Introduction
Oral squamous-cell carcinoma (OSCC) is the most
common type of cancer of the head and neck. There were 364,872
(147,897 females and 216,975 males) head and neck cancer-associated
mortalities in Japan in 2013, according to the latest survey by the
National Cancer Center (1). Also,
~60% head and neck cancer cases diagnosed are patients in the
advanced stages (stage III and stage IV) (2,3). Despite
modern advances in cancer therapy and clinical assessment, the
survival rate has not improved substantially in recent years.
Therefore, investigations must continue for the identification of
suitable biomarkers to ensure early diagnosis or prognostic
prediction for OSCC. The development of useful biomarkers may lead
to the development of a novel therapeutic strategy or
chemopreventive agents.
Carbonyl reductase (CBR) is an enzyme that catalyzes
the reduction of numerous carbonyl compounds by using
NADPH-dependent oxidoreductase activity (4,5). There
are two monomeric CBR genes (cbr1 and cbr3) in the
human genome. They exhibit high homology in their amino acid
sequence (6). CBR1 has been studied
regarding its ability to reduce a variety of carbonyl compounds,
including antitumor anthracycline antibiotics, daunorubicin and
doxorubicin, and prostaglandins (6,7).
Recently, it was reported that CBR1 serves an important role in
regulating the malignant potential of cancer cells. In addition,
the loss of CBR1 expression promotes tumor growth and metastasis
(8,9). Furthermore, reduced CBR1 expression is
associated with lymph node metastasis and poor prognosis in ovarian
cancer and uterine cervical cancer (9,10).
However, these reports did not indicate how decreased CBR1 levels
may affect the malignant behavior of cancer. Furthermore, the role
of CBR1 in OSCC has not yet been examined. Herein, the association
between CBR1 expression patterns, lymph node metastasis and
clinical prognosis in OSCC tissues was examined; the molecular
mechanism by which CBR1 affects cancer cell invasion and metastasis
in OSCC was also assessed.
Materials and methods
Clinical characteristics of patients
and patient samples
Primary oral cancer tissue samples were obtained
from 90 patients (46 males and 44 females; age range, 18–96 years)
with OSCC who were treated at Yamaguchi University Hospital (Ube,
Japan) from April 2006 to March 2012. All patients received
curative surgery. Prior to primary treatment, tissue specimens were
obtained from all 90 patients via biopsy. All tissue samples were
fixed in phosphate-buffered 10% formalin and were paraffin-embedded
(FFPE).
All tumors were staged according to the TNM
classification of the UICC (2002) (11), and the degree of differentiation was
determined according to the grade classification system of the
World Health Organization (12).
Approval for this study protocol was obtained from
the Institutional Review Board (IRB) and Ethical Committee of
Yamaguchi University Hospital (Ref. H27-123). Informed consent was
waived by the IRB as this was a retrospective study.
Immunohistochemical staining and the
analysis of staining
Tissue biopsy samples obtained prior to surgery were
used for immunohistochemical analyses. FFPE samples were heated in
an autoclave at 121°C for 15 min in 10 µM citrate buffer solution
at pH 9.0. After quenching the endogenous peroxidase activity, the
sections were treated for 2 h at room temperature with 10% normal
goat serum (cat. no. ab7481; Abcam, Cambridge, UK). The tissue
sections were then incubated overnight at 4°C with
rabbit-monoclonal anti-carbonyl reductase 1 (CBR1) antibody
(dilution, 1:100; cat. no. ab156590; Abcam). After washing the
tissue sections in PBS for 10 min at room temperature, the
antibodies were detected using the Dako REAL™ EnVision™ Detection
system (cat. no. K5007; Agilent Technologies, Inc., Santa Clara,
CA, USA), according to the manufacturer's protocol. Tissues were
washed in PBS for 5 min at room temperature, and then
counterstained with hematoxylin at room temperature for 1 min. The
tissue sections were subsequently dehydrated in (70–100% v/v)
graded ethyl alcohol, dewaxed using xylene for 10 min at room
temperature, and then mounted with glass coverslips using DPX
mounting medium (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany).
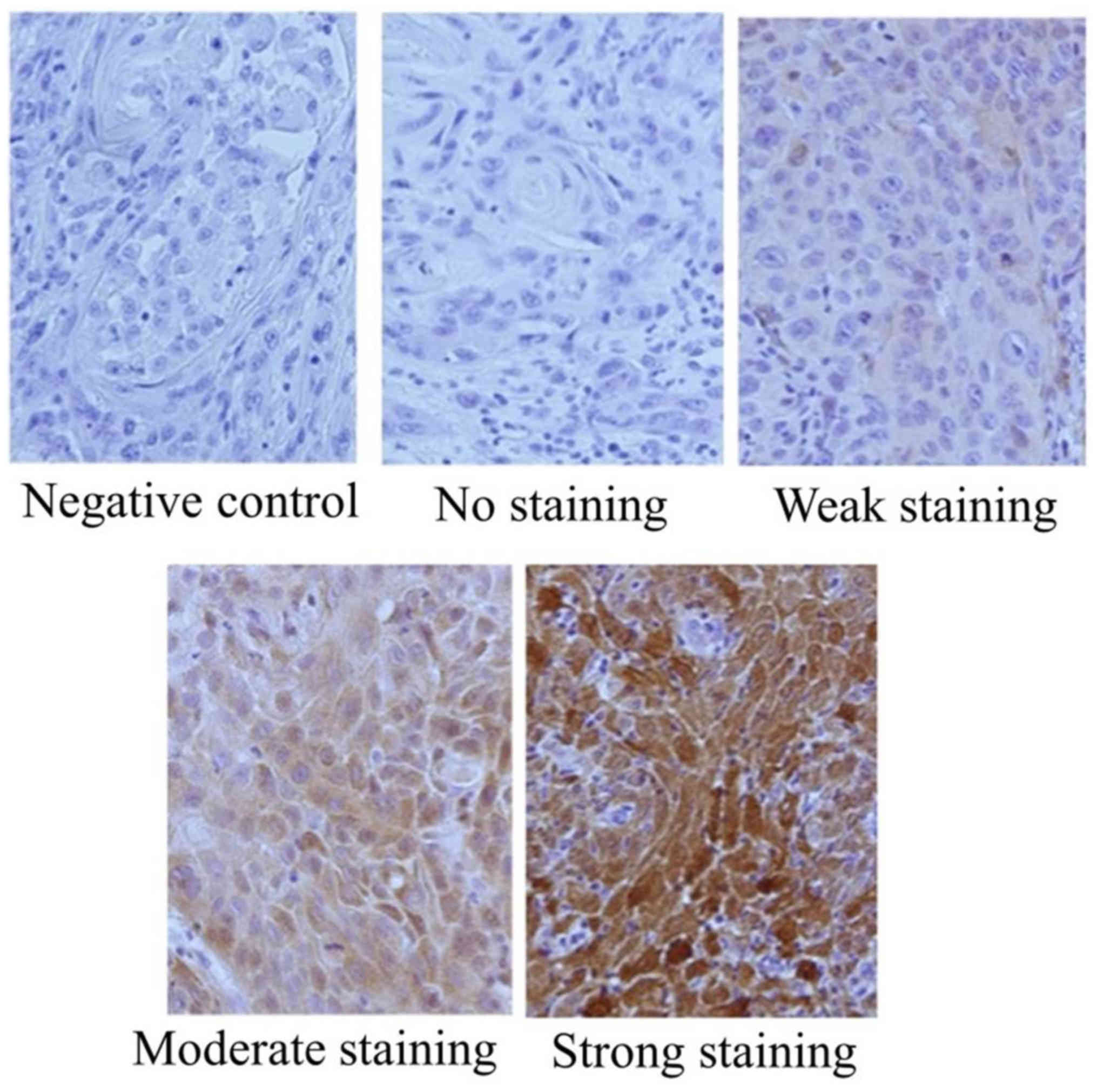
CBR1 expression was evaluated by the method
described by Murakami et al (13) with minor modifications. Briefly, the
intensity of CBR1 staining in cancer cells from cancerous tissues
was compared with that in the normal adjacent oral epithelium. The
immunoreactivity score was calculated as the sum of: (a) The
percentage of positive cells (0, 0% immunopositive cells; 1,
<50% positive cells; 2, ≥50% positive cells); and (b) the
staining intensity (0, negative; 1, weak; 2, moderate; 3, high).
The sum of the assigned values of the positive cells percentage (a)
and the staining intensity (b) was 0–5. Scores from 0–2 were
regarded as low expression, whereas scores of 3–5 were regarded as
high expression (Fig. 1).
Immunoreactivity for CBR1 expression was evaluated by three
authors, who were blinded to the patient's clinical status.
Cell lines and cell culture
OSCC cell lines (HSC2, HSC3, HSC4, SAS and Ca9-22)
and HaCaT were obtained from the Cell Bank (RIKEN BioResource
Center, Tsukuba, Japan). Cells were cultured in Dulbecco's modified
Eagle's medium (DMEM; Sigma-Aldrich; Merck KGaA) supplemented with
10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), 100 µg/ml streptomycin and 100 U/ml penicillin
(Thermo Fisher scientific) at 37°C in a humidified atmosphere
containing 5% CO2.
Small interfering (si)RNA
transfection
The siRNA for CBR1 and non-targeting negative
control siRNA were obtained from Thermo Fisher Scientific, Inc. The
sequences of siRNA used for the study were as follows: CBR1 siRNA,
sense, 5′-CAAGGUUGCUGAUCCCACATT-3′ and antisense,
5′-UGUGGGAUCAGCAACCUUGAA-3′. Silencer Select Negative Control No.1
siRNA (Thermo Fisher Scientific, Inc.) was used as a nonspecific
control. HSC2 cells were incubated with antibiotic-free DMEM
supplemented with 10% FBS, then transfected with 75 pM CBR1 siRNA
or negative control siRNA using a Lipofectamine® 3000
Transfection kit (Thermo Fisher Scientific, Inc.) to generate CBR1
siRNA and negative control siRNA cell lines, respectively. Also,
HSC2 cells were incubated with Lipofectamine® 3000
without any siRNA to generate a Lipofectamine cell line. The cells
were transfected for 48 h prior to use for each experiment. As the
HSC2 cell line has a better tumorigenic capacity than the other
OSCC cell lines (14), only HSC2 was
used for siRNA transfection with the aim of using the transfected
cells for future in vivo mouse xenograft experiments.
Western blot analysis
HSC2, HSC3, HSC4, SAS, Ca9-22 and HaCaT cells were
collected, centrifuged at 4°C and 390 × g for 5 min, and lysed with
radioimmunoprecipitation buffer (Wako Pure Chemical Industries,
Ltd., Osaka, Japan). The quantity of protein from the whole cell
lysates was quantified using a NanoDrop™ 1000 spectrophotometer
(Thermo Fisher Scientific, Inc.). A total of 20 µl protein sample
containing 50 µg/ml protein was loaded into each well of a NuPAGE™
4–12% Bis-Tris gel (Thermo Fisher Scientific, Inc.) and separated
using electrophoresis (200V, 110–125 mA gel, 40 min), then
transferred onto a polyvinylidene fluoride membrane using
iBlot® gel transfer stacks (Thermo Fisher Scientific,
Inc.). A blocking solution and a primary antibody dilution solution
were made from WesternBreeze® Blocker/Diluent part A and
B (cat. no. WB7050; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. Following blocking at room temperature
for 30 min, the membranes were incubated with a rabbit-monoclonal
anti-carbonyl reductase 1 (CBR1) antibody (dilution, 1:500; cat.
no. ab156590; Abcam) or an anti-α-tubulin mouse monoclonal antibody
(dilution, 1:500; cat. no. sc-5286; Santa Cruz Biotechnology, Inc.)
at 4°C overnight. Subsequently, the membranes were washed using 1X
WesternBreeze® wash solution (Thermo Fisher Scientific,
Inc.) three times at room temperature (5 min/wash), followed by
incubation with Novex® alkaline-phosphatase conjugated
goat anti-rabbit (cat. no. WP2007; Thermo Fisher Scientific, Inc.)
or goat anti-mouse immunoglobulin G secondary antibodies (cat. no.
WP20006; Thermo Fisher Scientific, Inc.) at room temperature for 30
min. Following washing of the membranes three times with the wash
solution at room temperature, protein bands were detected upon
incubation of the membranes with Novex® AP Chromogenic
substrate (cat. no. WP20001; Thermo Fisher Scientific, Inc.) at
room temperature for 5–15 min. ImageJ v.1.51 h software (National
Institutes of Health, Bethesda, MD, USA; http://imagej.nih.gov/ij/) was used to quantify the
average intensities of each standard protein band, which were
compared with the band intensities of the control protein,
α-tubulin.
In vitro cell viability assay
HSC2 cells (4×103 cells/well) were seeded
onto 96-well plates (BD Biosciences, Franklin lakes, NJ, USA) in
DMEM supplemented with 10% FBS. After 48 h, MTT was added to each
well (25 µl/well) and incubated for 4 h. After removing the MTT
solution, 100 µl dimethyl sulfoxide was added to each well and the
absorbance was measured with a spectrophotometer (Bio Rad
Laboratories, Inc., Hercules, CA, USA) at 490 nm. All assays were
run in triplicate.
Cell migration assay
A cell migration assay was performed using a Boyden
chamber set (Neuro Probe, Inc., Gaithersburg, MD, USA), according
to the manufacturer's instructions. A total of 5×103
HSC2 cells in 50 µl DMEM without FBS were seeded onto a
gelatin-coated polycarbonate membrane. In the lower chambers, 25 µl
DMEM with 10% FBS was added as a chemoattractant. After the cells
were incubated for 24 h at 37°C in a 5% CO2 atmosphere,
the polycarbonate membranes were washed with PBS and the cells on
the top surface of the polycarbonate membrane were removed with a
cotton swab. Cells adhering to the lower surface were fixed with
99.8% methanol at room temperature, counterstained with hematoxylin
solution at room temperature and counted under a light microscope
in five predetermined fields (magnification, ×200). All assays were
independently repeated ≥3 times.
Wound healing assay
HSC2 cells (1,5103 cells per well) were
seeded into a 24-well plate (BD Biosciences) and cultured in DMEM
with 10% FBS and 1% penicillin/streptomycin until a monolayer
formed. A 200-µl pipette tip was used to gently wound the cell
layer through the central axis of the plate. The migration of cells
into the wounded area was examined after 24 h using a light
microscope (BX-51-33-FLD2; Olympus Corporation, Tokyo, Japan). This
assay was repeated three times.
Statistical analysis
For the analysis of CBR1 expression levels and
associations between the clinicopathological variables in OSCC
tissues obtained from patients, the chi-squared test was used.
Overall survival (OS) of the patients with OSCC was defined as the
time from treatment initiation to the date of mortality due to any
cause. The Kaplan-Meier method was used to estimate the probability
of OS in patients with OSCC as a function of time, and statistical
differences in the survival time between patient subgroups were
compared using the log-rank test. A multivariate survival analysis
was performed using a Cox regression model to study the effects of
CBR1 expression on OS of patients with OSCC. The Student's t-test
was used to calculate the statistical significance among the
various cells in the in vitro cell viability and migration
assays. All P-values were based on two-tailed statistical analysis.
P<0.05 and P<0.01 were considered to indicate statistically
significant differences. All statistical analyses were conducted
using StatView v.5.0J software (SAS Institute, Inc., Cary, NC,
USA).
Results
Patients and tumor
characteristics
The characteristics of the cohort of patients
with OSCC are summarized in Table I.
This study included a total of 90 patients with OSCC. The median
follow-up time was 3.4 years, and the median patient age was 68
years (range, 18–96 years). For OSCC, clinical stages I, II, III
and IV were identified in 19, 38, 7 and 26 patients, respectively,
at the time of diagnosis.
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
|
| Total (n=90) |
|---|
|
|
|
|---|
| Characteristics | No. patients | % of patients |
|---|
| Sex |
|
|
| Male | 46 | 51.1 |
|
Female | 44 | 48.9 |
| T classification |
|
|
| 1 | 20 | 22.2 |
| 2 | 43 | 47.8 |
| 3 | 5 | 5.6 |
| 4 | 22 | 24.4 |
| N classification |
|
|
| 0 | 70 | 77.8 |
| 1 | 11 | 12.2 |
| 2 | 7 | 7.8 |
| 3 | 2 | 2.2 |
| Stage |
|
|
| I | 19 | 21.1 |
| II | 38 | 42.2 |
| III | 7 | 7.8 |
| IV | 26 | 28.9 |
| Outcome |
|
|
|
Alive | 85 | 94.4 |
|
Mortality | 5 | 55.6 |
| CBR1 expression
in |
|
|
| tumor cell
cytoplasm |
|
|
| Low | 26 | 28.9 |
| High | 64 | 71.1 |
|
| Median | Min-max |
| Age (years) | 68.0 | 18–96 |
CBR1 expression in tumor cells and
clinicopathological features
Table II details the
correlations between CBR1 expression levels in tumor cells and
certain clinicopathological features of patients. Among the 90
patients with OSCC, CBR1 expression in tumor cells was low in 26
patients (28.9%) and high in 64 patients (71.1%). CBR1 positivity
in tumor cells was significantly associated with the N
classification (P<0.0001), a higher clinical stage (P=0,0033)
and a fatal outcome (P=0.0095). On the other hand, CBR1 positivity
was not significantly correlated with sex, age and T classification
at the time of diagnosis.
 | Table II.Correlation of CBR1 expression and
clinicopathological factors in OSCC. |
Table II.
Correlation of CBR1 expression and
clinicopathological factors in OSCC.
|
| CBR1 expression in
tumor cell |
|
|
|---|
|
|
|
|
|
|---|
| Characteristic | Low expression
(n=26, 28.9%) | High expression
(n=64, 71.1%) | Total (n=90) | aP-value |
|---|
| Sex |
|
|
| 0.4897 |
|
Male | 15 | 31 | 46 |
|
|
Female | 11 | 33 | 44 |
|
| Age |
|
|
| 0.6409 |
|
≥65 | 17 | 38 | 55 |
|
|
<65 | 9 | 26 | 35 |
|
| T
classification |
|
|
| 0.1302 |
| 1 +
2 | 15 | 48 | 63 |
|
| 3 +
4 | 11 | 16 | 27 |
|
| N
classification |
|
|
|
<0.0001a |
| 0 | 12 | 59 | 71 |
|
| 1 + 2 +
3 | 14 | 5 | 19 |
|
| Stage |
|
|
| 0.0018a |
| I +
II | 10 | 47 | 57 |
|
| III +
IV | 16 | 17 | 33 |
|
| Outcome |
|
|
| 0.0095a |
|
Alive | 22 | 63 | 85 |
|
|
Mortality | 4 | 1 | 5 |
|
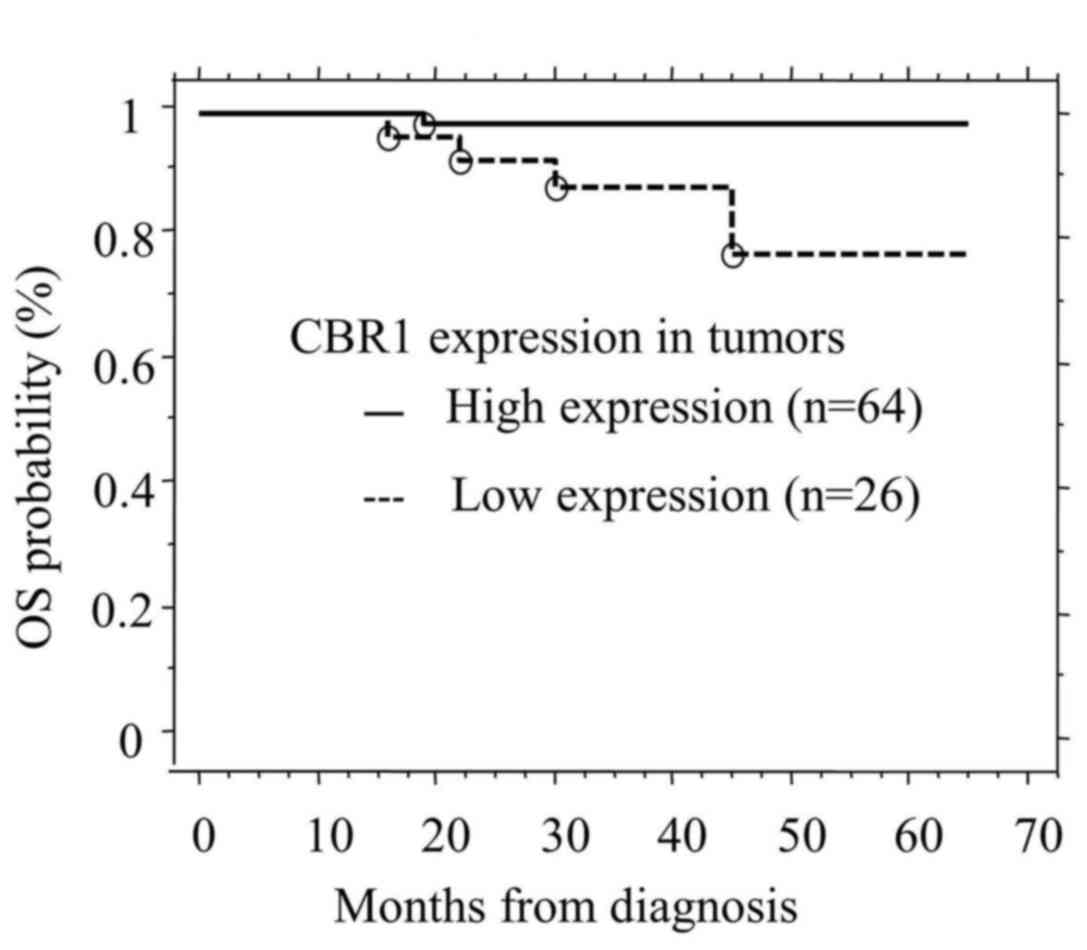
CBR1 expression in OSCCs and survival
time
The overall median follow-up of the cohort was 3.4
years and mortality occurred in five cases. CBR1 positivity on
tumor cell was associated with OS (P=0.0171; Fig. 2). Furthermore, multivariate analysis
revealed that low expression levels of CBR1 (P=0.0485) and Stage
III+IV (P=0.0466) are predictors of reduced survival, although no
other variables were identified (Table
III).
 | Table III.Risk factors affecting overall
survival rate determined by Cox's proportional hazards model. |
Table III.
Risk factors affecting overall
survival rate determined by Cox's proportional hazards model.
| Variable | Hazard ratio | 95% CI | aP-value |
|---|
| T
classification |
|
| 0.1537 |
| T3+T4
vs. T1+T2 | 3.68 | 0.614–22.054 |
|
| N
classification |
|
| 0.2709 |
| N+ vs.
N- | 2.753 | 0.456–16.399 |
|
| Stage |
|
| 0.0466 |
| Stage
III+IV vs. Stage I +II | 6.178 | 1.028–37.131 |
|
| CBR1
expression |
|
| 0.0485 |
| High
vs. low | 0.109 | 0.012–0.986 |
|
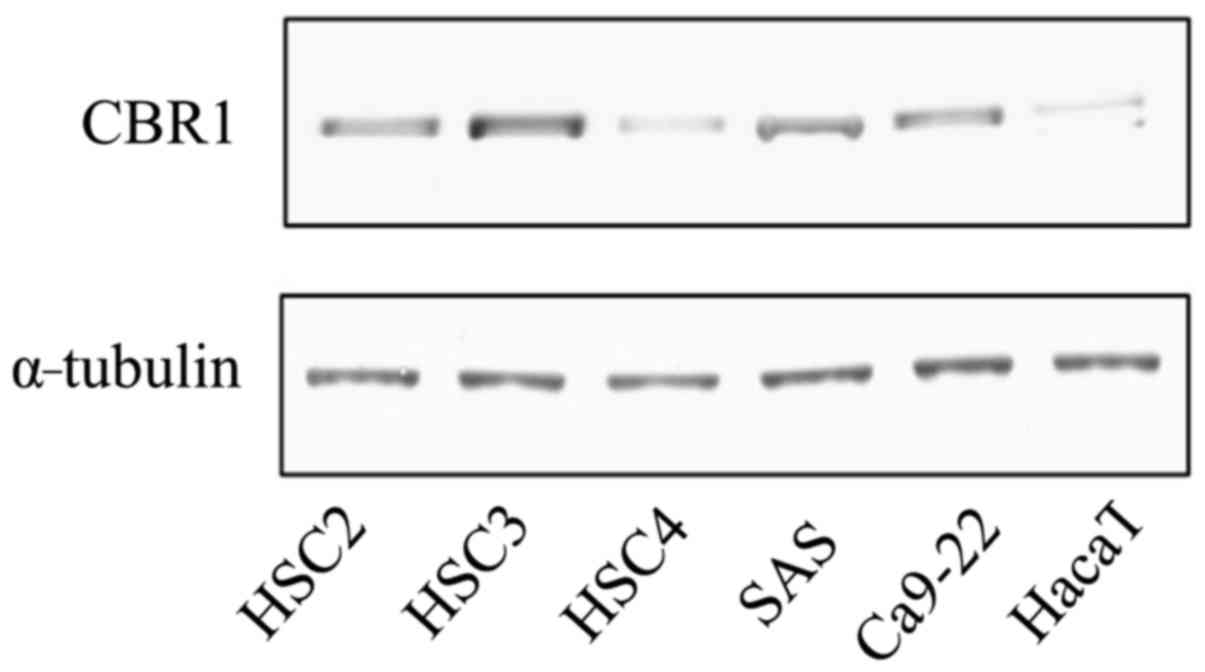
Analyses of protein expression of CBR1
in OSCC-derived cell lines
The level of CBR1 protein expression in OSCC-derived
cell lines (HSC2, HSC3, HSC4, SAS and Ca9-22) and HaCaT was
evaluated by western blot analysis. The highest expression of CBR1
was observed in HSC3 cells, the second highest expression was
observed in HSC2 cells and the lowest was recorded in HaCaT cells,
each when compared with all other cell lines (Fig. 3).
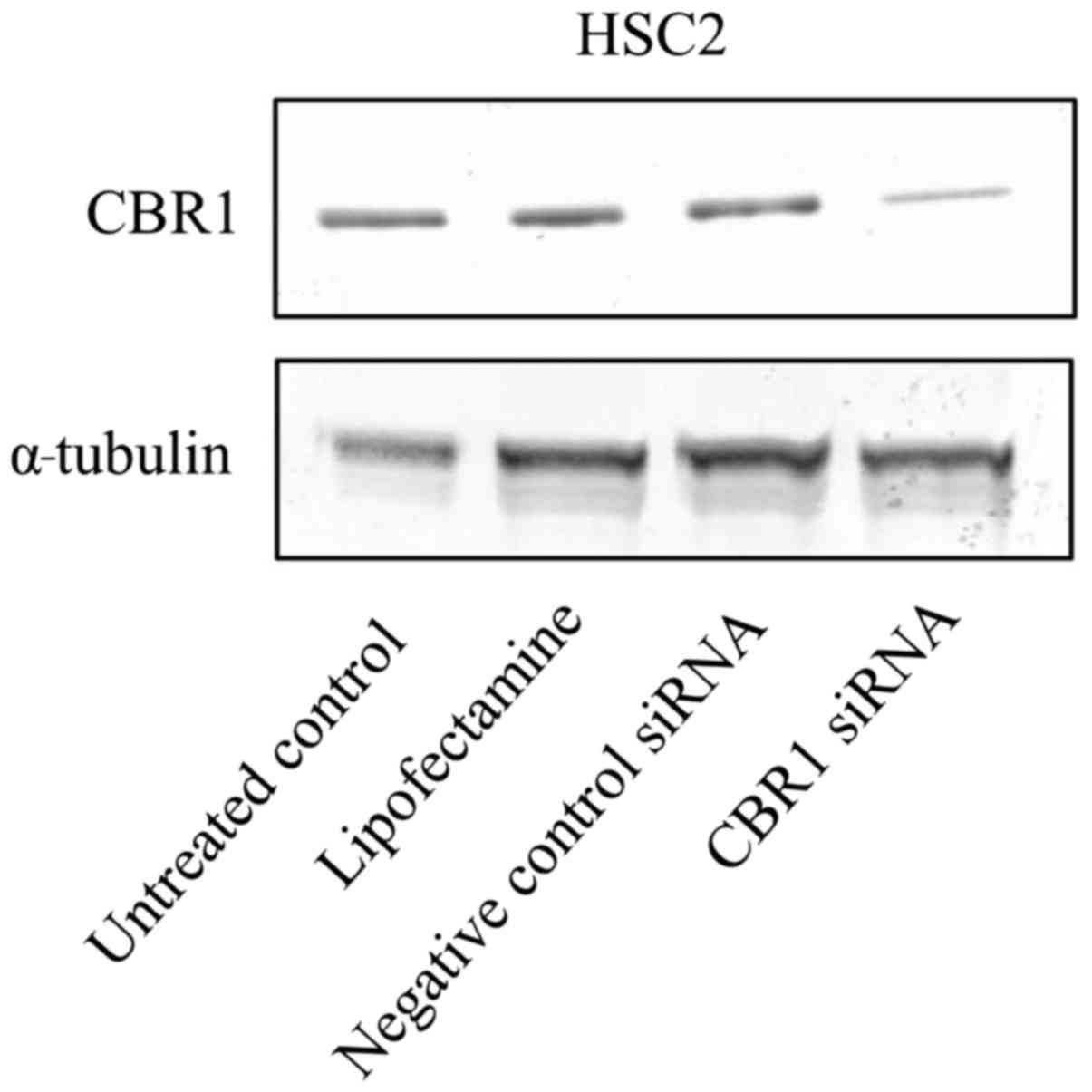
Downregulation of CBR1 by siRNA
To examine the role of CBR1 downregulation in the
OSCC-derived cell line HSC2, downregulation experiments were
carried out by transfecting HSC2 cells with CBR1 siRNA. HSC2 cells
were used only for siRNA transfection with the aim of using these
transfected cells for future in vivo mouse xenograft
experiments, as HSC2 cells have a better tumorigenic capacity than
other OSCC cell lines according to our previous study (14). Western blot analysis confirmed a
significant reduction (4~6.1-fold) in CBR1 protein expression in
the transfected CBR1 siRNA cells, as compared with in the untreated
control, Lipofectamine and negative control siRNA cells (Fig. 4).
Downregulation of CBR1 enhances the
cell growth ability
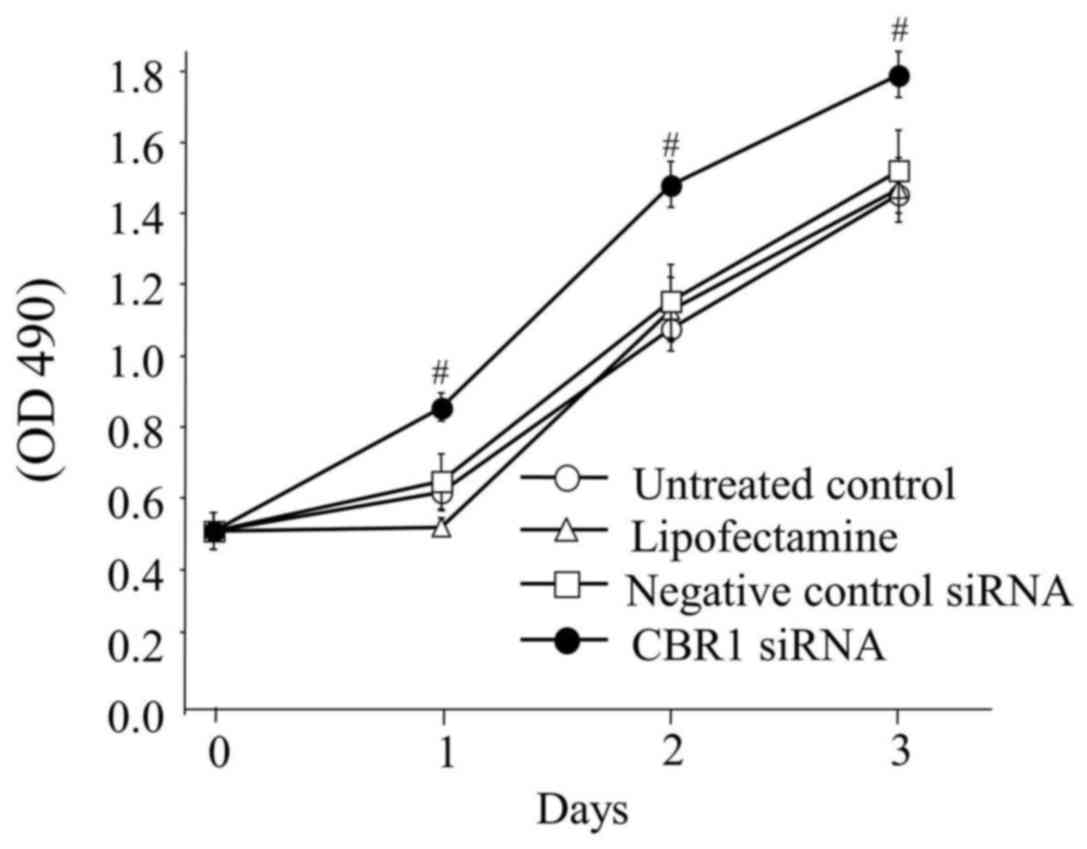
An MTT assay was performed with the untreated
control HSC2 cells, Lipofectamine, CBR1 siRNA and negative control
siRNA cells to check the viability of the cells, in order to
evaluate the proliferation rate of the cells. The cells with low
CBR1 expression (CBR1 siRNA cells) exhibited significantly enhanced
growth when compared with the other cells. These results suggest
that CBR1 may be a critical factor affecting the growth of OSCC
cells (Fig. 5).
Downregulation of CBR1 enhances the
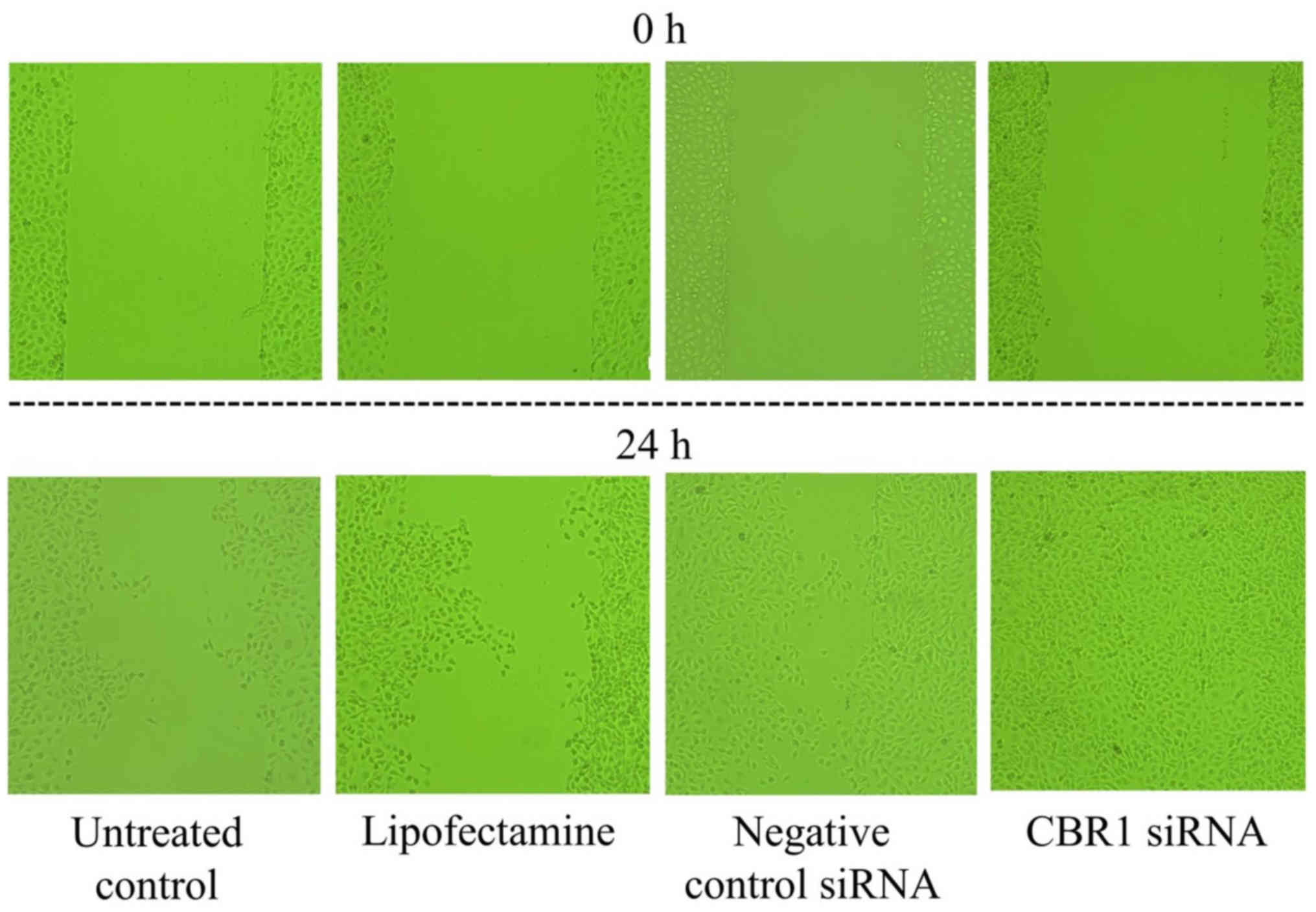
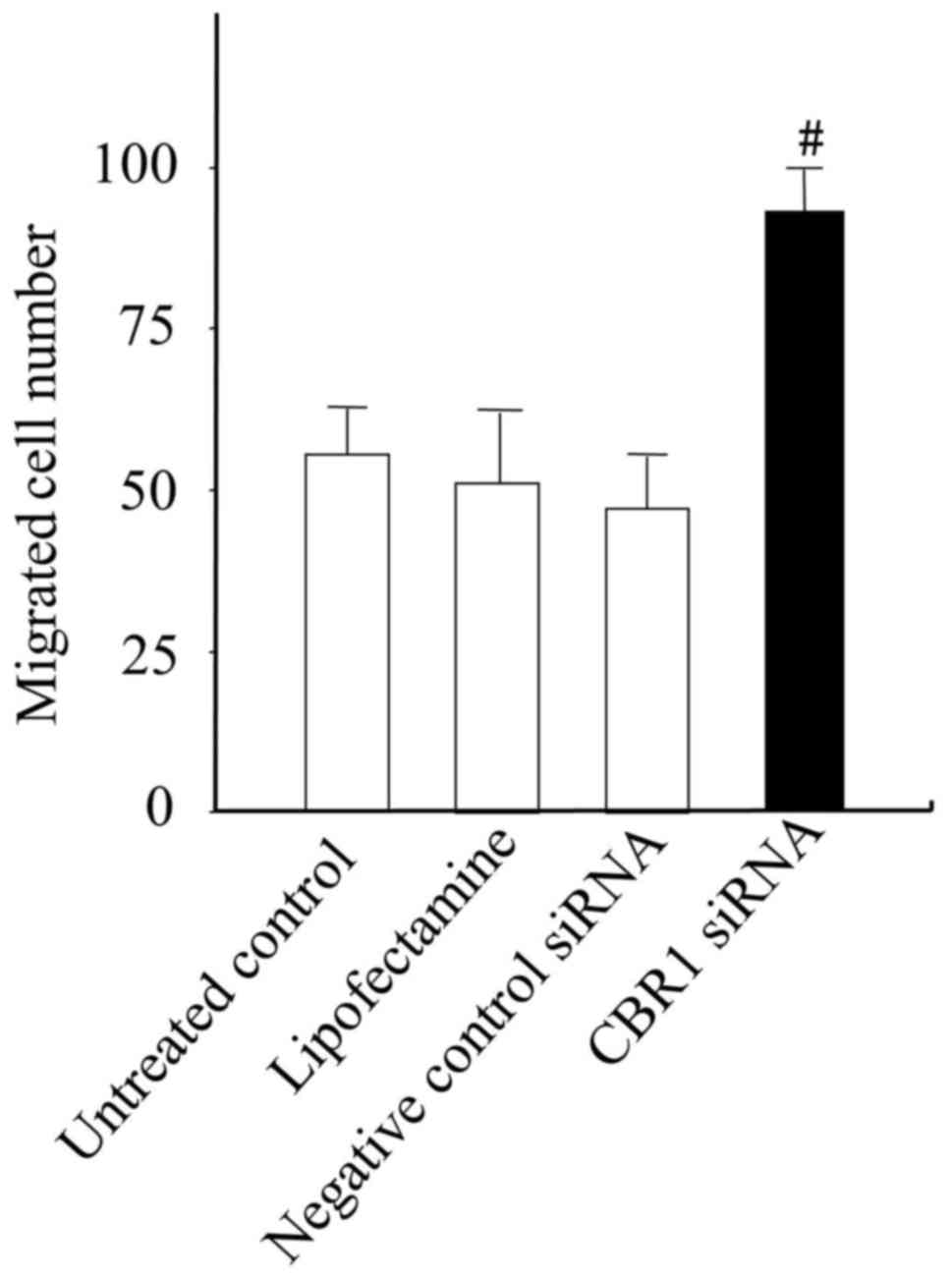
cell wound healing and migration ability
Low expression of CBR1 is associated with cancer
progression and metastasis. Therefore, a wound-healing assay was
performed to examine the migration capabilities of the CBR1 siRNA
and negative control siRNA cells. The migration activity of the
transfected cells was also measured using a Boyden chamber assay.
CBR1 siRNA cells exhibited higher wound healing and migration
abilities compared with the untreated control, Lipofectamine and
negative control siRNA cells (Figs.
6 and 7).
Discussion
In this study, it was demonstrated that decreased
CBR1 expression is associated with a poor prognosis in OSCC, and
therefore could be a useful prognostic factor for patients with
OSCC. According to the findings, the usefulness of reduced CBR1
expression as a prognostic factor is almost equal to that of lymph
node positive (N+) factor (Table
III). Murakami et al (13) reported that reduced CBR1 expression
could be a more useful predictive factor for poor prognosis than
other pathological risk factors in patients with endometrial
cancer. Murakami et al (13)
also reported that decreased CBR1 expression was significantly
associated with pelvic lymph node metastasis and a poor prognosis
in uterine cervical cancer (9). In
addition, Osawa et al (15)
and Umemoto et al (10)
reported that decreased CBR1 expression was correlated with growth,
proliferation, lymph node metastasis and poor prognosis in ovarian
cancer (15,10). These studies suggest that using
reduced CBR1 expression as a prognostic factor may be beneficial in
gynecological cancer cases.
Similarly, it is possible that reduced CBR1
expression is important for OSCC as a prognostic factor as,
histopathologically, the majority of gynecological cancer cases are
squamous cell carcinoma (16).
However, Miura et al (17)
reported that CBR1 inhibits growth of ovarian cancer via tumor
necrosis factor receptor signaling. Therefore, the role of CBR1
could vary between case studies of same type of cancer.
The present data also suggest that the
downregulation of CBR1 expression serves an important role in the
progression of OSCC cells, as the suppression of CBR1 expression
increased cancer cell proliferation, wound healing and migration
abilities (Figs. 5–7). Furthermore, it was observed that the
expression of E-cadherin was decreased, and the secretion of matrix
metalloproteinases (MMPs) was increased in cells with low CBR1
expression (data not presented). Reduced CBR1 expression may be
associated with cervical lymph node metastasis and advanced stage
of disease through the aforementioned mechanisms, as the loss of
E-cadherin and the activation of MMPs induce cancer cell invasion
and lymph node metastasis (18).
Notably, Murakami et al (9)
also reported that MMP activities are increased by CBR1 suppression
through an increase of cytochrome c oxidase subunit 2
(COX-2) in uterine squamous cell carcinoma cells. These reports
support the findings that indicated the clinical significance of
reduced CBR1 expression in OSCC.
It may be assumed that low expression or
downregulation of CBR1 may correlate with malignant behavior in
OSCC cells, consistent with the clinical data obtained (Fig. 2; Tables
II and III). CBR1 expression
is higher in cancer cells than in normal cells. However, CBR1
expression may be gradually reduced via the acquisition of
malignant behaviors, including EMT, angiogenesis, and invasive and
metastatic capacity (8,13). EMT serves a significant role in tumor
progression, invasion and metastasis, and E-cadherin loss is
crucial to the EMT process (19).
Decreased CBR1 expression is associated with lower E-cadherin
expression and the promotion of EMT, which causes poor prognosis in
endometrial cancer (9,13). However, CBR1 expression levels in
different OSCC cell lines frequently used for in vitro
studies may not always correlate with the aggressiveness or
invasive nature of the cells. In certain prior reports, HSC3 cells
were characterized by poor differentiation and a high level of
aggressiveness, whereas HSC2 cells were reported to have a high
rate of differentiation and low invasive potential; HSC4 cells are
reported to have a moderate differentiation rate with a low
metastatic potential (20,21). However, in the present study, high
expression of CBR1 protein was observed in HSC3 cells, while HSC2
cells exhibited moderate expression and HSC4 cells exhibited low
CBR1 protein expression (Fig. 3).
Therefore, further investigations with OSCC cells are required to
verify whether low CBR1 expression promotes tumor progression and
metastasis.
In future studies, to clarify the importance of CBR1
expression in OSCC, the authors intend to perform further siRNA
studies using different OSCC cell lines, in addition to evaluating
the growth, proliferation, migration and invasion capability of the
transfected OSCC cell lines using more sensitive experiments, such
as a bromodeoxyuridine assay, colony formation assay and a Boyden
chamber assay. It is also necessary to carry out mouse xenograft
experiments with these transfected cell lines in order to
understand the effects of low CBR1 expression in vivo in the
progression of tumors.
Based on the data, it is concluded that OSCC cells
may frequently exhibit low levels of CBR1 expression, which may
serve an important role in enhancing the metastatic potential of
OSCC and disease progression. The data indicate that CBR1
expression may be useful for predicting the prognosis of OSCC.
Immunohistochemical analyses of CBR1 expression may assist in
determining whether adjuvant treatment is required or not.
Furthermore, the present study provides a novel insight into the
mechanisms involved in the malignant behavior of OSCC cells, and
also suggests that CBR1 could be a novel target molecule to
regulate cancer cell invasion and metastasis by molecular targeting
therapy.
Acknowledgements
The present study was supported in part by a
Grant-in-Aid Scientific Research (grant no. 15K11292) from the
Ministry of Education, Culture, Sports, Science and Technology of
Japan.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
http://ganjoho.jp/reg_stat/statistics/brochure/backnumber/2014_jp.htmlCancer
statistics in Japan-2014. March 14–2016
|
|
2
|
National Comprehensive Cancer Network, .
NCCN clinical practice guidelines in oncology: Head and neck
cancers. Version 1. 2012.http://www.nccn.org/clinical.aspMarch
14–2016
|
|
3
|
Inagi K, Takahashi H, Okamoto MA, Nakayama
M, Makoshi T and Nagai H: Treatment effects in patients with
squamous cell carcinoma of the oral cavity. Acta Oto-Laryngol.
122:25–29. 2002. View Article : Google Scholar
|
|
4
|
Penning TM and Drury JE: Human aldo-keto
reductases: Function, gene regulation, and single nucleotide
polymorphisms. Arch Biochem Biophys. 464:241–250. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mindnich RD and Pennning TM: Aldo-Keto
Reductase (Akr) superfamily: Genomics and annotation. Hum Genomics.
3:362–370. 2009.PubMed/NCBI
|
|
6
|
Miura T, Nishinaka T and Terada T:
Different functions between human monomeric carbonyl reductase 3
and carbonyl reductase 1. Mol Cell Biochem. 315:113–121. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gonzales-Covarrubias V, Ghosh D, Lakhman
SS, Pendyala L and Blanco JG: A functional genetic polymorphism on
human carbonyl reductase 1 (CBR1 V88I) impacts on catalytic
activity and NADPH binding affinity. Drug Metab Dispos. 35:973–980.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ismail E, Al-Mulla F, Tsuchida S, Suto K,
Motley P, Harrison PR and Birnie GD: Carbonyl reductase: A novel
metastasis-modulating function. Cancer Res. 60:1173–1176.
2000.PubMed/NCBI
|
|
9
|
Murakami A, Fukushima C, Yoshidomi K,
Sueoka K, Nawata S, Yokoyama Y, Tsuchida S, Ismail E, Al-Mulla F
and Sugino N: Suppression of carbonyl reductase expression enhances
malignant behaviour in uterine cervical squamous cell carcinoma:
Carbonyl reductase predicts prognosis and lymph node metastasis.
Cancer Lett. 311:77–84. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Umemoto M, Yokoyama Y, Sato S, Tsuchida S,
Al-Mulla F and Saito Y: Carbonyl reductase as a significant
predictor of survival and lymph node metastasis in epithelial
ovarian cancer. Brit J Cancer. 85:1032–1036. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Leslie SH and Christian W: TNM
Classification of Malignant Tumours. 6th. International union
against cancer (UICC). Wiley-Blackwell; pp. 2642002
|
|
12
|
Pindborg JJ, Reichart PA, Smith CJ and van
der Waal I: Histological typing of cancer and precancer of the oral
mucosa. 2nd. World health organization (WHO). Springer-verlag; pp.
871997
|
|
13
|
Murakami A, Yakabe K, Yoshidomi K, Sueoka
K, Nawata S, Yokoyama Y, Tsuchida S, Al-Mulla F and Sugino N:
Decreased carbonyl reductase 1 expression promotes malignant
behaviours by induction of epithelial mesenchymal transition and
its clinical significance. Cancer Lett. 323:69–76. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Harada K, Ferdous T and Ueyama Y:
Establishment of 5-fluorouracil-resistant oral squamous cell
carcinoma cell lines with epithelial to mesenchymal transition
changes. Int J Oncol. 44:1302–1308. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Osawa Y, Yokoyama Y, Shigeto T, Futagami M
and Mizunuma H: Decreased expression of carbonyl reductase 1
promotes ovarian cancer growth and proliferation. Int J Oncol.
46:1252–1258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cervical Cancer Treatment (PDQ®)-Health
Professional Version, . National Cancer Institute. https://www.cancer.gov/types/cervical/hp/cervical-treatment-pdq#section/August
31–2016
|
|
17
|
Miura R, Yokoyama Y, Shigeto T, Futagami M
and Mizunuma H: Inhibitory effect of carbonyl reductase 1 on
ovarian cancer growth via tumor necrosis factor receptor signaling.
Int J Oncol. 47:2173–2180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schipper JH, Frixen UH, Behrens J, Unger
A, Jahnke K and Birchmeier W: E-cadherin expression in squamous
cell carcinomas of head and neck: Inverse correlation with tumor
dedifferentiation and lymph node metastasis. Cancer Res.
51:6328–6337. 1991.PubMed/NCBI
|
|
19
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Momose F, Araida T, Negishi A, Ichijo H,
Shioda S and Sasaki S: Variant sublines with different metastatic
potentials selected in nude mice from human oral squamous cell
carcinomas. J Oral Pathol Med. 18:391–395. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Takahashi H, Shigeta T, Umeda M and Komori
T: A new in vitro invasion model for oral cancer using an acellular
allogenic dermal matrix (Alloderm): The relationship among in vitro
invasion activity, in vivo invasion and metastasis. Kobe J Med Sci.
57:E128–E136. 2012.PubMed/NCBI
|