Introduction
Breast cancer accounts for 25.2% of all female
cancers worldwide. It has the highest incidence among female
cancers (1), displaying a rapidly
increasing trend. Among imaging medical diagnostic methods used for
early diagnosis of breast cancer, mammography is the only
clinically proven test method (2).
However, mammography has problems such as false negative diagnosis
and excessive additional examination according to false positive
diagnosis, unnecessary biopsy, psychological burden and radiation
exposure. The sensitivity of mammography ranges from 62.2 to 89.5%
with a specificity of 62.7%. More women have dense breasts in Korea
than in western countries. The sensitivity of mammography is
significantly lower for dense breasts. Thus, the need for auxiliary
test is being emphasized (3).
Ultrasonography of the breast is mainly used as an auxiliary test
for mammography. However, breast ultrasound used as an auxiliary
test has the following problems. First, it has a high dependence on
test equipment and testers. Second, scientific and objective
evidence for the mortality rate from breast cancer has not been
established yet. Third, false positives cannot be avoided, and
additional tests and biopsy tests are required. Fourth, early
diagnosis of breast cancer is difficult due to calcified lesions
(4). Therefore, it is necessary to
develop a new and auxiliary test for diagnosing breast cancer in
order to solve these limitations of mammography.
Invasive biopsy has disadvantages in that the
patient suffers severely. It also has side effects due to infection
that might require a recovery period after hospitalization and
examination. On the other hand, a non-invasive method using blood
(blood collection) has the advantages of being very simple and
painless. It requires no recovery period after hospitalization and
examination. In addition, such liquid biopsy has the advantage of
avoiding side effects of tissue biopsy. Early diagnosis is possible
even for potential patients who have not developed cancer. It is
advantageous to periodically observe the progress of treatment of
patients who have already developed the disease (5-7).
Therefore, screening high-risk groups of patients for breast cancer
using liquid biopsy and the development of early diagnosis methods
can compensate for problems of existing tissue biopsy and greatly
contribute to the reduction of medical expenses.
Exosome is a group of small membranous vesicles that
are shed into body fluids or extracellular environment by tumoral
or nontumoral cells. These vesicles play pivotal roles in cellular
communication through shuttling between donor and recipient cells
(8). The lumen of an exosome has
different components such as DNA, RNA, lipids and proteins,
representing bioactive molecules in donor cells. miRNAs are among
cargos of exosomes. They are involved in different processes such
as angiogenesis and metastasis of cancer (9). Exosomes are nano-vesicles present in
the circulation. They are involved in cell-to-cell communication
and the regulation of different biological processes. miRNAs as
cargos of exosomes are potential biomarkers (10). Due to interesting features of
exosomal miRNAs, they can be promising biomarkers for cancer
diagnosis (11). Due to the
presence of exosomes in various body fluids and the stability of
miRNAs in exosomes, exosomal miRNAs might be a new class of
biomarkers for early and minimally invasive cancer diagnosis
(12).
Recently, the emergence of miRNA, a small
non-protein-coding RNA that plays an important role in tumor
initiation and progression, has opened up new opportunities for
early cancer diagnosis (13,14).
miRNAs are 19-25 nucleotides regulatory non-coding RNA molecules
that regulate expression levels of a wide variety of genes by
sequence-specific base pairing for 39 untranslated regions of
target mRNA, resulting in mRNA degradation or inhibition of
translation. Evidence suggests that miRNA expression profiles can
cluster similar tumor types together more accurately than
expression profiles of protein-coding mRNA genes (15). Furthermore, miRNA expression
signatures have been used to predict prognosis (16,17).
As a screening tool that is easily accepted by the general
population, it would be desirable to detect cancer accurately,
without resorting to an invasive procedure. Recently, several
reports have suggested that circulating miRNAs are stable and
detectable in serum/plasma and that levels of some miRNAs in breast
cancer patients are specifically elevated (18,19).
Canonically, biogenesis of miRNAs starts in cell
nucleus where DNA containing miRNAs is transcribed by RNA
polymerase II to generate primary miRNAs (pri-miRNAs) (20). These pri-miRNAs are then processed
by a microprocessor complex consisting of RNase type III
endonuclease Drosha and an essential cofactor (DiGeorge syndrome
critical region 8)/Pasha (protein containing two double-stranded
RNA binding domains) to generate precursor miRNA (pre-miRNAs)
(21,22). Pre-miRNAs are then exported to the
cytoplasm by an exchange factor of guanine Ran nucleotide
(GTP-binding nuclear protein Ran) and an exportine-5 receptor. They
are then processed by another RNase type III endonuclease known as
Dicer, releasing ~22-nucleotide miRNA duplex. One strand of the RNA
duplex is selected to be subsequently loaded into the RNA-induced
silencing complex (RISC), along with argonaut (AGO2) and GW182
(23,24). Whether incorporation of miRNA into
exosomes occurs at pre-miRNA or mature miRNA level remains unclear.
However, it has been reported that precursor miRNA contains a
higher ratio of mature miRNA (25).
Based on years of research experience comparing mature miRNA and
pre-miRNA expression in the same sample from our team (data not
shown), the aim of this study was to analyse pre-miRNA expression,
rather than mature miRNA, as a biomarker for early diagnosis of
breast cancer.
To obtain accurate results in real-time PCR, it is
important to accurately combine templates and primers.
Dumbbell-like structural primer for pre-miRNA amplification is our
team's original technique that can minimise real-time PCR
nonspecific reactions (26). Thus,
it was used in the present study. In this study, we tried to
construct a set of multiple pre-miRNA biomarkers that are optimal
for developing new blood-based early diagnostic assays for breast
cancer.
Materials and methods
Cohorts and plasma samples
In this study, 226 breast cancer patients and 146
healthy control plasma were used. Specifically, 146 breast cancer
patients and 90 healthy control serum were used in the initial
discovery study. Then 80 breast cancer patients and 56 healthy
control plasma were used to validate the classification model of
this study. Plasma samples from breast cancer patients with cancers
were obtained from Korea Regional Biobank of Busan National
University Hospital, Inje University Busan Paik Hospital and
Chonnam National University Hwasun Hospital. These breast cancer
samples were obtained before any therapeutic approaches were
performed. This study was ethically approved by our Institutional
review board from the IRB (BIOINFRA Life Science Institutional
Review Board). The member was of this review board Jung Bo Kyung,
Kim Chul Woo, Shin Yong Sung, and Kim Hee Yoon. Samples were stored
at -80˚C until analysed. Plasma samples from asymptomatic healthy
donors were obtained from Korea Regional Biobank of Ajou University
Hospital, Wonkwang University Hospital, Jeonbuk National University
Hospital, Chonnam National University Hospital and Kyungpook
National University Hospital. Healthy controls with a known history
of cancer, high-grade dysplasia, autoimmune disease, chronic kidney
disease, pregnancy, or inflammatory conditions that needed medical
management were excluded. The clinical stage of cancer was
determined by the final pathological diagnosis after resection
according to the 7th edition of the Union for International Cancer
Control tumor-node-metastasis classification.
Isolation of RNA from plasma
Total RNAs were extracted from plasma samples (300
µl) using a nucleic acid automatic extraction equipment (Smart Lab
Assist-24, Korea KETT) and finally eluted with 150 µl RNase-free
water. Concentration of extracted total RNAs were measured.
Measured concentrations were analysed to correct concentration
values of all samples.
Removal of genomic DNA and cDNA
synthesis by reverse transcription (RT)
The gDNA was removed from the extracted total RNA.
Total RNA was then used to synthesise cDNA which was then
quantified using an internal control primer. gDNA removal and RT
were performed using PrimeScript™ RT reagent kit with gDNA Eraser
(product code RR047A, Takara).
Analysis of miRNA gene expression by
quantitative real time polymerase chain reaction (qPCR)
In the first multiplex PCR, a total of nine miRNA
primers (2.5-30 pmol) (1 µl) were mixed with 4 µl of template cDNA
and 5 µl of 2X multiplex PCR master mix. Primer concentrations for
each miRNA for secondary real-time PCR were in the range 4-10 pmol.
The primary PCR product was diluted 1:10 and used for secondary
real-time PCR analysis. One µl of each primer, 4 µl of the primary
PCR product and 5 µl of 2X SYBR master mix were mixed to make a
final volume of 10 µl for PCR. Primer sequences used for PCR are
shown below. X in primer sequence is inosine. miR-223 (NR_029637.1)
forward primer 5'-GACCAXXXXXAGTTGGACACTCCATGTGGTC-3' and reverse
primer 5'-AGTGCXXXXXTGGTAAGCATGTGCCGCACT-3'; miR-1246 (NR_031648.1)
forward primer 5'-CAGGTXXXXXTGGAGCAGGAGTGGACACCTG-3' and reverse
primer 5'-CAATCXXXXXATTGCTAGCCTATGGATTG-3'; miR-206 (NR_029713.1)
forward primer 5'-AGCATXXXXXTGCTTCCCGAGGCCACATGCT-3' and reverse
primer 5'-AAGTGXXXXXACTTGCCGAAACCACACACTT-3'; miR-24 (NR_029497.1)
forward primer 5'-CTGTGXXXXXGTGCCTACTGAGCTGAAACACAG-3' and reverse
primer 5'-CACTGXXXXXGTTCCTGCTGAACTGAGCCAGTG-3'; miR-373
(NR_029866.1) forward primer 5'-CAGACXXXXXCGCTTTCCTTTTTGTCTG-3' and
reverse primer 5'-GTGCTXXXXXGACACCCCAAAATCGAAGCAC-3'; miR-21
(NR_029493.1) forward primer 5'-CAGTCXXXXXGTCGGGTAGCTTATCAGACTG-3'
and reverse primer 5'-CAGTCXXXXXCAGACAGCCCATCGACTG-3'; miR-6875
(NR_106935.1) forward primer 5'-CTTCTXXXXXGACCCAGGACAGGAGAAG-3' and
reverse primer 5'-GTGATXXXXXGCAGGAAGAATGCAAATCAC-3'; miR-202
(NR_030170.1) forward primer
5'-GGCCAXXXXXGCATATACTTCTTTGAGGATCTGGCC-3' and reverse primer
5'-CATGGXXXXXGACCGCCCCGTTTTCCCATG-3'; miR-219B (NR_039815.1)
forward primer 5'-ACATCXXXXXGGAGCTCAGCCACAGATGT-3' and reverse
primer 5'-GTTTGXXXXXGCGCCACTGATTGTCCAAAC-3'.
Statistical methods
Mann-Whitney U test (Wilcoxon rank-sum test) was
used to determine whether these nine candidate microRNA markers
might be significantly different between breast cancer patients and
healthy controls. P<0.01 was considered to indicate a
statistically significant difference. Spearman correlation test
between markers was also performed to evaluate the independence of
candidate markers. Next, to minimise the influence of outliers in
biomarker measurement values, a log10 transformation was
performed for measurement values. This study included 226 total
breast cancer samples and 146 healthy control samples. To select an
optimal marker panel, a training data set (146 breast cancer and 90
healthy controls) and a validation data set (80 breast cancer and
56 healthy controls) were used. To select the optimal marker panel,
511 biomarker panel sets were generated. This number was the number
of all possible combinations for these nine candidate biomarkers.
For the training data set, a classification model corresponding to
511 biomarker panel sets was generated using Random Forest (RF)
algorithm, one non-linear classification technique. The
classification model was then verified using the validation set.
Criteria for selecting the optimal marker panel should be excellent
in performance such that the area-under-the-curve (AUC) of the
receiver operation characteristic (ROC) calculated at model
generation is close to 1. It was confirmed that the performance was
similar for the validation set. All analysis procedures were
performed using R statistical package version 3.5.1 (https://www.r-project.org/ and https://ftp.harukasan.org/CRAN/index.html), a
statistical analysis tool.
Results
Performance value of each candidate
miRNA biomarkers in the training set
As summarised in Table
I, nine candidate miRNAs were selected based on reference
search (27-40).
First, expression levels of these nine candidate miRNAs (miR-223,
1246, 206, 24, 373, 21, 6875, 202 and 219B) predicted to be
important for screening normal and breast cancer patients were
analysed. Samples used in the analysis included 146 healthy
controls (31 from the Korea Regional Biobank of Ajou University
Hospital, 25 from Wonkwang University Hospital, 16 from Jeonbuk
National University Hospital, 23 from Chungnam National University
Hospital, 27 from Kyungpook National University Hospital and 24
from Ajou University Hospital), together with 226 breast cancer
patients (147 from the Korea Regional Biobank of Busan National
University Hospital, 39 from Inje University Busan Paik Hospital
and 40 from Chonnam National University Hwasun Hospital). At this
time, all normal plasma samples were female samples. The expression
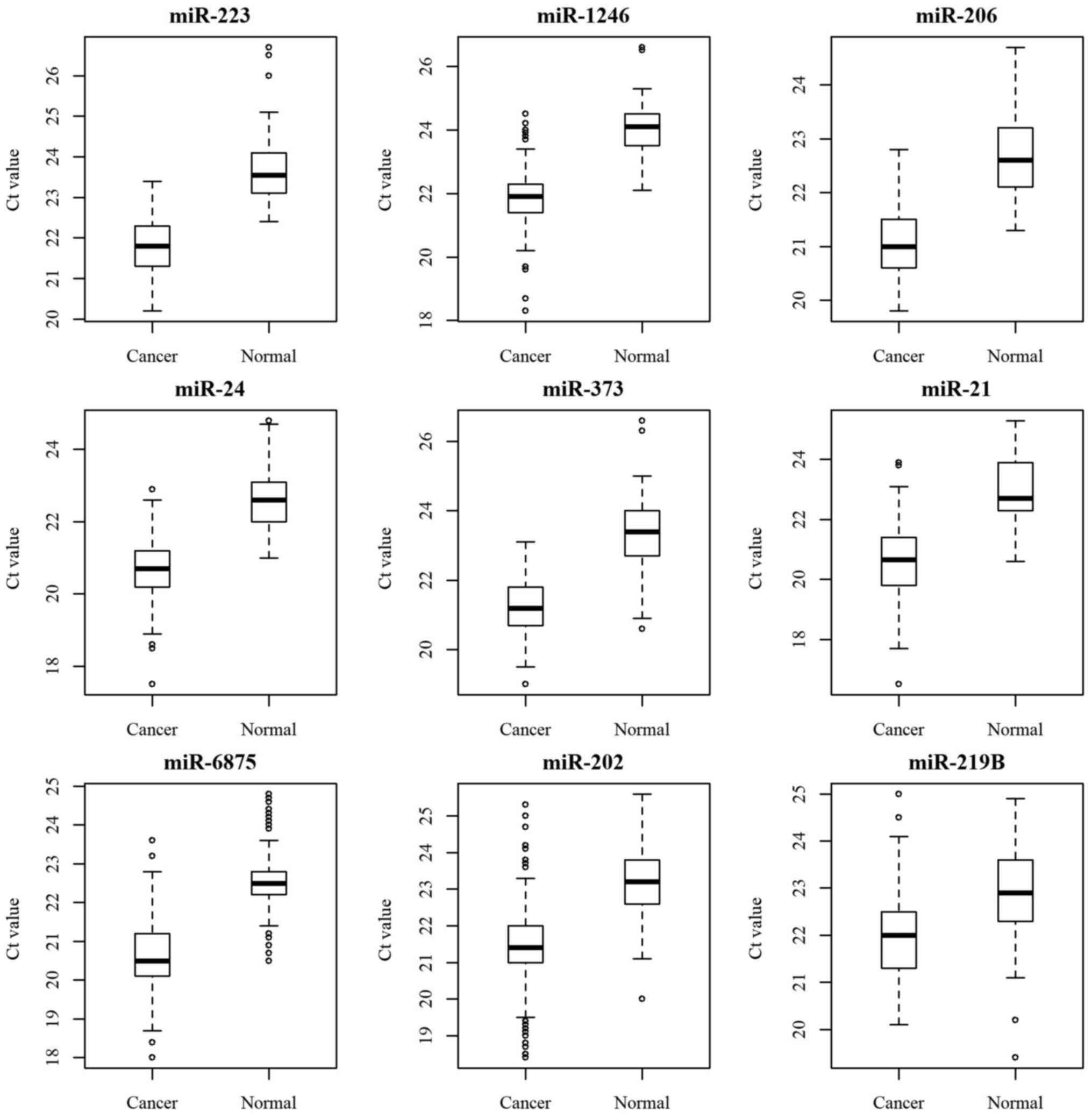
level of each miRNA was a cross point between a miRNA amplification
curve and a threshold line in a threshold cycle (Ct), meaning a
relative measurement of the target miRNA concentration in a
real-time PCR reaction. All nine candidate miRNAs were found to be
meaningful for distinguishing between normal and breast cancer
patients (Table II). This was also
confirmed in a bar graph boxplot (Fig.
1).
 | Table IPreclinical study report of nine
candidate miRNAs. |
Table I
Preclinical study report of nine
candidate miRNAs.
| Reference name and
year | Biomarker | Function | Refs. |
|---|
| Pinatel et
al 2014; Yoshikawa et al 2018 | miR-223 | miR-223 is a
coordinator of breast cancer progression; The expression level is
higher in the patients IDC than with DCIS | (27,28) |
| Fu et al
2016; Li et al 2017 | miR-1246 | miR-382-3p,
-598-3p, -1246, and - 184 are all involved in the development of
breast cancer, and are promising biomarkers for breast cancer
detection; Exosomal microRNA miR-1246 promotes cell proliferation,
invasion and drug resistance by targeting CCNG2 in breast
cancer | (29,30) |
| Zhou et al
2019 | miR-206 | miR-206 promotes
cancer progression by targeting full-length Neurokinin-1 receptor
in breast cancer | (31) |
| Khodadadi-Jamayran
et al 2018; Lu et al 2015 | miR-24 | Prognostic role of
elevated mir-24-3p in breast cancer and its association with the
metastatic process; miRNA-24-3p promotes cell proliferation and
inhibits apoptosis in human breast cancer by targeting p27Kip1 | (32,33) |
| Eichelser et
al 2013; Piasecka et al 2018 | miR-373 | miR-373 is known to
be relevant for cancer development, progression, and metastasis;
mR-373 is associated with EMT/CSC and invasion | (34,35) |
| Asaga et al
2011; Corcoran et al 2011 | miR-21 | Circulating miR-21
has diagnostic and prognostic potential in breast cancer | (36,37) |
| Shimomura et
al 2016 | miR-6875 | A combination of
miR-1246, miR-1307-3p, miR-4634, miR-6861-5p, and miR-6875-5p
measured from serum can be used to detect breast cancer in the
early stages, and to differentiate breast cancer from
pancreas/biliary tract/prostate benign diseases or other
cancers | (38) |
| Schrauder et
al 2017 | miR-202 | miR-202 was
significantly upregulated in whole blood samples of early-stage
breast cancer patients | (39) |
| Zhao et al
2017 | miR-219B | Gga-miR-219b
targeting BCL11B suppresses proliferation, migration, and invasion
of Marek's disease tumor cell MSB1 | (40) |
 | Table IIResult of the U-test analysis of
single miRNA biomarkers. |
Table II
Result of the U-test analysis of
single miRNA biomarkers.
| Rank | miRNA
biomarker | P-value |
|---|
| 1 | miR-223 |
1.28x10-53 |
| 2 | miR-1246 |
1.47x10-53 |
| 3 | miR-206 |
1.17x10-49 |
| 4 | miR-24 |
1.22x10-49 |
| 5 | miR-373 |
1.58x10-49 |
| 6 | miR-21 |
1.53x10-46 |
| 7 | miR-6875 |
2.24x10-43 |
| 8 | miR-202 |
1.73x10-39 |
| 9 | miR-219B |
4.99x10-23 |
Correlation analysis for nine miRNA
biomarkers
Results of correlation analysis for these nine miRNA
biomarkers (miR-223, 1246, 206, 24, 373, 21, 6875, 202 and 219B)
are shown in Table III.
Correlations among nine miRNA biomarkers were analysed using
Spearman's correlation analysis. The degree of correlation was
generally expressed as follows: Correlation coefficient R=1, same;
R≥0.9, very high correlation; 0.7≤R<0.9, high correlation;
0.4≤R<0.7, slightly higher correlation; 0.2≤R<0.4, low
correlation; and R≤0.2, little correlation. No miRNA had a very
high or high correlation. It was confirmed that correlations of
miR-223 with miR-373 and miR-202 were slightly higher. On the other
hand, miR-373 had a somewhat higher correlation with miR-223 and
miR-206. These results show that it is possible to select miRNAs
that are not highly correlated when selecting an optimal biomarker
combination. However, when each miRNA plays an important function
in breast cancer onset and progression, these miRNAs should be
included in the combination set, even if there is a rather high
correlation.
 | Table IIIAnalysis of correlation between the
nine miRNAs. |
Table III
Analysis of correlation between the
nine miRNAs.
| Correlation | miR-223 | miR-1246 | miR-206 | miR-24 | miR-373 | miR-21 | miR-6875 | miR-202 | miR-219B |
|---|
| miR-223 | 1.00 | 0.28 | 0.45 | 0.12 | 0.60 | 0.27 | 0.38 | 0.59 | 0.13 |
| miR-1246 | 0.28 | 1.00 | 0.14 | 0.29 | 0.38 | 0.36 | 0.19 | 0.43 | 0.13 |
| miR-206 | 0.45 | 0.14 | 1.00 | 0.34 | 0.56 | 0.17 | 0.41 | 0.23 | 0.07 |
| miR-24 | 0.12 | 0.29 | 0.34 | 1.00 | 0.27 | 0.14 | 0.28 | 0.16 | 0.00 |
| miR-373 | 0.60 | 0.38 | 0.56 | 0.27 | 1.00 | 0.32 | 0.41 | 0.51 | 0.12 |
| miR-21 | 0.27 | 0.36 | 0.17 | 0.14 | 0.32 | 1.00 | 0.17 | 0.36 | 0.34 |
| miR-6875 | 0.38 | 0.19 | 0.41 | 0.28 | 0.41 | 0.17 | 1.00 | 0.20 | 0.15 |
| miR-202 | 0.59 | 0.43 | 0.23 | 0.16 | 0.51 | 0.36 | 0.20 | 1.00 | 0.01 |
| miR-219B | 0.13 | 0.13 | 0.07 | 0.00 | 0.12 | 0.34 | 0.15 | 0.01 | 1.00 |
Combination of multiple miRNA
biomarkers for breast cancer diagnosis
To create a classification model, total samples were
divided into samples for model generation (training set) and
samples for model verification (validation set). Data for model
generation and verification were distributed at a ratio of ~2:1
(training: Validation set) (Fig. 2
and Table IV). Samples for model
generation and verification were randomised. At this time, age
information was not reflected. Table
IV shows the accuracy of a single miRNA in training and
validation sets. Table V shows
examples of a representative set of multiple miRNA biomarkers that
meet all rules. Tables IV and
V confirmed that the performance of
multiple biomarkers was improved compared to that of a single
biomarker. In addition to examples given in Tables V and SI-III
of Supplementary Information (SI) shows examples of two to four
combinations of miRNA biomarkers out of 511 biomarker panel sets
that met all rules. Results showed that multiple markers had higher
accuracy for early diagnosis of breast cancer than single
markers.
 | Table IVAUC values of a single biomarker of
breast cancer using statistical methods in the training and
validation sets. |
Table IV
AUC values of a single biomarker of
breast cancer using statistical methods in the training and
validation sets.
| Index | Biomarker | Training set
AUC | Validation set
AUC |
|---|
| 1 | miR-223 | 0.963 | 0.958 |
| 2 | miR-1246 | 0.954 | 0.962 |
| 3 | miR-206 | 0.932 | 0.935 |
| 4 | miR-24 | 0.942 | 0.962 |
| 5 | miR-373 | 0.914 | 0.933 |
| 6 | miR-21 | 0.912 | 0.922 |
| 7 | miR-6875 | 0.904 | 0.880 |
| 8 | miR-202 | 0.892 | 0.859 |
| 9 | miR-219B | 0.786 | 0.809 |
 | Table VPerformance of classification models
in the training and validation sets. |
Table V
Performance of classification models
in the training and validation sets.
| A, Training
set |
|---|
| Example of
combination | Values |
|---|
| Biomarkers | AUC | Accuracy (%) | Specificity
(%) | Sensitivity
(%) | Early stage (stage
0-2) Sensitivity (%) | Late stage (stage
3-4) Sensitivity (%) |
|---|
| miR-1246 | 0.955 | 88.0 | 93.0 | 85.0 | 83.0 | 95.0 |
| miR-206 | 0.932 | 80.0 | 93.0 | 71.0 | 67.0 | 100.0 |
| miR-24 | 0.938 | 79.0 | 93.0 | 70.0 | 66.0 | 95.0 |
| miR-373 | 0.914 | 73.0 | 93.0 | 60.0 | 56.0 | 89.0 |
|
miR-1246+miR-206 | 0.979 | 91.0 | 93.0 | 89.0 | 87.0 | 100.0 |
|
miR-1246+miR-24 | 0.984 | 94.0 | 93.0 | 94.0 | 94.0 | 95.0 |
|
miR-1246+miR-373 | 0.968 | 94.0 | 93.0 | 94.0 | 93.0 | 100.0 |
| miR-206+miR-24 | 0.976 | 90.0 | 92.0 | 88.0 | 87.0 | 95.0 |
|
miR-206+miR-373 | 0.964 | 87.0 | 93.0 | 83.0 | 80.0 | 100.0 |
| miR-24+miR-373 | 0.982 | 92.0 | 93.0 | 92.0 | 91.0 | 95.0 |
|
miR-1246+miR-206+miR-24 | 1.000 | 97.0 | 93.0 | 100.0 | 100.0 | 100.0 |
|
miR-1246+miR-206+miR-373 | 0.985 | 94.0 | 93.0 | 95.0 | 94.0 | 100.0 |
|
miR-1246+miR-24+miR-373 | 0.990 | 94.0 | 93.0 | 95.0 | 95.0 | 95.0 |
|
miR-206+miR-24+miR-373 | 0.989 | 92.0 | 93.0 | 90.0 | 90.0 | 95.0 |
|
miR-1246+miR-206+miR-24+miR-373 | 0.993 | 96.0 | 93.0 | 97.0 | 97.0 | 100.0 |
| B, Validation
set |
| Example of
combination | Values |
| Biomarkers | AUC | Accuracy (%) | Specificity
(%) | Sensitivity
(%) | Early stage (stage
0-2) Sensitivity (%) | Late stage (stage
3-4) Sensitivity (%) |
| miR-1246 | 0.963 | 90.0 | 96.0 | 86.0 | 84.0 | 100.0 |
| miR-206 | 0.935 | 86.0 | 96.0 | 79.0 | 75.0 | 100.0 |
| miR-24 | 0.965 | 81.0 | 96.0 | 70.0 | 65.0 | 100.0 |
| miR-373 | 0.935 | 73.0 | 95.0 | 57.0 | 53.0 | 83.0 |
|
miR-1246+miR-206 | 0.988 | 96.0 | 98.0 | 95.0 | 94.0 | 100.0 |
|
miR-1246+miR-24 | 0.987 | 96.0 | 96.0 | 96.0 | 96.0 | 100.0 |
|
miR-1246+miR-373 | 0.983 | 93.0 | 95.0 | 92.0 | 91.0 | 100.0 |
| miR-206+miR-24 | 0.973 | 91.0 | 98.0 | 86.0 | 84.0 | 100.0 |
|
miR-206+miR-373 | 0.981 | 91.0 | 98.0 | 86.0 | 84.0 | 100.0 |
| miR-24+miR-373 | 0.977 | 96.0 | 95.0 | 96.0 | 96.0 | 100.0 |
|
miR-1246+miR-206+miR-24 | 0.977 | 93.0 | 86.0 | 98.0 | 97.0 | 100.0 |
|
miR-1246+miR-206+miR-373 | 0.991 | 96.0 | 95.0 | 96.0 | 96.0 | 100.0 |
|
miR-1246+miR-24+miR-373 | 0.989 | 97.0 | 96.0 | 98.0 | 97.0 | 100.0 |
|
miR-206+miR-24+miR-373 | 0.987 | 93.0 | 95.0 | 91.0 | 90.0 | 100.0 |
|
miR-1246+miR-206+miR-24+miR-373 | 0.992 | 97.0 | 96.0 | 98.0 | 97.0 | 100.0 |
Discussion
To improve the performance of breast cancer
treatment, discovery and accurate diagnosis are very important.
Representative imaging methods used for breast examination are
mammography, breast ultrasound and breast magnetic resonance
imaging (MRI). Mammography is the effective diagnostic tool and
screening test for breast cancer and has been scientifically proven
to be able to lower breast cancer and mortality. However,
mammography can lead to false negative diagnosis and false positive
diagnosis, resulting in excessive additional examination and
unnecessary biopsy with disadvantages such as inpatient treatment,
psychological burden and radiation exposure. Moreover, sensitivity
of mammography is lower for young women and women with dense
breasts. Assisted mammography methods include breast ultrasound and
magnetic resonance imaging. However, these methods also have
limitations (41-43).
For this reason, many attempts have been made recently to select
high-risk patients for breast cancer. Liquid biopsy has been used
as an aid to early diagnosis of breast cancer. A liquid biopsy has
the advantage of being non-invasive for early detection of cancer.
It also enables multiple repetitions and easy monitoring of the
disease (44).
Several previous studies have demonstrated the value
of circulating miRNAs in breast cancer diagnosis. A number of new
breast cancer related miRNAs have been identified (45,46).
However, there have not been comprehensive reports of numerous
miRNAs using plasma samples of breast cancer patients. In this
study, we analysed expression levels of a number of plasma miRNAs
expected to be valuable for early diagnosis of breast cancer in an
attempt to obtain an optimal combination of multiple miRNAs.
Dumbbell-like structural primer for pre-miRNA amplification by
real-time PCR as our team's proprietary technology that could
minimise non-specific PCR in real time was used in the present
study.
Results of this study showed AUC values of
0.809-0.962 for the classification model with a single miRNA alone
(Table IV). However, it has been
demonstrated that diagnostic performance can be increased by
combining multiple significant miRNAs under the same conditions.
Therefore, two, three and four combinations of miRNAs among nine
candidate miRNAs were used from comparative analysis in this study
and a diagnostic panel of miRNAs was developed. The performance of
the diagnostic panel was verified using a validation cohort. For
example, a combination of four miRNAs (miR-1246, miR-206, miR-24
and miR-373) showed AUC of 0.992 (Table
V). When choosing a set of biomarker combinations, usually
biomarkers having the highest AUC are selected. However, the AUC
value may change as the number of samples increases. In addition,
the correlation of markers and the function of each marker must be
considered. A large number of biomarkers may not be an optimal set
of biomarker combinations. Many factors need to be considered for
commercialization.
Compared to a previous study (47) as an example, the combination of
plasma exosome miR-1246 and miR-21 (AUC=0.7266) was a better
indicator of breast cancer than individual miRNAs (AUC: 0.6914 and
0.6875, respectively). However, the number of breast cancer patient
samples used in that study (47)
was <16 and the accuracy was lower than that of our study.
Another example was a study (38)
that verified the accuracy of using a combination of miR-1246,
miR-1307-3p, miR-4634, miR-6861-5p and miR-6875-5p measured from
serum for early diagnosis of breast cancer. That combination had a
sensitivity of 97.3%, a specificity of 82.9% and an accuracy of
89.7% for breast cancer in the test cohort. Among these five miRNAs
used in that study (38), miR-1246
and miR-6875 were miRNAs that overlapped with our study. Different
from our study, sera samples of Japanese breast cancer patients
were used and mature miRNA expression was analysed by microarray in
that study (38).
In the present study, considering that results could
be influenced by storage conditions of specimens, our study used
independent clinical trials from multiple institutions. Plasma
samples of breast cancer patients were stored at -80˚C for up to 5
years. Whether such multi-miRNA set developed in this study can
distinguish benign breast disease from breast cancer remains
unknown. In the future, studies should be conducted using samples
of patients with benign breast diseases so that benign breast
diseases can be distinguished from breast cancer.
In conclusion, a set of biomarkers developed in this
study showed high accuracy for early diagnosis of breast cancer.
This set was prepared using a combination of two or more of nine
miRNAs measured in plasma samples. It can be used to detect breast
cancer at an early stage. We hope that these diagnostic indicators
can be implemented to address problems of existing assistive
methods for conventional mammography and effectively detect breast
cancer at an early stage.
Supplementary Material
RF model analysis result including 2
combinations of nine miRNAs.
RF model analysis result including 3
combinations of nine miRNAs.
model analysis result including 4
combinations of nine miRNAs.
Acknowledgements
Not applicable.
Funding
Funding from BIOINFRA Life Science Inc was
received.
Availability of data and materials
Datasets used and/or analysed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
JYJ and YSK conceived and designed the study,
acquired data, directed drafting of the manuscript and revised it.
KNK performed statistical analysis. KHK and YJP managed plasma
samples and performed experiments. CWK conceived and designed the
study and provided financial support. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
This study was ethically approved by the
Institutional review board from the IRB (BIOINFRA Life Science
Institutional Review Board). Plasma samples were provided by Korean
Biobank through established procedures.
Patient consent for publication
Subjects whose plasma samples were used in this
study provided written informed consent for the publication of
their data.
Competing interests
The authors confirm that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386.
2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang L: Early diagnosis of breast cancer.
Sensors. 17(1572)2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Korean Breast Cancer Society. Early
screening of breast cancer in Korea. J Korean Breast Cancer Soc.
5:225–234. 2002.
|
|
4
|
Devolli-Disha E, Manxhuka-Kërliu S, Ymeri
H and Kutllovci A: Comparative accuracy of mammography and
ultrasound in women with breast symptoms according to age and
breast density. Bosn J Basic Med Sci. 9:131–136. 2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Marrugo-Ramírez J, Mir M and Samitier J:
Blood-Based cancer biomarkers in liquid biopsy: A promising
non-invasive alternative to tissue biopsy. Int J Mol Sci.
19(2877)2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Bai Y and Zhao H: Liquid biopsy in tumors:
Opportunities and challenges. Ann Transl Med. 6(S89)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Castro-Giner F, Gkountela S, Donato C,
Alborelli I, Quagliata L, Ng CKY, Piscuoglio S and Aceto N: Cancer
diagnosis using a liquid biopsy: Challenges and expectations.
Diagnostics (Basel). 8(31)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Simpson RJ, Lim JW, Moritz RL and
Mathivanan S: Exosomes: Proteomic insights and diagnostic
potential. Expert Rev Proteomics. 6:267–283. 2009.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen X, Liang H, Zhang J, Zen K and Zhang
CY: Horizontal transfer of microRNAs: Molecular mechanisms and
clinical applications. Protein Cell. 3:28–37. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Sanz-Rubio D, Martin-Burriel I, Gil A,
Cubero P, Forner M, Khalyfa A and Marin JM: Stability of
circulating Exosomal miRNAs in healthy subjects. Sci Rep.
8(10306)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Salehi M and Sharifi M: Exosomal miRNAs as
novel cancer biomarkers: Challenges and opportunities. J Cell
Physiol. 233:6370–6380. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
He L, He X, Lim LP, de Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Calin GA, Ferracin M, Cimmino A, Di Leva
G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et
al: A MicroRNA signature associated with prognosis and progression
in chronic lymphocytic leukemia. N Engl J Med. 353:1793–1801.
2005.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Heneghan HM, Miller N, Lowery AJ, Sweeney
KJ, Newell J and Kerin MJ: Circulating microRNAs as novel minimally
invasive biomarkers for breast cancer. Ann Surg. 251:499–505.
2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhao H, Shen J, Medico L, Wang D,
Ambrosone CB and Liu S: A pilot study of circulating miRNAs as
potential biomarkers of early stage breast cancer. PLoS One.
5(e13735)2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139.
2009.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Han J, Lee Y, Yeom KH, Kim YK, Jin H and
Kim VN: The Drosha-DGCR8 complex in primary microRNA processing.
Genes Dev. 18:3016–3027. 2004.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Suzuki HI and Miyazono K: Emerging
complexity of microRNA generation cascades. J Biochem. 149:15–25.
2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Salido-Guadarrama I, Romero-Cordoba S,
Peralta-Zaragoza O, Hidalgo-Miranda A and Rodríguez-Dorantes M:
MicroRNAs transported by exosomes in body fluids as mediators of
intercellular communication in cancer. Onco Targets Ther.
7:1327–1338. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lim SS and Kim YS:
WO2016137031-Dumbell-structure oligonucleotide, nucleic acid
amplification primer comprising same, and nucleic acid
amplification method using same. KR Patent WO/2016/137031. DIOGENE
Inc. Filed February 25, 2015; issued September 1, 2016.
|
|
27
|
Pinatel EM, Orso F, Penna E, Cimino D,
Elia AR, Circosta P, Dentelli P, Brizzi MF, Provero P and Taverna
D: miR-223 is a coordinator of breast cancer progression as
revealed by bioinformatics predictions. PLoS One.
9(e84859)2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yoshikawa M, Iinuma H, Umemoto Y,
Yanagisawa T, Matsumoto A and Jinno H: Exosome-encapsulated
microRNA-223-3p as a minimally invasive biomarker for the early
detection of invasive breast cancer. Oncol Lett. 15:9584–9592.
2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Fu L, Li Z, Zhu J, Wang P, Fan G, Dai Y,
Zheng Z and Liu Y: Serum expression levels of microRNA-382-3p,
-598-3p, -1246 and -184 in breast cancer patients. Oncol Lett.
12:269–274. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li XJ, Ren ZJ, Tang JH and Yu Q: Exosomal
MicroRNA miR-1246 Promotes Cell Proliferation, Invasion and Drug
Resistance by Targeting CCNG2 in Breast Cancer. Cell Physiol
Biochem. 44:1741–1748. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhou Y, Wang M, Tong Y, Liu X, Zhang L,
Dong D, Shao J and Zhou Y: miR-206 promotes cancer progression by
targeting Full-Length Neurokinin-1 Receptor in breast cancer.
Technol Cancer Res Treat. 18(1533033819875168)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Khodadadi-Jamayran A, Akgol-Oksuz B,
Afanasyeva Y, Heguy A, Thompson M, Ray K, Giro-Perafita A, Sánchez
I, Wu X, Tripathy D, et al: Prognostic role of elevated mir-24-3p
in breast cancer and its association with the metastatic process.
Oncotarget. 9:12868–12878. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lu K, Wang J, Song Y, Zhao S, Liu H, Tang
D, Pan B, Zhao H and Zhang Q: miRNA-24-3p promotes cell
proliferation and inhibits apoptosis in human breast cancer by
targeting p27Kip1. Oncol Rep. 34:995–1002.
2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Eichelser C, Flesch-Janys D, Chang-Claude
J, Pantel K and Schwarzenbach H: Deregulated Serum Concentrations
of Circulating Cell-Free MicroRNAs miR-17, miR-34a, miR-155 and
miR-373 in Human Breast Cancer Development and Progression. Clin
Chem. 59:1489–1496. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Piasecka D, Braun M, Kordek R, Sadej R and
Romanska H: MicroRNAs in regulation of triple-negative breast
cancer progression. J Cancer Res Clin Oncol. 144:1401–1411.
2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Asaga S, Kuo C, Nguyen T, Terpenning M,
Giuliano AE and Hoon DS: Direct serum assay for microRNA-21
concentrations in early and advanced breast cancer. Clin Chem.
57:84–91. 2011.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Corcoran C, Friel AM, Duffy MJ, Crown J
and O'Driscoll L: Intracellular and Extracellular MicroRNAs in
Breast Cancer. Clin Chem. 57:18–32. 2011.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Shimomura A, Shiino S, Kawauchi J,
Takizawa S, Sakamoto H, Matsuzaki J, Ono M, Takeshita F, Niida S,
Shimizu C, et al: Novel combination of serum microRNA for detecting
breast cancer in the early stage. Cancer Sci. 107:326–334.
2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Schrauder MG, Strick R, Schulz-Wendtland
R, Strissel PL, Kahmann L, Loehberg CR, Lux MP, Jud SM, Hartmann A,
Hein A, et al: Circulating micro-RNAs as potential blood-based
markers for early stage breast cancer detection. PLoS One.
7(e29770)2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhao C, Li X, Han B, You Z, Qu L, Liu C,
Song J, Lian L and Yang N: Gga-miR-219b targeting BCL11B suppresses
proliferation, migration and invasion of Marek's disease tumor cell
MSB1. Sci Rep. 7(4247)2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Løberg M, Lousdal ML, Bretthauer M and
Kalager M: Benefits and harms of mammography screening. Breast
Cancer Res. 17(63)2015.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Guo R, Lu G, Qin B and Fei B: Ultrasound
imaging technologies for breast cancer detection and management.
Ultrasound Med Biol. 44:37–70. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Lebron-Zapata L and Jochelson MS: Overview
of breast cancer screening and diagnosis. PET Clin. 13:301–323.
2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Alimirzaie S, Bagherzadeh M and Akbari MR:
Liquid biopsy in breast cancer: A comprehensive review. Clin Genet.
95:643–660. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Hamam R, Hamam D, Alsaleh KA, Kassem M,
Zaher W, Alfayez M, Aldahmash A and Alajez NM: Circulating
microRNAs in breast cancer: Novel diagnostic and prognostic
biomarkers. Cell Death Dis. 8(e3045)2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
He Y, Deng F, Yang S, Wang D, Chen X,
Zhong S, Zhao J and Tang J: Exosomal microRNA: A novel biomarker
for breast cancer. Biomark Med. 12:177–188. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hannafon BN, Trigoso YD, Calloway CL, Zhao
YD, Lum DH, Welm AL, Zhao ZJ, Blick KE, Dooley WC and Ding WQ:
Plasma exosome microRNAs are indicative of breast cancer. Breast
Cancer Res. 18(90)2016.PubMed/NCBI View Article : Google Scholar
|