Introduction
Down syndrome (DS) or trisomy 21 is the most common
genetic cause of mental retardation. In 95% of the cases, DS is
caused by an error occurring during meiosis, mainly of maternal
origin (1–3). Despite extensive research, the
molecular mechanisms underlying the meiotic non-disjunction are
poorly understood. However, several studies suggested that genomic
DNA hypomethylation may be associated with chromosomal instability
and abnormal segregation (4,5). Moreover, impairment of folate
metabolism has been causally associated with DNA hypomethylation
(6–8). Thus, polymorphisms involved in folate
metabolism have been investigated as maternal risk factors for DS.
Several key enzymes, including methylenetetrahydrofolate reductase
(MTHFR), methionine synthase (MTR) and cystathionine b-synthase
(CBS) are involved in the folate metabolic pathway. MTHFR converts
tetrahydrofolate to 5,10-methylenetetrahydrofolate, which is vital
for nucleic acid metabolism (9,10). C677T
and A1298C, two common variants in the MTHFR gene, have been
investigated in two previous meta-analyses for their role as
maternal risk factors for DS (11,12).
The MTR enzyme, encoded by the MTR gene, catalyzes
the remethylation of homocysteine to methionine, which is required
for the production of the universal methyl donor adenosylmethionine
(13,14). CBS acts in the transsulfuration of
homocysteine to cystathionine, playing a critical role in linking
the folate and methionine cycles in regulating homocysteine levels
(15,16). In addition to the metabolism,
folate-transporting proteins, including reduced folate carrier-1
(RFC-1), are also crucial for the maintenance of DNA methylation.
RFC-1 is responsible for folate uptake from the jejunum and its
subsequent translocation across biological membranes in a variety
of cells (17). The A80G
polymorphism of the RFC-1 gene has been recently demonstrated to
affect plasma folate and homocysteine levels (18,19).
To date, a number of studies have investigated the
association between RFC-1 A80G/MTR A2756G/CBS 844ins68
polymorphisms and the risk of DS in the offspring (20–34);
however, it remains inconclusive whether these polymorphisms in the
mother are causal in determining DS susceptibility in the
offspring. Therefore, a meta-analysis of all relevant studies was
conducted to quantitatively assess the effect of these three
polymorphisms on the risk of DS.
Data collection methods
Search strategy
Literature databases, including PubMed, Embase,
China National Knowledge Infrastructure (CNKI) and Chinese
Biomedicine, were searched for relevant studies (the last search
was updated in April 2013). The following search terms were used:
(‘cystathionine-beta-synthase’, ‘CBS’, ‘methionine synthase’,
‘MTR’, ‘rs1805087’, ‘reduced folate carrier 1’, or ‘RFC 1’,
‘rs1051266’), (‘Down syndrome’ or ‘trisomy 21’), and (polymorphism
OR variant). The search was limited to studies published in English
or Chinese language. In addition, a snowball search was conducted
to identify additional potentially relevant studies in the
references of reviews and retrieved articles.
Inclusion and exclusion criteria
The studies included in the present meta-analysis
were required to meet the following criteria: i) RFC-1 A80G or MTR
A2756G or CBS 844ins68 polymorphisms and maternal risk for DS; ii)
case-control design iii) sufficient maternal genotype data for
calculation of odds ratio (OR) with 95% confidence interval (CI);
iv) published in English or Chinese. Studies were excluded for the
following reasons: i) Data duplication; ii) no usable maternal
genotype data provided; and iii) abstracts, comments and
reviews.
Data extraction
The following information was extracted from each
study by two investigators independently: Name of the first author,
year of publication, source of control subjects, country of origin,
ethnicities of the individuals involved, number of cases and
controls, and number of genotypes for the three polymorphisms in
cases and controls. Disagreements were resolved by discussion
between the two investigators.
Statistical analysis
STATA software, version 12 (StataCorp LP, College
Station, TX, USA) was used to perform all the statistical analyses.
P-values <0.05 were considered to indicate statistically
significant differences. The distribution of genotypes in the
control group of each study was assessed for Hardy-Weinberg
equilibrium (HWE) and P<0.05 was considered as significant
disequilibrium.
The association between the RFC-1 A80G polymorphism
and maternal risk for DS was evaluated using OR and 95% CI
(35) under the codominant, dominant
and recessive genetic models, as well as the allele model. For the
MTR A2756G and CBS 844ins68 polymorphisms, the four genetic models
were also used. In addition, subgroup analysis for the RFC-1 A80G
and MTR A2756G polymorphisms was performed based on ethnicity,
sample size and source of controls.
All the meta-analyses were evaluated for
heterogeneity using the Chi-squared-based Q test and the
I2 test (36). A
random-effects model was used when the heterogeneity test result
was P<0.10; otherwise, the fixed-effects model was used
(37). Moreover, sensitivity
analysis was performed to evaluate the stability of the results
following sequential removal of each study. Cumulative
meta-analyses of associations for each polymorphism were also
performed through assortment of studies with publication time.
Finally, publication bias was assessed via Egger's test and funnel
plots: P<0.05 was considered to indicate statistical
significance and the funnel plot should be asymmetric when there
was publication bias (38–40).
Results
Characteristics of the included
studies
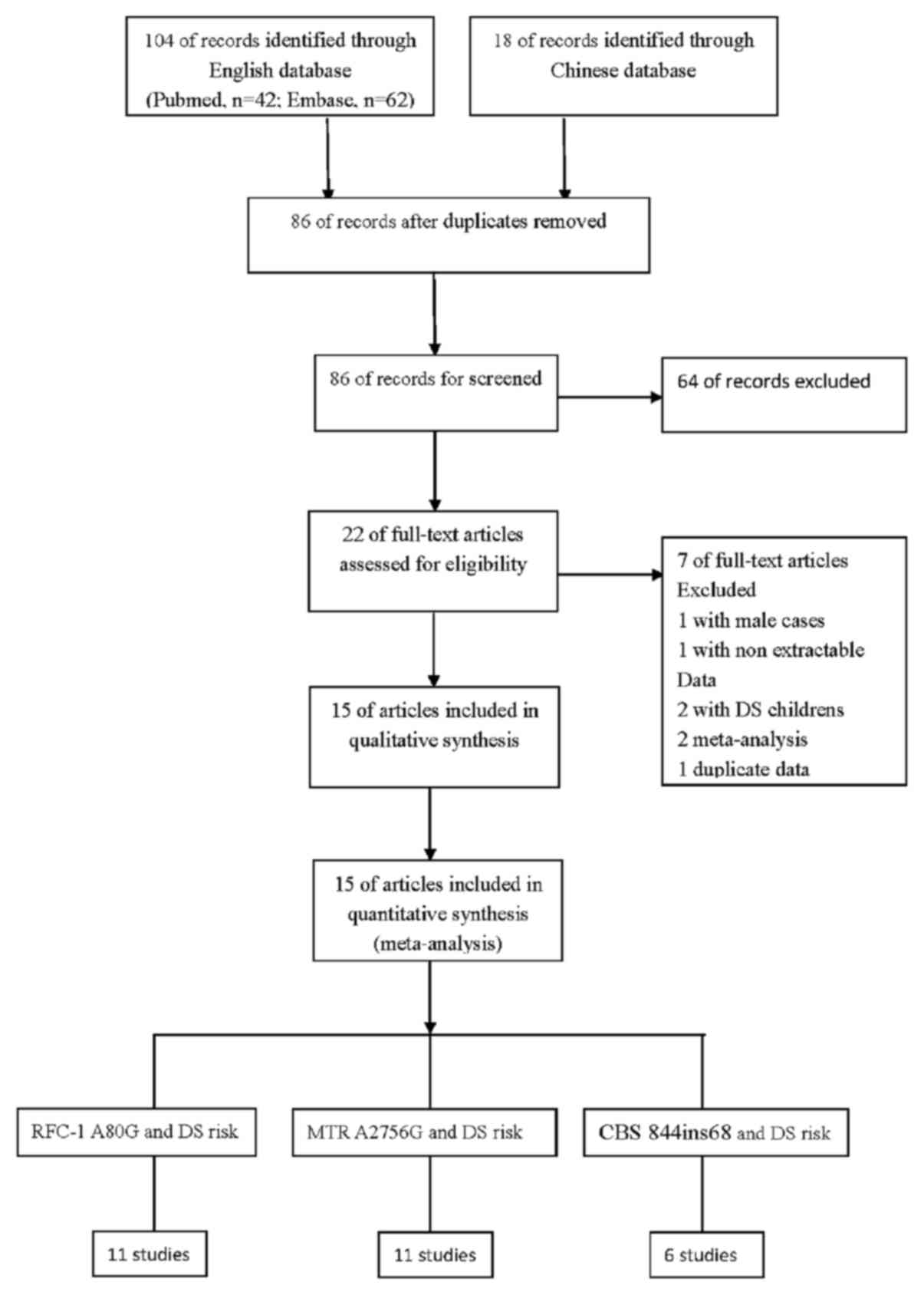
A total of 122 titles in the PubMed, Embase, CNKI
and Chinese Biomedicine databases were found to be relevant to the
search terms. Subsequently, the abstracts and full articles of the
retrieved studies were read to assess their eligibility for
inclusion in the meta-analysis. Finally, 15 studies investigating
the association between any of the RFC-1 A80G, MTR A2756G and CBS
844ins68 polymorphisms and DS were included in the meta-analysis.
Among these studies, 2 and 4 studies investigated only MTR A2756G
and RFC-1 A80G polymorphisms, respectively; 3 studies included both
RFC-1 A80G and MTR A2756G polymorphisms; 2 studies included both
MTR A2756G and CBS 844ins68 polymorphisms; and 4 studies
investigated RFC-1 A80G, MTR A2756G and CBS 844ins68 polymorphisms.
The flow chart of the study selection process is shown in Fig. 1.
There were 11 studies with 2,389 mothers (997 cases
and 1,392 controls) for the RFC-1 A80G polymorphism, 11 studies
with 2,717 mothers (1,162 cases and 1,555 controls) for the MTR
A2756G polymorphism and 6 studies with 1,859 mothers (825 cases and
1,034 controls) for the CBS 844ins68 polymorphism. The studies
included in the present meta-analysis were conducted on different
ethnic populations: 6 studies included a Caucasian population, 3
studies included an Asian population, and 6 studies included a
mixed population. The studies were published between 2005 and 2013.
Additionally, the distribution of genotypes in the control groups
deviated from HWE in the study of Neagos et al (22). The characteristics of all the
included studies are summarized in Tables I and II.
 | Table I.Distribution of the RFC-1 A80G and
MTR A2756G genotypes among DS mothers (cases and controls) included
in the meta-analysis. |
Table I.
Distribution of the RFC-1 A80G and
MTR A2756G genotypes among DS mothers (cases and controls) included
in the meta-analysis.
|
|
|
|
|
|
| Genotype
distribution |
|
|---|
|
|
|
|
|
|
|
|
|
|---|
|
|
|
|
|
|
| Cases | Controls |
|
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| Authors | Year | Source of
controls | Country | Ethnicity | GG | AG | AA | GG | AG | AA |
PHWE | (Refs.) |
|---|
| RFC-1 A80G
polymorphism |
|
|
|
|
|
|
|
|
|
|
|
|
| Wang
et al | 2013 | HB | China | Asian | 16 | 41 | 47 | 13 | 71 | 100 | 0.93 | (20) |
| Neagos
et al | 2010 | PB | Romania | Caucasian | 9 | 16 | 1 | 11 | 30 | 5 | 0.03 | (22) |
|
Brandalize et al | 2010 | HB | Brazil | Mixed | 65 | 101 | 73 | 42 | 91 | 64 | 0.37 | (21) |
|
Fintelman-Rodrigues et
al | 2009 | PB | Brazil | Mixed | 25 | 64 | 25 | 26 | 55 | 29 | 0.99 | (26) |
| Coppedè
et al | 2007 | HB | Italy | Caucasian | 15 | 17 | 1 | 7 | 14 | 9 | 0.73 | (23) |
| Coppedè
et al | 2006 | PB | Italy | Caucasian | 27 | 29 | 13 | 31 | 42 | 20 | 0.42 | (24) |
| Chango
et al | 2005 | PB | France | Caucasian | 29 | 66 | 24 | 26 | 52 | 16 | 0.25 | (25) |
| Biselli
et al | 2008 | HB | Brazil | Mixed | 20 | 33 | 14 | 34 | 49 | 30 | 0.16 | (28) |
| Scala
et al | 2006 | HB | Italy | Caucasian | 26 | 41 | 27 | 48 | 113 | 102 | 0.10 | (27) |
| Liao
et al | 2010 | PB | China | Asian | 14 | 22 | 24 | 16 | 40 | 12 | 0.14 | (31) |
| Biselli
et al | 2008 | HB | Brazil | Mixed | 21 | 36 | 15 | 50 | 94 | 50 | 0.67 | (28) |
| MTR A2756G
polymorphism |
|
|
|
|
|
|
|
|
|
|
|
|
|
Brandalize et al | 2010 | HB | Brazil | Mixed | 9 | 71 | 159 | 6 | 61 | 130 | 0.72 | (21) |
|
Fintelman-Rodrigues et
al | 2009 | PB | Brazil | Mixed | 7 | 28 | 79 | 2 | 37 | 71 | 0.25 | (26) |
| Coppedè
et al | 2007 | HB | Italy | Caucasian | 0 | 8 | 23 | 1 | 8 | 24 | 0.74 | (23) |
| Chango
et al | 2005 | PB | France | Caucasian | 2 | 33 | 84 | 1 | 32 | 87 | 0.29 | (25) |
| Scala
et al | 2006 | HB | Italy | Caucasian | 1 | 21 | 72 | 3 | 73 | 183 | 0.15 | (27) |
| Liao
et al | 2010 | PB | China | Asian | 1 | 6 | 53 | 1 | 10 | 57 | 0.48 | (31) |
| Biselli
et al | 2008 | HB | Brazil | Mixed | 4 | 21 | 47 | 8 | 57 | 129 | 0.59 | (28) |
|
Zampieri et al | 2012 | HB | Brazil | Mixed | 5 | 38 | 62 | 9 | 49 | 127 | 0.15 | (33) |
| da
Silva et al | 2005 | HB | Brazil | Mixed | 6 | 51 | 97 | 4 | 46 | 108 | 0.73 | (32) |
| Coppedè
et al | 2009 | PB | Italy | Caucasian | 2 | 22 | 66 | 3 | 30 | 78 | 0.95 | (30) |
| Wang
et al | 2012 | HB | China | Asian | 0 | 12 | 72 | 0 | 14 | 106 | 0.50 | (49) |
 | Table II.Distribution of the CBS 844ins68
genotype among DS mothers (cases and controls) included in the
meta-analysis. |
Table II.
Distribution of the CBS 844ins68
genotype among DS mothers (cases and controls) included in the
meta-analysis.
|
|
|
|
|
| Genotype
distribution |
|
|
|---|
|
|
|
|
|
|
|
|
|
|---|
|
|
|
|
|
| Cases | Controls |
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| Authors | Year | Source of
controls | Country | Ethnicity | ii | iw | 22 | ii | iw | 22 |
PHWE | (Refs.) |
|---|
| Brandalize et
al | 2010 | HB | Brazil | Mixed | 4 | 28 | 207 | 3 | 25 | 169 | 0.08 | (21) |
| Fintelman-Rodrigues
et al | 2009 | PB | Brazil | Mixed | 3 | 31 | 80 | 4 | 21 | 85 | 0.08 | (26) |
| Chango et
al | 2005 | HB | France | Caucasian | 0 | 10 | 109 | 1 | 13 | 106 | 0.41 | (25) |
| Scala et
al | 2006 | HB | Italy | Caucasian | 0 | 11 | 83 | 0 | 35 | 229 | 0.25 | (27) |
| Zampieri et
al | 2012 | HB | Brazil | Mixed | 4 | 18 | 83 | 2 | 38 | 145 | 0.78 | (33) |
| da Silva et
al | 2005 | HB | Brazil | Mixed | 2 | 42 | 110 | 3 | 38 | 117 | 0.97 | (32) |
RFC-1 A80G polymorphism
A total of 11 case-control studies on the
association between RFC-1 A80G polymorphism and maternal risk for
DS were included (Table III). The
results of the combined analyses revealed a significantly increased
maternal DS risk for the RFC-1 A80G polymorphism: G vs. A: OR=1.19,
P=0.04; and recessive model: OR=1.28, P=0.01 (Fig. 2A). When stratified by ethnicity, a
significantly elevated maternal risk for DS was observed among
Caucasians in the recessive model (OR=1.38, P=0.046), but not among
Asians or in the mixed population (Table III).
 | Table III.Effect of the RFC-1 A80G polymorphism
on DS risk. |
Table III.
Effect of the RFC-1 A80G polymorphism
on DS risk.
|
| G vs. A | GG vs. AA | GA vs. AA | GG+GA vs. AA | GG vs. GA+AA |
|---|
|
|
|
|
|
|
|
|---|
| Variables | OR (95% CI) |
P-valuea | OR (95% CI) |
P-valuea | OR (95% CI) |
P-valuea | OR (95% CI) |
P-valuea | OR (95% CI) |
P-valuea |
|---|
| Overall | 1.19
(1.004–1.40) | 0.05 | 1.34
(0.98–1.96) | 0.06 | 1.10
(0.89–1.29) | 0.50 | 1.18
(0.89–1.58) | 0.04 | 1.28
(1.05–1.56) | 0.46 |
| All in HWE | 1.18
(0.99–1.40) | 0.04 | 1.36
(0.96–1.93) | 0.05 | 1.06
(0.88–1.28) | 0.46 | 1.16
(0.87–1.56) | 0.03 | 1.27
(1.04–1.55) | 0.39 |
| Ethnicity |
|
|
|
|
|
|
|
|
|
|
|
Caucasian | 1.35
(0.97–1.86) | 0.07 | 1.80
(0.85–3.79) | 0.05 | 1.20
(0.84–1.69) | 0.43 | 1.45
(0.82–2.57) | 0.11 | 1.38
(1.005–1.88) | 0.31 |
|
Mixed | 1.14
(0.96–1.35) | 0.97 | 1.30
(0.92–1.82) | 0.97 | 1.10
(0.84–1.45) | 0.89 | 1.21
(0.92–1.59) | 0.93 | 1.16
(0.87–1.53) | 0.71 |
|
Asian | 1.00
(0.43–2.33) | 0.01 | 1.10
(0.19–6.34) | 0.01 | 0.74
(0.31–1.78) | 0.05 | 0.71
(0.16–3.08) | 0.02 | 1.56
(0.89–2.73) | 0.13 |
| Sample size |
|
|
|
|
|
|
|
|
|
|
|
>200 | 1.20
(1.03–1.40) | 0.297 | 1.45
(1.05–1.99) | 0.29 | 1.08
(0.88–1.33) | 0.97 | 1.25
(1.01–1.55) | 0.70 | 1.29
(1.02–1.63) | 0.25 |
|
≤200 | 1.21
(0.80–1.83) | 0.02 | 1.52
(0.61–3.78) | 0.03 | 1.02
(0.68–1.53) | 0.08 | 1.32
(0.51–3.40) | 0.004 | 1.27
(0.89–1.81) | 0.54 |
| Source of
controls |
|
|
|
|
|
|
|
|
|
|
| HB | 1.34
(1.12–1.61) | 0.24 | 1.76
(1.12–2.60) | 0.19 | 1.14
(0.91–1.42) | 0.63 | 1.37
(1.06–1.79) | 0.31 | 1.47
(1.14–1.88) | 0.41 |
| PB | 0.97
(0.77–1.23) | 0.25 | 0.93
(0.58–1.50) | 0.29 | 0.91
(0.64–1.29) | 0.29 | 0.88
(0.50–1.55) | 0.06 | 1.03
(0.75–1.42) | 0.76 |
In the stratified analyses by sample size, a
significantly increased risk was observed among large-sample
studies (>200 subjects) (G vs. A: OR=1.20, P=0.02; GG vs. AA:
OR=1.45, P=0.02; dominant model: OR=1.25, P=0.04; and recessive
model: OR=1.29, P=0.03); however, an increased risk was not
observed in small-sample studies (≤200 subjects) (Table III). Moreover, when subgroup
analysis was performed by source of controls, a significantly
increased DS risk was found among hospital-based (HB) controls (G
vs. A: OR=1.34, P=0.001; GG vs. AA: OR=1.76, P<0.001; dominant
model: OR=1.37, P=0.02; recessive model: OR=1.47, P<0.001), but
not among population-based (PB) controls. Interestingly, a
statistical correlation between homozygotes and the dominant model,
observed in large-sample and HB control groups, was not found in
the overall comparison.
MTR A2756G and CBS 844ins68
polymorphisms
The analysis of the MTR A2756G polymorphism and its
association with maternal DS risk revealed that the fixed-effects
pooled OR for the recessive model: GG vs. GA+AA was
non-significant: OR=1.24, P=0.37. (Fig.
2B) Additionally, the allele, dominant and codominant models
revealed no significant association. Finally, for the CBS 844ins68
polymorphism, no statistically significant association with
maternal DS risk was observed in any of the comparisons (Table IV). Fig.
2C shows the recessive model comparison for the CBS 844ins68
polymorphism. Additionally, a subgroup analysis by ethnicity,
source of controls and sample size was performed for the two
polymorphisms; however, no significant associations were
observed.
 | Table IV.Association of the MTR A2756G
polymorphism with DS risk. |
Table IV.
Association of the MTR A2756G
polymorphism with DS risk.
|
| G vs. A | GG vs. AA | GA vs. AA | GG+GA vs. AA | GG vs. GA+AA |
|---|
|
|
|
|
|
|
|
|---|
| Variables | OR (95% CI) |
P-valuea | OR (95% CI) |
P-valuea | OR (95% CI) |
P-valuea | OR (95% CI) |
P-valuea | OR (95% CI) |
P-valuea |
|---|
| Overall | 1.05
(0.9–1.22) | 0.89 | 1.33
(0.83–2.15) | 0.98 | 1.00
(0.89–1.13) | 1.00 | 1.02
(0.86–1.22) | 0.76 | 1.32
(0.82–2.11) | 0.96 |
| Ethnicity |
|
|
|
|
|
|
|
|
|
|
|
Caucasian | 0.90
(0.68–1.20) | 0.78 | 0.89
(0.30–2.70) | 0.85 | 0.97
(0.78–1.20) | 0.99 | 0.89
(0.65–1.21) | 0.80 | 0.92
(0.30–2.77) | 0.85 |
|
Mixed | 1.12
(0.93–1.35) | 0.80 | 1.48
(0.86–2.55) | 0.87 | 1.02
(0.86–1.20) | 0.91 | 1.10
(0.89–1.37) | 0.47 | 1.44
(0.85–2.46) | 0.78 |
|
Asian | 0.99
(0.54–1.81) | 0.41 | 1.08
(0.66–17.63) | – | 1.00
(0.72–1.37) | 0.80 | 0.98
(0.52–1.87) | 0.36 | 1.14
(0.07–18.56) | – |
| Sample size |
|
|
|
|
|
|
|
|
|
|
|
>250 | 1.08
(0.9–1.31) | 0.49 | 1.28
(0.73–2.25) | 0.99 | 1.02
(0.87–1.20) | 0.93 | 1.08
(0.87–1.34) | 0.35 | 1.22
(0.70–2.14) | 0.98 |
|
≤250 | 0.98
(0.76–1.26) | 0.94 | 1.49
(0.61–3.65) | 0.7 | 0.97
(0.81–1.13) | 1.00 | 0.93
(0.70–1.24) | 0.91 | 1.58
(0.65–3.84) | 0.66 |
| Source of
controls |
|
|
|
|
|
|
|
|
|
|
| HB | 1.08
(0.91–1.29) | 0.71 | 1.22
(0.70–2.12) | 0.97 | 1.02
(0.88–1.18) | 0.99 | 1.09
(0.89–1.33) | 0.60 | 1.17
(0.68–2.02) | 0.97 |
| PB | 0.96
(0.73–1.28) | 0.86 | 1.73
(0.66–4.52) | 0.71 | 0.96
(0.77–1.19) | 0.98 | 0.89
(0.65–1.22) | 0.82 | 1.85
(0.72–4.80) | 0.67 |
Heterogeneity analysis
There was significant heterogeneity in the three
genetic models for the RFC-1 A80G polymorphism: G vs. A: P=0.04, GG
vs. AA: P=0.03 and GG+GA vs. AA: P=0.01 (Table III). The source of heterogeneity
was assessed by ethnicity (Caucasian/Asian/mixed), publication year
(prior to or during 2009/after 2009), source of controls (HB/PB)
and sample size (≤200/>200 subjects). The subgroup analyses
revealed removed heterogeneities in several subgroups, including
the Caucasian and mixed population, and the large-sample group.
However, meta-regression analyses did not reveal any sources
contributing to the substantial heterogeneity. For the MTRR A2756G
and CBS 844ins68 polymorphisms and their association with DS risk,
no statistically significant heterogeneity was observed in any of
the genetic models.
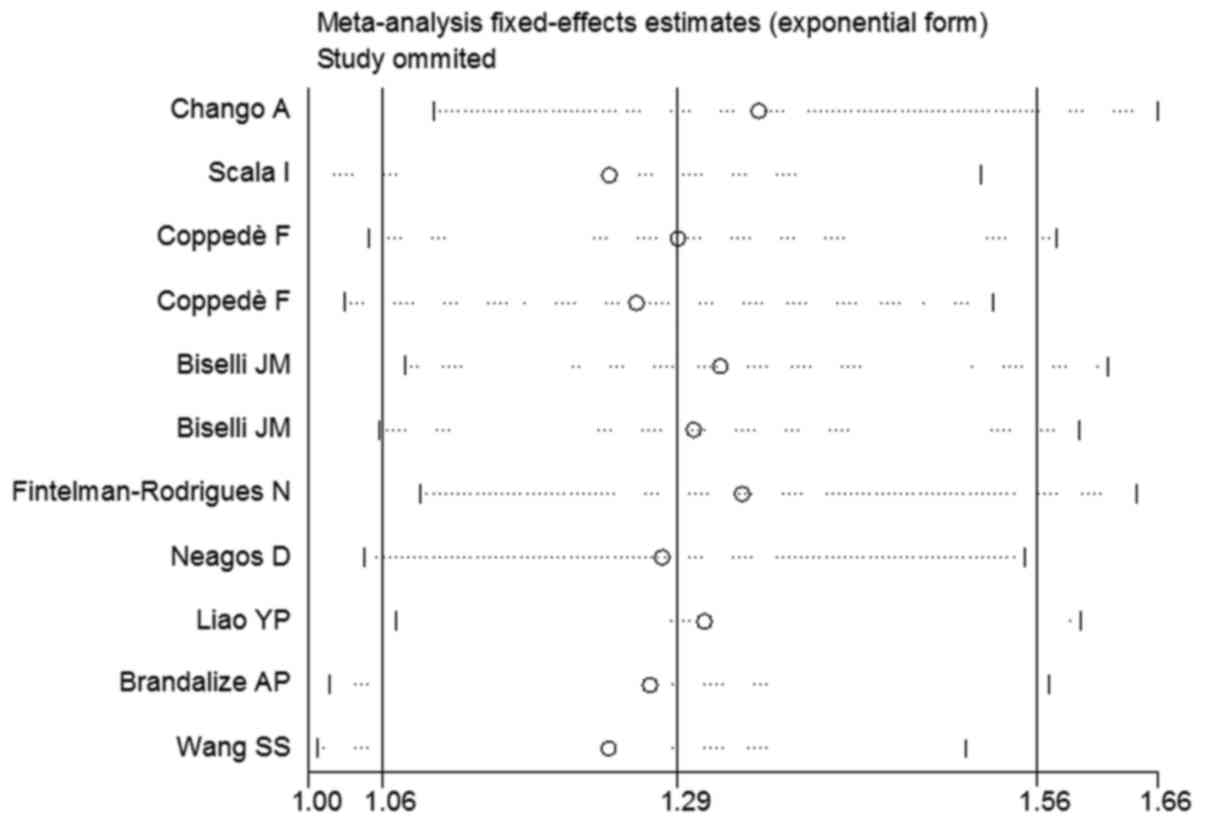
Sensitivity analysis
Sensitivity analysis was performed following
sequential removal of each eligible study. As regards the
association of the RFC-1 A80G polymorphism with maternal DS risk,
no single study qualitatively affected the pooled ORs, indicating
that the results of the meta-analysis were highly stable (Fig. 3). There were 2 studies that departed
from HWE for the CBS 844ins68 polymorphism but, when they were
excluded, the pooled OR remained unaffected (Table V).
 | Table V.Effect of the CBS 844ins68
polymorphism on DS risk. |
Table V.
Effect of the CBS 844ins68
polymorphism on DS risk.
|
| i vs. w | ii vs. ww | iw vs. ww | ii+iw vs. ww | ii vs. iw+ww |
|---|
|
|
|
|
|
|
|
|---|
| Variables | OR (95% CI) |
P-valuea | OR (95% CI) |
P-valuea | OR (95% CI) |
P-valuea | OR (95% CI) |
P-valuea | OR (95% CI) |
P-valuea |
|---|
| Overall | 1.03
(0.82–1.29) | 0.82 | 1.10
(0.51–2.34) | 0.61 | 1.02
(0.79–1.32) | 0.65 | 1.03
(0.80–1.31) | 0.76 | 1.07
(0.50–2.28) | 0.56 |
| All in HWE | 0.99
(0.74–1.13) | 0.71 | 1.29
(0.44–3.80) | 0.30 | 0.95
(0.69–1.3) | 0.75 | 0.97
(0.71–1.32) | 0.78 | 1.30
(0.44–3.80) | 0.28 |
Cumulative meta-analysis
Cumulative meta-analyses of the 3 associations were
also conducted via the assortment of studies by publication time.
The results of the association of the RFC-1A80G polymorphism with
DS risk for cumulative meta-analysis in chronological order are
shown in Fig. 4. The pooled ORs
tended to be stable, and the associations exhibited a trend towards
significance with the accumulation of more data over time.
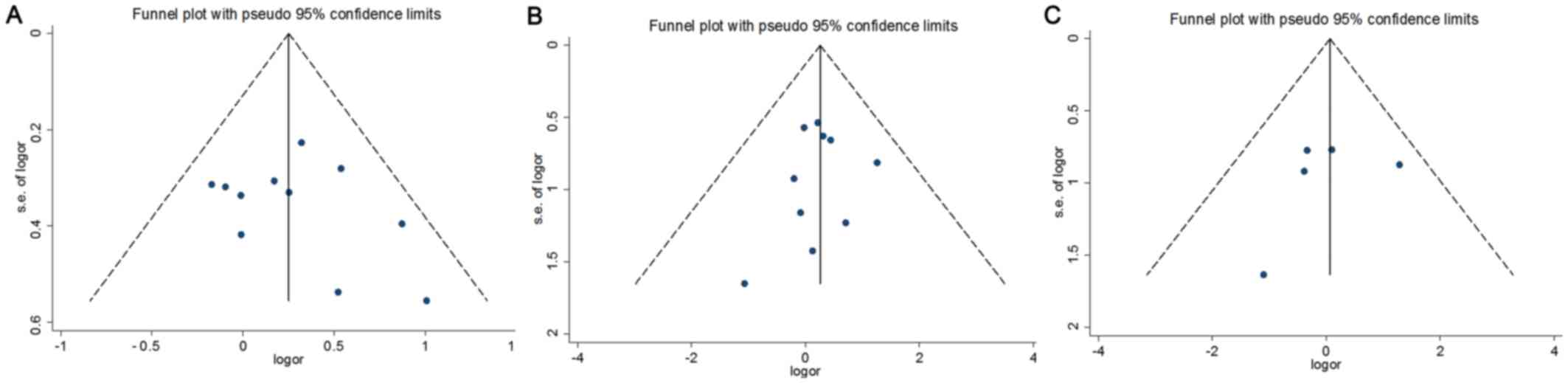
Publication bias
Egger's test and funnel plots were applied to
evaluate potential publication bias for the RFC-1 A80G, MTR A2756G
and CBS 844ins68 polymorphisms (Table
VI). The results revealed no evidence of publication bias. The
funnel plot investigating the maternal RFC-1 A80G/MTR A2756G/CBS
844ins68 polymorphisms and DS risk is shown in Fig. 5.
 | Table VI.Egger's publication bias test for
RFC-1 A80G, MTR A2756G and CBS 844ins68 polymorphisms. |
Table VI.
Egger's publication bias test for
RFC-1 A80G, MTR A2756G and CBS 844ins68 polymorphisms.
| Comparisons | Coefficient | Standard error | t | P>|t| | 95% CI |
|---|
| RFC-1 A80G
polymorphism |
|
|
|
|
|
| G vs.
A | 0.81 | 1.63 | 0.50 | 0.63 | −2.88, 4.51 |
| GG vs.
AA | 1.08 | 1.20 | 0.90 | 0.39 | −1.63, 3.78 |
| GA vs.
AA | 0.79 | .74 | 1.07 | 0.314 | −0.89, 2.47 |
| GG+GA
vs. AA | 0.75 | 1.15 | 0.65 | 0.53 | −1.84, 3.34 |
| GG vs.
GA+AA | 1.05 | 1.26 | 0.83 | 0.43 | −1.81, 3.92 |
| MTR A2756G
polymorphism |
|
|
|
|
|
| G vs.
A | −0.87 | 0.73 | −1.19 | 0.27 | −2.52, 0.79 |
| GG vs.
AA | −0.39 | 0.51 | −0.77 | 0.46 | −1.58, 0.79 |
| GA vs.
AA |
−.122 | 0.59 | −0.21 | 0.84 | −1.45, 1.21 |
| GG+GA
vs. AA | −0.73 | 0.94 | −0.78 | 0.46 | −2.86, 1.40 |
| GG vs.
GA+AA | −0.27 | 0.56 | −0.49 | 0.64 | −1.56, 1.02 |
| CBS 844ins68
polymorphism |
|
|
|
|
|
| i vs.
w | −2.37 | 1.04 | −2.28 | 0.08 | −5.26, 0.51 |
| ii vs.
ww | −1.25 | 1.74 | −0.72 | 0.52 | −6.78, 4.28 |
| iw vs.
ww | −2.13 | 2.07 | −1.03 | 0.36 | −7.88, 3.62 |
| ii+iw
vs. ww | −2.31 | 1.58 | −1.46 | 0.22 | −6.72, 2.09 |
| ii vs.
iw+ww | −1.16 | 1.85 | −0.63 | 0.58 | −7.06, 4.74 |
Discussion
The folate metabolism pathway plays an important
role in DNA methylation, DNA synthesis and cell division (41). Recent meta-analyses suggested that
the MTHFR C667T polymorphism of the folate metabolism gene may be
associated with an increased occurrence of congenital heart defects
or neural tube defects (42,43). Abnormal folate metabolism and
variants of key enzymes in the folate cycle have been described as
possible risk factors for DS through impairing DNA methylation
(44–46). A80G polymorphism in the RFC-1 gene
may impair folate membrane transport. Recently, several studies
were performed to evaluate the effects of the RFC-1 A80G/MTR
A2756G/CBS 844ins68 variant on the risk of DS, but the results were
inconclusive. For example, Brandalize et al (21) and Wang et al (20) reported opposite results on the
correlation between the RFC-1 A80G polymorphism and DS risk.
In the present meta-analysis, a significant
correlation was observed between the RFC-1 A80G polymorphism and
maternal DS risk. A significantly increased DS risk was observed in
G vs. A and in the recessive model (OR=1.19, P=0.04; and OR=1.28,
P=0.01, respectively). However, for MTR A2756G/CBS 844ins68
variants, no significant association between the two polymorphisms
and maternal DS risk was observed; the available evidence did not
support these two polymorphisms as independent risk factors for DS,
which may be attributed to the fact that the interaction of gene
polymorphisms rather than any individual polymorphism may be a
major determinant of disease risk (47).
In the subgroup analysis stratified by ethnicity, a
significant increased DS risk was not observed among Caucasians,
Asians and the mixed population for the MTR A2756G and CBS 844ins68
polymorphisms. Thus, the data of the present study suggest that the
interactions of these polymorphisms with ethnic variations may not
significantly affect DS risk. However, in the recessive model for
the RFC-1 A80G variant, a statistically significant increased risk
was observed (OR=1.38, P=0.046). Further large-scale studies are
required to investigate the possible ethnic differences in the
association of the RFC-1 A80G polymorphism with DS risk.
In the stratification analysis by study sample size
for the RFC-1 A80G variant, a significantly increased DS risk was
only observed in large-sample studies (>200 subjects). In
addition, a statistically significant correlation was found between
the RFC-1 A80G polymorphism and DS risk for the dominant model and
homozygotes, which was not observed in the overall study sample.
These results indicate that large-sample studies may offer quite
different outcomes compared with small-sample studies, which is
possibly due to the fact that small and underpowered studies may be
unable to identify true genetic associations (48). Therefore, the use of a proper,
large-sample study is crucial for reducing biases in such genotype
association studies. In the subgroup analysis stratified by source
of controls, a significantly increased DS risk was observed in the
HB groups, but not among PB groups. However, HB controls may not
always be truly representative of the general population,
particularly when the polymorphisms under investigation are
expected to affect disease conditions, which may be observed in the
HB controls, indicating the presence of possible selection bias.
Thus, further studies using proper controls with strict matching
criteria are crucial for reducing such selection biases.
In the present meta-analysis, all the available
relevant publications in English and Chinese were searched.
Furthermore, the Q-test and I2 statistics were applied
to test the significance of heterogeneity. Heterogeneity was
observed in three genetic models; subsequently, meta-regression
analysis and subgroup analyses were conducted. As a result,
heterogeneities were removed in the subgroup analysis on the
Caucasian and mixed populations, as well as on the large-sample
group. Moreover, publication bias, an important factor considered
in meta-analyses, was evaluated by funnel plots and Egger's tests.
Neither the shape of the funnel plots nor the statistical results
demonstrated publication bias, indicating the robustness and
reliability of the results.
There were certain limitations to this
meta-analysis: First, folate metabolism is complex and involves
several regulatory mechanisms. Coppedè et al (24) indicated that the presence of both
RFC-1 80GG and MTHFR 677GG was associated with a significantly
increased risk of a DS offspring (OR=6, 95% CI: 1.00–35.9), which
was also reported by other studies (26,27).
However, such gene-gene interactions were not addressed in the
present meta-analysis due to the lack of sufficient data. Second, a
subgroup analysis by maternal age was not performed in the present
meta-analysis. Maternal age is the major risk factor for DS,
although several children with DS are born to mothers aged <35
years. Recently, Scala et al (27) demonstrated that the 80G allele may
increase DS risk in mothers aged >34 years at conception.
However, only two studies in the present meta-analysis reported the
association between genetic polymorphisms and maternal age. Third,
the primary articles included in the present meta-analysis only
provided data on Caucasian, Asian and mixed ethnicities. The
majority of the studies included Caucasians and mixed populations,
whereas data on other ethnicities were not available. Finally,
although there was no publication bias observed in the funnel plot
and Egger's test, selection bias may have occurred, as only studies
in English or Chinese were selected.
In summary, the present meta-analysis suggests that
the RFC-1 A80G polymorphism may be a maternal genetic risk for DS,
particularly in the large-sample and HB control groups. However,
there was no evidence of an association between the MTR A2756G/CBS
844ins68 polymorphism and maternal DS risk. Further well-designed
large studies are required to investigate gene-environment
interactions, gene-maternal age interactions and combinations of
gene polymorphisms.
References
|
1
|
Antonarakis SE, Petersen MB, McInnis MG,
Adelsberger PA, Schinzel AA, Binkert F, Pangalos C, Raoul O,
Slaugenhaupt SA, Hafez M, et al: The meiotic stage of
nondisjunction in trisomy 21: Determination by using DNA
polymorphisms. Am J Hum Genet. 50:544–550. 1992.PubMed/NCBI
|
|
2
|
Jyothy A, Kumar KS, Mallikarjuna GN, Rao V
Babu, Devi B Uma, Sujatha M and Reddy PP: Parental age and the
origin of extra chromosome 21 in Down syndrome. J Hum Genet.
46:347–350. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Oliver TR, Feingold E, Yu K, Cheung V,
Tinker S, Yadav-Shah M, Masse N and Sherman SL: New insights into
human nondisjunction of chromosome 21 in oocytes. PLoS Genet.
4:e10000332008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen RZ, Pettersson U, Beard C,
Jackson-Grusby L and Jaenisch R: DNA hypomethylation leads to
elevated mutation rates. Nature. 395:89–93. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fenech M: Micronutrients and genomic
stability: A new paradigm for recommended dietary allowances
(RDAs). Food Chem Toxicol. 40:1113–1117. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu JJ and Ward RL: Folate and one-carbon
metabolism and its impact on aberrant DNA methylation in cancer.
Adv Genet. 71:79–121. 2010.PubMed/NCBI
|
|
7
|
McKay JA, Groom A, Potter C, Coneyworth
LJ, Ford D, Mathers JC and Relton CL: Genetic and non-genetic
influences during pregnancy on infant global and site specific DNA
methylation: Role for folate gene variants and vitamin B12. PLoS
One. 7:e332902012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
James SJ, Melnyk S, Jernigan S, Pavliv O,
Trusty T, Lehman S, Seidel L, Gaylor DW and Cleves MA: A functional
polymorphism in the reduced folate carrier gene and DNA
hypomethylation in mothers of children with autism. Am J Med Genet
B Neuropsychiatr Genet 153B. 1209–1220. 2010.
|
|
9
|
Bailey LB: New standard for dietary folate
intake in pregnant women. Am J Clin Nutr. 71 5 Suppl:1304S–1307S.
2000.PubMed/NCBI
|
|
10
|
Goyette P, Sumner JS, Milos R, Duncan AM,
Rosenblatt DS, Matthews RG and Rozen R: Human
methylenetetrahydrofolate reductase: Isolation of cDNA mapping and
mutation identification. Nat Genet. 7:5511994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu X, Wang X, Chan Y, Jia S, Luo Y and
Tang W: Folate metabolism gene polymorphisms MTHFR C677T and A1298C
and risk for Down syndrome offspring: A meta-analysis. Eur J Obstet
Gynecol Reprod Biol. 167:154–159. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zintzaras E: Maternal gene polymorphisms
involved in folate metabolism and risk of Down syndrome offspring:
A meta-analysis. J Hum Genet. 52:943–953. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
van der Put NM, van der Molen EF,
Kluijtmans LA, Heil SG, Trijbels JM, Eskes TK, Van
Oppenraaij-Emmerzaal D, Banerjee R and Blom HJ: Sequence analysis
of the coding region of human methionine synthase: Relevance to
hyperhomocysteinaemia in neural-tube defects and vascular disease.
QJM. 90:511–517. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Leclerc D, Campeau E, Goyette P, Adjalla
CE, Christensen B, Ross M, Eydoux P, Rosenblatt DS, Rozen R and
Gravel RA: Human methionine synthase: cDNA cloning and
identification of mutations in patients of the cblG complementation
group of folate/cobalamin disorders. Hum Mol Genet. 5:1867–1874.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsai MY, Bignell M, Schwichtenberg K and
Hanson NQ: High prevalence of a mutation in the cystathionine
beta-synthase gene. Am J Hum Genet. 59:1262–1267. 1996.PubMed/NCBI
|
|
16
|
Zhang J, Handy DE, Wang Y, Bouchard G,
Selhub J, Loscalzo J and Carey MC: Hyperhomocysteinemia from
trimethylation of hepatic phosphatidylethanolamine during
cholesterol cholelithogenesis in inbred mice. Hepatology.
54:697–706. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lopreato FR, Stabler SP, Carvalho FR,
Hirata RD, Hirata MH, Robi DL, Sampaio-Neto LF, Allen RH and
Guerra-Shinohara EM: Relationships between gene polymorphisms of
folate-related proteins and vitamins and metabolites in pregnant
women and neonates. Clin Chim Acta. 398:134–139. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sukla KK and Raman R: Association of MTHFR
and RFC1 gene polymorphism with hyperhomocysteinemia and its
modulation by vitamin B12 and folic acid in an Indian population.
Eur J Clin Nutr. 66:111–118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chango A, Emery-Fillon N, de Courcy GP,
Lambert D, Pfister M, Rosenblatt DS and Nicolas JP: A polymorphism
(80G->A) in the reduced folate carrier gene and its associations
with folate status and homocysteinemia. Mol Genet Metab.
70:310–315. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang SS, Wang C, Qiao FY, Lv JJ and Feng
L: Polymorphisms in genes RFC-1/CBS as maternal risk factors for
Down syndrome in China. Arch Gynecol Obstet. 288:273–277. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brandalize AP, Bandinelli E, Dos Santos PA
and Schüler-Faccini L: Maternal gene polymorphisms involved in
folate metabolism as risk factors for Down syndrome offspring in
Southern Brazil. Dis Markers. 29:95–101. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Neagos D, Cretu R, Tutulan-Cunita A,
Stoian V and Bohiltea LC: RFC-1 Gene Polymorphism and the Risk of
Down Syndrome in Romanian Population. Maedica (Buchar). 5:280–285.
2010.PubMed/NCBI
|
|
23
|
Coppedè F, Colognato R, Bonelli A, Astrea
G, Bargagna S, Siciliano G and Migliore L: Polymorphisms in folate
and homocysteine metabolizing genes and chromosome damage in
mothers of Down syndrome children. Am J Med Genet A.
143A:2006–2015. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Coppedè F, Marini G, Bargagna S, Stuppia
L, Minichilli F, Fontana I, Colognato R, Astrea G, Palka G and
Migliore L: Folate gene polymorphisms and the risk of Down syndrome
pregnancies in young Italian women. Am J Med Genet A.
140:1083–1091. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chango A, Fillon-Emery N, Mircher C,
Bléhaut H, Lambert D, Herbeth B, James SJ, Réthoré MO and Nicolas
JP: No association between common polymorphisms in genes of folate
and homocysteine metabolism and the risk of Down's syndrome among
French mothers. Br J Nutr. 94:166–169. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fintelman-Rodrigues N, Corrêa JC, Santos
JM, Pimentel MM and Santos-Rebouças CB: Investigation of CBS MTR,
RFC-1 and TC polymorphisms as maternal risk factors for Down
syndrome. Dis Markers. 26:155–161. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Scala I, Granese B, Sellitto M, Salomé S,
Sammartino A, Pepe A, Mastroiacovo P, Sebastio G and Andria G:
Analysis of seven maternal polymorphisms of genes involved in
homocysteine/folate metabolism and risk of Down syndrome offspring.
Genet Med. 8:409–416. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Biselli JM, Brumati D, Frigeri VF,
Zampieri BL, Goloni-Bertollo EM and Pavarino-Bertelli EC: A80G
polymorphism of reduced folate carrier 1 (RFC1) and C776G
polymorphism of transcobalamin 2(TC2) genes in Down's syndrome
etiology. Sao Paulo Med J. 126:329–332. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Biselli JM, Goloni-Bertollo EM, Zampieri
BL, Haddad R, Eberlin MN and Pavarino-Bertelli EC: Genetic
polymorphisms involved in folate metabolism and elevated plasma
concentrations of homocysteine: Maternal risk factors for Down
syndrome in Brazil. Genet Mol Res. 7:33–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Coppedè F, Grossi E, Migheli F and
Migliore L: Polymorphisms in folate-metabolizing genes, chromosome
damage, and risk of Down syndrome in Italian women: Identification
of key factors using artificial neural networks. BMC Med Genomics.
3:422010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liao YP, Bao MS, Liu CQ, Liu H and Zhang
D: Folate gene polymorphism and the risk of Down syndrome
pregnancies in young Chinese women. Yi Chuan. 32:461–466. 2010.(In
Chinese). View Article : Google Scholar : PubMed/NCBI
|
|
32
|
da Silva LR, Vergani N, Lde C Galdieri,
Porto MP Ribeiro, Longhitano SB, Brunoni D, D'Almeida V and Perez
AB Alvarez: Relationship between polymorphisms in genes involved in
homocysteine metabolism and maternal risk for Down syndrome in
Brazil. Am J Med Genet A. 135:263–267. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zampieri BL, Biselli JM, Goloni-Bertollo
EM, Vannucchi H, Carvalho VM, Cordeiro JA and Pavarino EC: Maternal
risk for Down syndrome is modulated by genes involved in folate
metabolism. Dis Markers. 32:73–81. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Coppedè F, Migheli F, Bargagna S,
Siciliano G, Antonucci I, Stuppia L, Palka G and Migliore L:
Association of maternal polymorphisms in folate metabolizing genes
with chromosome damage and risk of Down syndrome offspring.
Neurosci Lett. 449:15–19. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zintzaras E, Doxani C, Koufakis T,
Kastanis A, Rodopoulou P and Karachalios T: Synopsis and
meta-analysis of genetic association studies in osteoporosis for
the focal adhesion family genes: The CUMAGAS-OSTEOporosis
information system. BMC Med. 9:92011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Higgins JP and Thompson SG: Quantifying
heterogeneity in a meta-analysis. Stat Med. 21:1539–1558. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Macaskill P, Walter SD and Irwig L: A
comparison of methods to detect publication bias in meta-analysis.
Stat Med. 20:641–654. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Begg CB and Mazumdar M: Operating
characteristics of a rank correlation test for publication bias.
Biometrics. 50:1088–1101. 1994. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Egger M, Smith G Davey, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Morrison K, Papapetrou C, Hol FA, Mariman
EC, Lynch SA, Burn J and Edwards YH: Susceptibility to spina
bifida; an association study of five candidate genes. Ann Hum
Genet. 62:379–396. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yan L, Zhao L, Long Y, Zou P, Ji G, Gu A
and Zhao P: Association of the maternal MTHFR C677T polymorphism
with susceptibility to neural tube defects in offsprings: Evidence
from 25 case-control studies. PLoS One. 7:e416892012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yin M, Dong L, Zheng J, Zhang H, Liu J and
Xu Z: Meta analysis of the association between MTHFR C677T
polymorphism and the risk of congenital heart defects. Ann Hum
Genet. 76:9–16. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Marucci GH, Zampieri BL, Biselli JM,
Valentin S, Bertollo EM, Eberlin MN, Haddad R, Riccio MF, Vannucchi
H, Carvalho VM and Pavarino EC: Polymorphism C1420T of Serine
hydroxymethyltransferase gene on maternal risk for Down syndrome.
Mol Biol Rep. 39:2561–2566. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Costa-Lima MA, Amorim MR and Orioli IM:
Association of methylenetetrahydrofolate reductase gene 677C > T
polymorphism and Down syndrome. Mol Biol Rep. 40:2115–2125. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Biselli JM, Zampieri BL, Goloni-Bertollo
EM, Haddad R, Fonseca MF, Eberlin MN, Vannucchi H, Carvalho VM and
Pavarino EC: Genetic polymorphisms modulate the folate metabolism
of Brazilian individuals with Down syndrome. Mol Biol Rep.
39:9277–9284. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hobbs CA, Cleves MA, Lauer RM, Burns TL
and James SJ: Preferential transmission of the MTHFR 677 T allele
to infants with Down syndrome: Implications for a survival
advantage. Am J Med Genet. 113:9–14. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hindorff LA, Sethupathy P, Junkins HA,
Ramos EM, Mehta JP, Collins FS and Manolio TA: Potential etiologic
and functional implications of genome-wide association loci for
human diseases and traits. Proc Natl Acad Sci USA. 106:9362–9367.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang XD, Gong J, Wu Y, Cao SY, Yang SY, Li
MR, et al: Genetic polymorphisms involved in folate metabolism as
maternal risk factors for Down syndrome. Chin J Birth Health Hered.
2012. 2012.
|