained from 47 patients, namely 31 cases with
NSCLCs (adenocarcinomas) and 16 cases with SCLCs. The median age of
the patients was 63 years, and 66.66% of the patients were male.
Epidemiological and clinical information was collected from patient
records and registries. All the cancer tissue samples were graded
by a pathologist and histologically classified. After diagnosis,
25.8% of the NSCLC patients were diagnosed as stage I (n=8), 29.03%
were stage II (n=9), 22.58% were stage III (n=7) and 22.58% were
stage IV (n=7).
KLF4 protein expression in normal and
tumor tissues
The profile of KLF4 protein expression in the
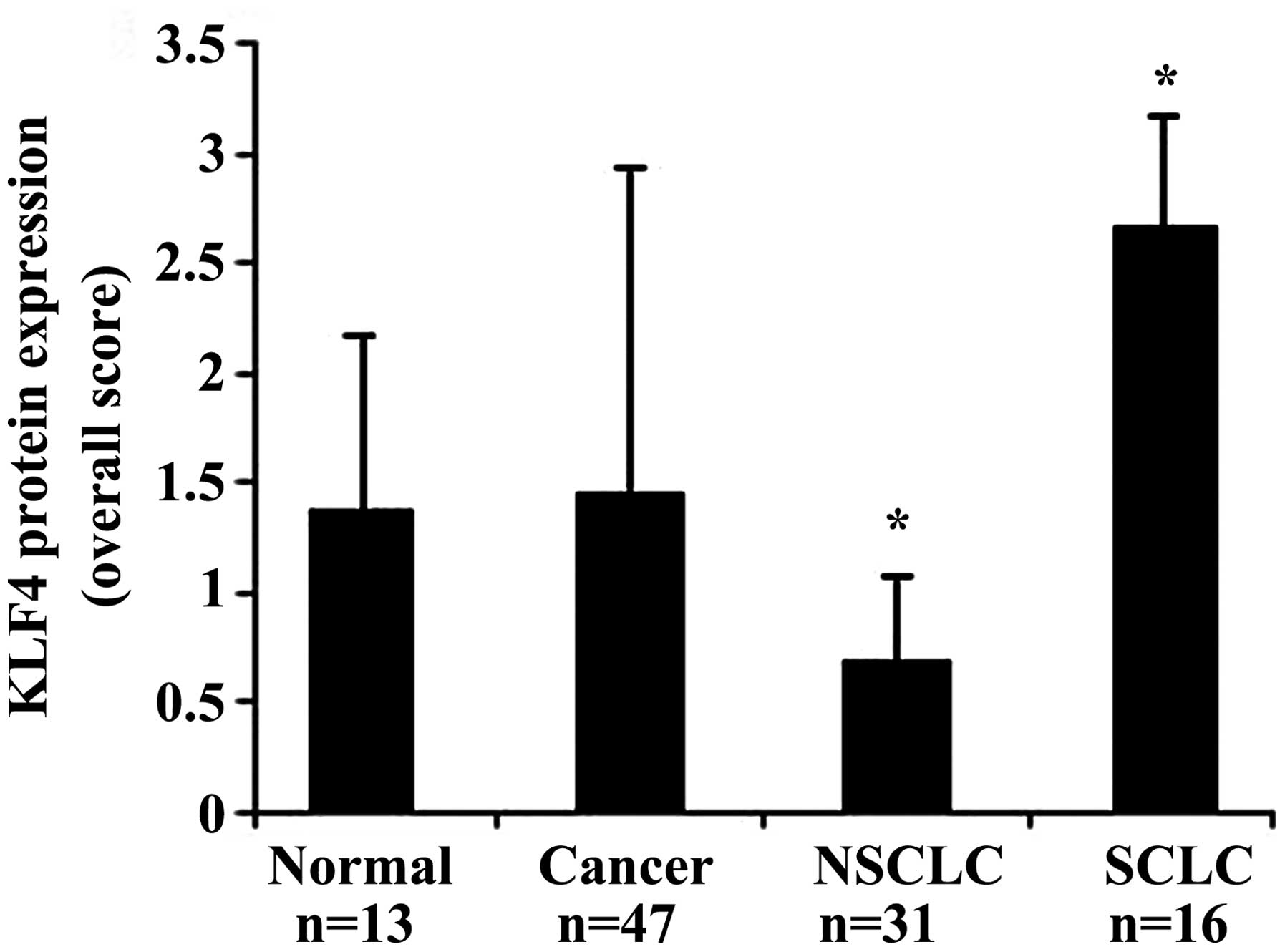
different types of tissues is presented in Table II and in Fig. 1. In normal tissues, KLF4 was expressed
in the nuclei with an overall score of 1.38±0.6 (mean ± standard
deviation). In all tumor tissue types, the overall score of KLF4
expression was 1.46±1.27. The statistical analysis revealed no
significant difference in expression between normal and cancer
tissues (P=0.995).
 | Table II.Mean KLF4 protein expression in normal
and lung cancer tissues. |
Table II.
Mean KLF4 protein expression in normal
and lung cancer tissues.
| Tissue type | Mean KLF4 protein
expression | P-value vs. normal
tissue |
|---|
| Normal (n=13) | 1.38±0.6 |
|
| Cancer (n=47) | 1.46±1.27 |
0.995 |
| NSCLC
(n=31) | 0.7±0.3 | 0.02 |
| SCLC
(n=16) | 2.68±0.46 |
0.00003 |
Histological classification of the lung cancer
tissues was performed in order to identify differential expression
of KLF4 between NSCLC and SCLC and between each tumor type and
normal tissue. The statistical analysis revealed a significant
difference in KLF4 expression between NSCLC and SCLC samples
(P<0.0001). Of the 31 NSCLC cases, 19 (61%) were negative for
KLF4 and 12 (39%) were positive. All 16 SCLC cases were positive
for KLF4 [11 cases exhibited intense staining (3+) and 5 cases
moderate staining (2+)]. The overall score of KLF4 expression was
0.7±0.3 and 2.68±0.46 in NSCLC and SCLC, respectively. A
significant difference was also observed between normal tissues and
each of the cancer tissue types (P=0.00003 for SCLC and P=0.02 for
NSCLC).
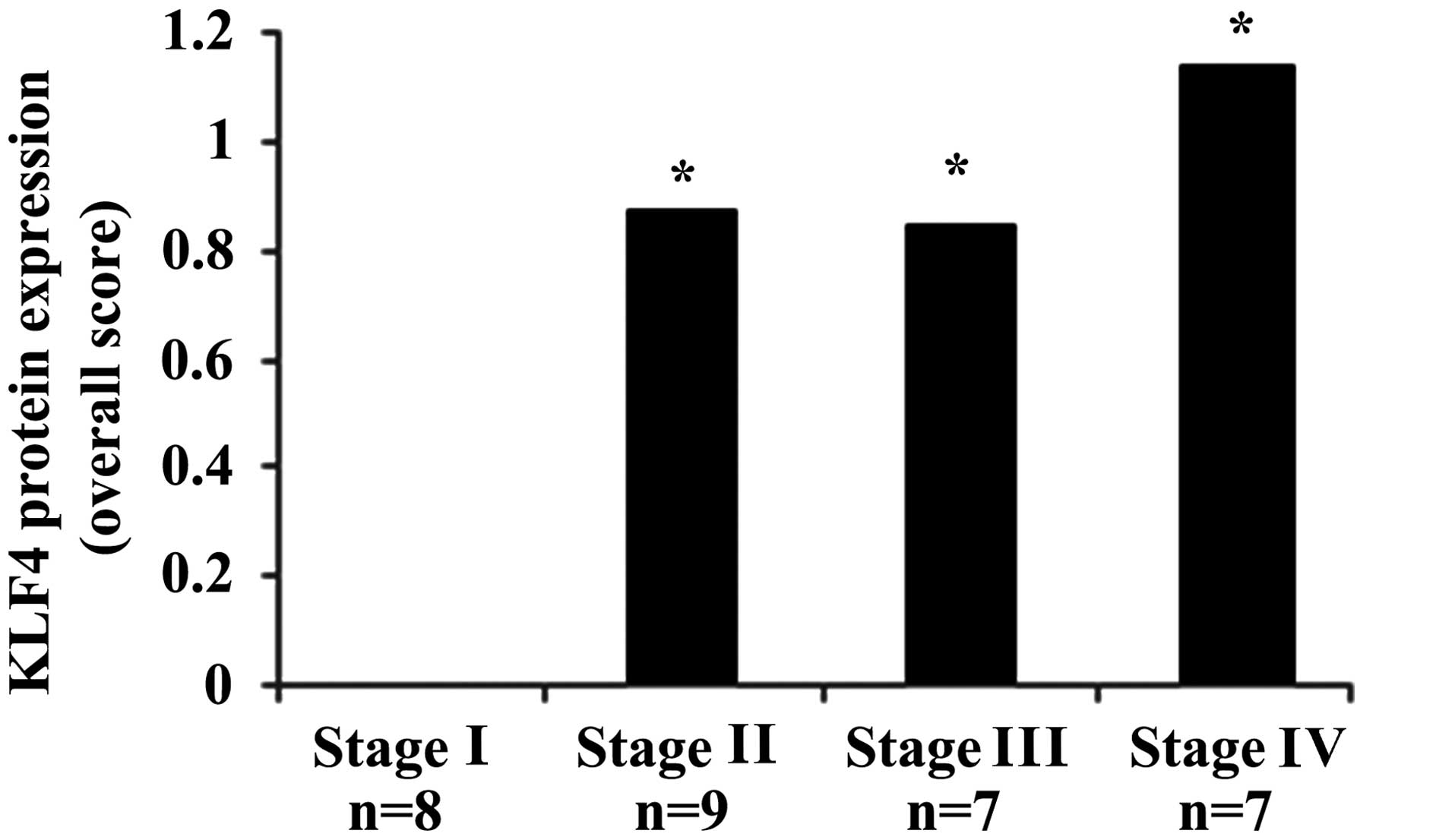
In NSCLC, a significant difference in KLF4
expression was observed between stage I and stages II, III and IV
(P<0.05) (Fig. 2). KLF4 expression
was significantly increased in tumor stages II, III and IV, whereas
all stage I cases (n=8) were negative for KLF4 expression. The
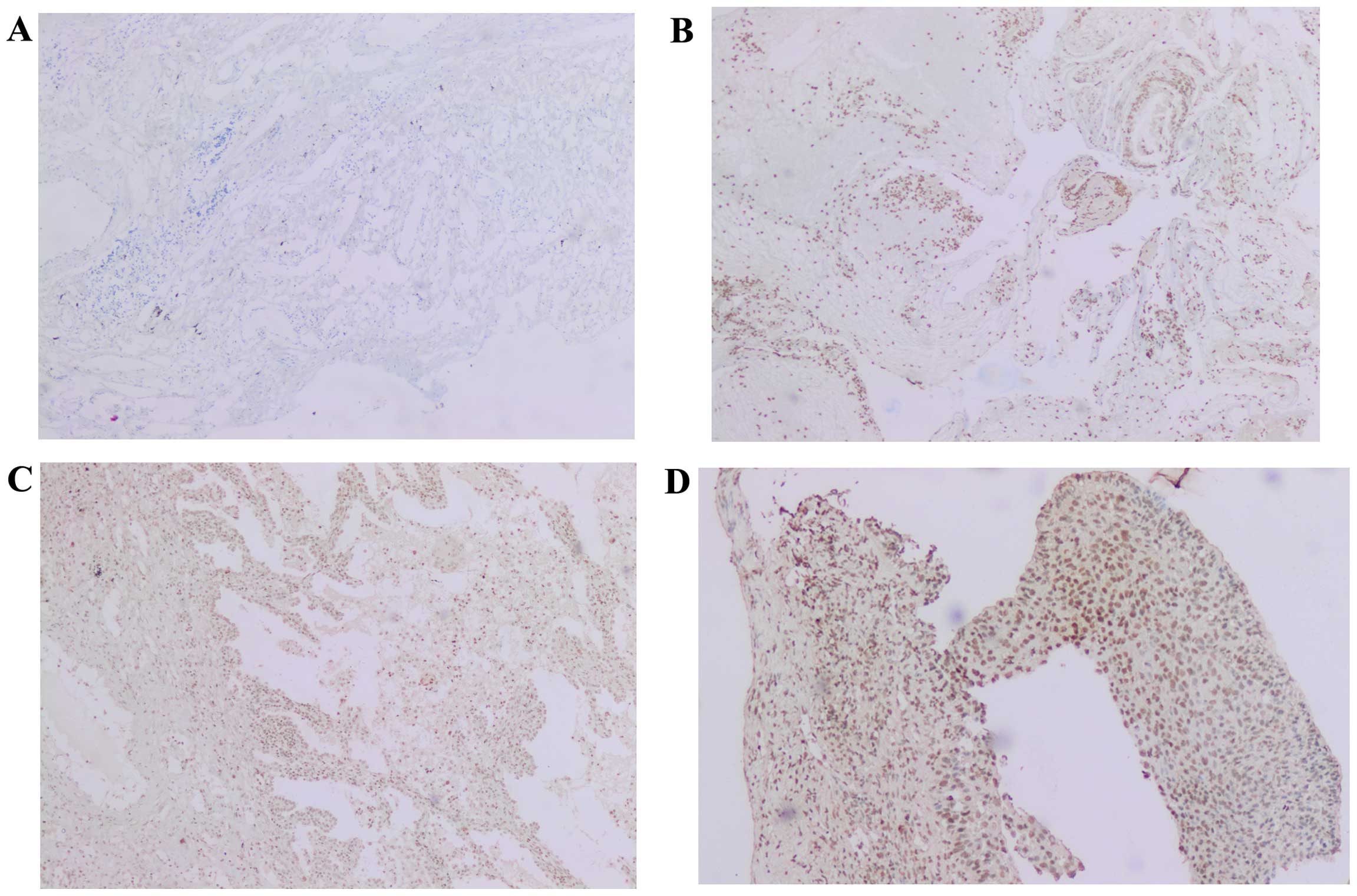
profile of KLF4 expression in each tumor stage is shown in Table III and examples of the
immunohistochemical staining for KLF4 are shown in Fig. 3.
 | Table III.Profile of Krüppel-like factor 4
protein expression in each tumor stage (cases per intensity of
expression). |
Table III.
Profile of Krüppel-like factor 4
protein expression in each tumor stage (cases per intensity of
expression).
|
| Expression, n
(%) |
|---|
|
|
|
|---|
| Stage | 0 (no staining) | 1+ (mild
staining) | 2+ (moderate
staining) | 3+ (intense
staining) |
|---|
| I
(n=8) | 8
(100.0) |
|
|
|
| II
(n=9) | 5 (55.0) |
| 4 (45.0) |
|
| III (n=7) | 3 (44.0) | 2 (28.0) | 2 (28.0) |
|
| IV (n=7) | 3 (420) | 1 (140) | 2 (28.0) | 1 (14.0) |
Factors associated with KLF4 protein
level
In order to determine any correlation of KLF4
expression with age and gender, a statistical analysis was
performed; KLF4 expression was not found to be significantly
associated with age or gender.
Discussion
The identification of proteins or transcription
factors with altered expression as a manifestation of human lung
carcinogenesis is important in the discovery of biomarkers for
early detection of lung cancer. Only a limited number of studies
have analyzed the expression and role of KLF4 in lung cancer
(16,23–26). In
the present study, we investigated the KLF4 protein expression in a
series of human lung tumors and normal tissues. As a result,
differential expression was observed between healthy tissue and
each of the two major lung cancer types (normal vs. NSCLC and
normal vs. SCLC) and also between NSCLC and SCLC. The protein
expression level of KLF4 was significantly decreased in NSCLC
compared with that in normal tissue, while significant
overexpression was detected in SCLC, which represents the
fast-growing nature of this type of lung cancer that is considered
highly lethal. These findings suggest that KLF4 may play a role in
the carcinogenic process. Stage II, III and IV lung adenocarcinomas
exhibited significantly higher rates of KLF4 expression compared
with stage I disease, where the expression of KLF4 was absent. The
absence of KLF4 expression may be explained by potential gene
silencing due to hypermethylation. However, the mechanisms
underlying this silencing require elucidation by future studies.
The increase in KLF4 expression in stage II, III and IV disease may
be associated with decreased tumor differentiation and increased
aggressiveness.
Our findings provide preliminary data regarding the
expression and the potential role of KLF4 in the proliferation of
lung tumors depending on the cell type and context. Moreover, to
the best of our knowledge, this is the first collected data showing
a significant difference in KLF4 protein expression between the two
major lung cancer types. However, due to the lack of patient
survival data, we were unable to investigate any correlation
between immunohistochemical findings and patient survival.
Our results were consistent with those reported by
Naranjo Gómez et al showing high expression of KLF4 in
neuroendocrine lung carcinomas, where KLF4 was positive in 23 of 35
large-cell neuroendocrine carcinomas, 10 of 10 tumorlets, 15 of 47
typical carcinoids and 18 of 18 SCLCs (25). Our results are also in agreement with
those of Zhang et al, who demonstrated a reduction of KLF4
protein expression in NSCLC tumor specimens, compared with the
expression in control tissues (26).
However, in their study, Zhang et al did not evaluate the
protein expression level of KLF4 in a SCLC tissue sample;
therefore, a comparison of the KLF4 profile between the two major
cancer types was lacking. The expression of KLF4 appears to exert a
dual effect on lung cancer, depending on the cell context and gene
network. Our actual in vitro study aims to determine the
underlying mechanisms and the potential factors that regulate the
gene or the protein expression of KLF4 in the two major
histological groups of lung cancer. DNA mutations, molecular
alterations, hypermethylation or microRNA expression may be
associated with altered KLF4 expression in lung cancer types.
Our observations that KLF4 was increased in SCLC
were not consistent with those of Hu et al, since our
observations indicate that KLF4 may function as a tumor-promoting
gene in lung cancer. The abovementioned studies have demonstrated
that the expression of KLF4 is downregulated in a number of primary
lung cancers and the ectopic expression of KLF4 suppresses lung
cancer cell proliferation and tumor growth in vivo (16). This discrepancy may be due to the
marginally larger tumor sample collection in our study, potentially
contributing to more relevant results, and to the different method
used to evaluate KLF4 expression. We examined the level of KLF4
protein expression by immunohistochemistry, whereas the level of
KLF4 protein was measured by western blot analysis in the other
study (16). However, the
downregulation of KLF4 in NSCLC may be associated with promoter
hypermethylation, a loss of heterozygosity of the KLF4 locus, or to
point mutations in the coding region. How KLF4 is differentially
expressed in lung cancers remains unclear. We hypothesized that
epigenetic control and the gene network may play a role in the
variable KLF4 expression levels in lung cancers and, therefore,
requires further investigation.
KLF4 expression and its role in the two major types
of lung cancer have not been extensively investigated to date.
Furthermore, the molecular mechanisms underlying the
tumor-suppressive or oncogenic function of KLF4 in lung cancer
remain to be determined. It is important that the regulation of
KLF4 expression in normal and tumor tissues is elucidated in future
studies.
Our data suggest that KLF4 protein expression level
in normal as well as tumor tissues may be a potential biomarker in
patients with lung cancer. Our findings may be useful for
determining prognostic factors associated with lung cancer and for
supporting their possible use in lung cancer case stratification.
In addition, SCLCs present with a more aggressive clinical course.
Investigating how the microenvironment and cell context affect KLF4
expression and, thus, tumorigenesis, tumor progression and
prognosis, is a major goal in future studies.
Acknowledgements
The present study was supported by a grant from the
Lebanese University.
References
|
1.
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small-cell lung cancer: Epidemiology, risk
factors, treatment and survivorship. Mayo Clin Proc. 83:584–594.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Travis WD, Travis LB and Devesa SS: Lung
cancer. Cancer. 75(1 Suppl): S191–S202. 1995. View Article : Google Scholar
|
|
3.
|
Preiss A, Rosenberg UB, Kienlin A, Seifert
E and Jäckle H: Molecular genetics of Krüppel, a gene required for
segmentation of the Drosophila embryo. Nature. 313:27–32. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Dang DT, Pevsner J and Yang VW: The
biology of the mammalian Krüppel-like family of transcription
factors. Int J Biochem Cell Biol. 32:1103–1121. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Black AR, Black JD and Azizkhan-Clifford
J: Sp1 and Krüppel-like factor family of transcription factors in
cell growth regulation and cancer. J Cell Physiol. 188:143–160.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Kaczynski J, Cook T and Urrutia R: Sp1-
and Krüppel-like transcription factors. Genome Biol. 4:2062003.
View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Garrett-Sinha LA, Eberspaecher H, Seldin
MF and de Crombrugghe B: A gene for a novel zinc-finger protein
expressed in differentiated epithelial cells and transiently in
certain mesenchymal cells. J Biol Chem. 271:31384–31390. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Shields JM, Christy RJ and Yang VW:
Identification and characterization of a gene encoding a
gut-enriched Krüppel-like factor expressed during growth arrest. J
Biol Chem. 271:20009–20017. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Zhao W, Hisamuddin IM, Nandan MO, Babbin
BA, Lamb NE and Yang VW: Identification of Krüppel-like factor 4 as
a potential tumor suppressor gene in colorectal cancer. Oncogene.
23:395–402. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Wei D, Gong W, Kanai M, Schlunk C, Wang L,
Yao JC, Wu TT, Huang S and Xie K: Drastic down-regulation of
Krüppel-like factor 4 expression is critical in human gastric
cancer development and progression. Cancer Res. 65:2746–2754. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Wei D, Kanai M, Huang S and Xie K:
Emerging role of KLF4 in human gastrointestinal cancer.
Carcinogenesis. 27:23–31. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Nakahara Y, Northcott PA, Li M, Kongkham
PN, Smith C, Yan H, Croul S, Ra YS, Eberhart C, Huang A, et al:
Genetic and epigenetic inactivation of Kruppel-like factor 4 in
medulloblastoma. Neoplasia. 12:20–27. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Yasunaga J, Taniguchi Y, Nosaka K, Yoshida
M, Satou Y, Sakai T, Mitsuya H and Matsuoka M: Identification of
aberrantly methylated genes in association with adult T-cell
leukemia. Cancer Res. 64:6002–6009. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Foster KW, Ren S, Louro ID, Lobo-Ruppert
SM, McKie-Bell P, Grizzle W, Hayes MR, Broker TR, Chow LT and
Ruppert JM: Oncogene expression cloning by retroviral transduction
of adenovirus E1A-immortalized rat kidney RK3E cells:
Transformation of a host with epithelial features by c-MYC and the
zinc finger protein GKLF. Cell Growth Differ. 10:423–434.
1999.PubMed/NCBI
|
|
15.
|
Wang N, Liu ZH, Ding F, Wang XQ, Zhou CN
and Wu M: Down-regulation of gut-enriched Kruppel-like factor
expression in esophageal cancer. World J Gastroenterol. 8:966–970.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Hu W, Hofstetter WL, Li H, Zhou Y, He Y,
Pataer A, Wang L, Xie K, Swisher SG and Fang B: Putative
tumor-suppressive function of Kruppel-like factor 4 in primary lung
carcinoma. Clin Cancer Res. 15:5688–5695. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Ohnishi S, Ohnami S, Laub F, Aoki K,
Suzuki K, Kanai Y, Haga K, Asaka M, Ramirez F and Yoshida T:
Downregulation and growth inhibitory effect of epithelial-type
Krüppel-like transcription factor KLF4, but not KLF5, in bladder
cancer. Biochem Biophys Res Commun. 308:251–256. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Foster KW, Frost AR, McKie-Bell P, Lin CY,
Engler JA, Grizzle WE and Ruppert JM: Increase of GKLF messenger
RNA and protein expression during progression of breast cancer.
Cancer Res. 60:6488–6495. 2000.PubMed/NCBI
|
|
19.
|
Pandya AY, Talley LI, Frost AR, Fitzgerald
TJ, Trivedi V, Chakravarthy M, Chhieng DC, Grizzle WE, Engler JA,
Krontiras H, et al: Nuclear localization of KLF4 is associated with
an aggressive phenotype in early-stage breast cancer. Clin Cancer
Res. 10:2709–2719. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Chen YJ, Wu CY, Chang CC, Ma CJ, Li MC and
Chen CM: Nuclear Krüppel-like factor 4 expression is associated
with human skin squamous cell carcinoma progression and metastasis.
Cancer Biol Ther. 7:777–782. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Foster KW, Liu Z, Nail CD, Li X,
Fitzgerald TJ, Bailey SK, Frost AR, Louro ID, Townes TM, Paterson
AJ, et al: Induction of KLF4 in basal keratinocytes blocks the
proliferation-differentiation switch and initiates squamous
epithelial dysplasia. Oncogene. 24:1491–1500. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Huang CC, Liu Z, Li X, Bailey SK, Nail CD,
Foster KW, Frost AR, Ruppert JM and Lobo-Ruppert SM: KLF4 and PCNA
identify stages of tumor initiation in a conditional model of
cutaneous squamous epithelial neoplasia. Cancer Biol Ther.
4:1401–1408. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Yu T, Chen X, Zhang W, Liu J, Avdiushko R,
Napier DL, Liu AX, Neltner JM, Wang C, Cohen D and Liu C: KLF4
regulates adult lung tumor-initiating cells and represses
K-Ras-mediated lung cancer. Cell Death Differ. 23:207–215. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Zhou Y, Hofstetter WL, He Y, Hu W, Pataer
A, Wang L, Wang J, Zhou Y, Yu L, Fang B and Swisher SG: KLF4
inhibition of lung cancer cell invasion by suppression of SPARC
expression. Cancer Biol Ther. 9:507–513. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Naranjo Gómez JM, Bernal JF, Arranz PG,
Fernández SL and Roman JJ: Alterations in the expression of p53,
KLF4 and p21 in neuroendocrine lung tumors. Arch Pathol Lab Med.
138:936–942. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Zhang Z, Wang Z, Liu X, Shi M, Chen G,
Zhang B, Li Z and Song L: Correlation of KLF4 and SPARC expression
with the clinical characteristics of non-small cell lung cancer.
Chin J Lung Cancer. 720–724. 2012.(In Chinese).
|