Introduction
Gliomas comprise a very heterogeneous group of primary tumors of the central nervous system (CNS), originally classified according to their histological similarity or the supposed origin of non-neoplastic glial cells (1). The common presentation of gliomas includes seizures, cranial nerve palsies, visual field defects, language dysfunction, or increased intracranial pressure. Gliomas are considered to originate from glial (progenitor) or stem cells that develop glial characteristics. Most gliomas are characterized by diffuse infiltrative growth of tumor cells in the preexisting parenchyma of the CNS (2). Gliomas are traditionally divided into two main categories: Diffuse gliomas (DGs) and non-diffuse gliomas (NDGs) (3).
DGs are one of the most frequent primary neoplasms (4). They represent approximately 81% of all malignant brain tumors and have a high mortality and morbidity rate (5). In these tumors, the degree of malignancy is assigned based on pathological features (6). The main classes of DGs are astrocytomas, oligodendrogliomas, oligoastrocytomas and glioblastoma multiforme, and are classified according to the criteria of the World Health Organization (WHO) as grades II, III or IV (6,7).
In 2016, WHO updated its classification and incorporated the detection of distinctive molecular alterations to traditional histological methods (8). The use of ‘integrated’ phenotypic and genotypic parameters for classification added greater objectivity in the diagnostic process (9). This has allowed defining more biologically homogeneous diagnostic entities, leading to better patient management and more precise indications of response to treatment. The 2016 WHO classification of DGs has incorporated two molecular biomarkers: The mutation status of the isocitrate dehydrogenase (IDH) gene (the IDH1 (codon 132) and IDH2 (codon 172) genes) and the presence or absence of the 1p/19q co-deletion (complete deletion of both the short arm of chromosome 1 and the long arm of chromosome 19) (10). When these molecular biomarkers could not be performed, the term NOS (not otherwise specified) should be added to indicate that the study has been inconclusive (8,11). The new glioma classification allows differentiating astrocytomas and glioblastomas according to the IDH1 or IDH2 mutation status and allows diagnosing tumors with 1p/19q co-deletion and IDH1 or IDH2 mutation as oligodendrogliomas (Fig. 1).
 |
Figure 1
Simplified algorithm for integrated diagnosis of diffuse gliomas (2016 World Health Organization classification). IDH, isocitrate dehydrogenase.
|
The IDH status (mutant or wild type) is very relevant for the diagnosis of gliomas. The most common variant (~88%) is a substitution of the amino acid Arginine to Histidine in codon 132 of the IDH1 gene (IDH1-R132H) (12). Less common variants such as R132C, R132M, and R132K, as well as a substitution in the IDH2 gene (R172H, R172C, R172M and others), have also been described (13). The IDH status allows identifying a subgroup of glial tumors that have a better survival prognosis (14).
The presence of the 1p/19q co-deletion is associated with greater survival and better response to chemotherapy (15). This co-deletion is observed mainly in oligodendrogliomas, and is useful to minimize the diagnosis of oligoastrocytomas, which are tumors exhibiting mixed histology (16). The WHO classification discourages this diagnosis as molecular genetic analyses have proved to be efficient to re-classify them.
In this study, we analyzed and evaluated This study aimed to evaluate the impact caused by the new diagnostic classification of gliomas in a series of cases studied in Argentina. In addition, we considered the possible associations between erroneous tumor classification, histological characteristics and survival data.
Materials and methods
Glioma samples and patient characteristics
A retrospective study of 139 cases of glial tumors was performed. Biopsy samples were obtained from archives of the Department of Pathological Anatomy, Emergency Hospital ‘Dr Clemente Alvarez’ (HECA) and Center for Pathological Diagnosis SRL (Gamma Group), Rosario, Santa Fe, Argentina. Inclusion criteria were adult patients undergoing a glial tumor biopsy between January 2015 and December 2017, with compatible histological diagnosis of DG, complete medical history, minimum follow-up of 24 months, and sufficient quantity and quality of material to perform molecular studies. Exclusion criteria were: Children, patients with histological diagnosis of NDG, midline glioma location, incomplete clinical history, presence of more than one primary cancer or post-mortem diagnosis and cases without enough material to perform all molecular studies. The variables included in the study were: The patient's age, the patient's sex, the date of diagnosis and the date of death. The degree of resection and the radiological variables of the surgical resection were not studied because it was not the objective of this work. Data on the Karnofsky performance status, type of surgery, location of the tumor and first-line treatment could not be obtained for most of the patients. For all cases, formalin-fixed paraffin-embedded tumor tissue was available for pathological and immunohistochemical analyses. The tumors included in this study were typed and graded according to the WHO 2007 classification.
Histological analysis and immunohistochemistry (IHC) staining
Formalin-fixed paraffin-embedded tissue was used for histological and immunohistochemical analysis. All the Hematoxylin and Eosin (H&E)-stained slides were revised by two independent neuropathologists, who made consensus diagnoses. These pathologists were kept blind to the results of the molecular genetic analysis.
Based on the resemblance of the tumor cells with non-neoplastic glial cells (astrocytes, oligodendroglial cells), most DGs can be typed as astrocytic, oligodendroglial, or mixed oligoastrocytic tumors (17). Astrocytes are stellate cells with an oval-to-elongate nucleus and scant eosinophilic cytoplasm (17). The astrocytic phenotype can be identified by Glial fibrillary acidic protein (GFAP) immunohistochemical staining. Oligodendroglial cells are characterized by a round nucleus with a dense chromatin pattern and a perinuclear halo and lack of staining for GFAP (2). Mixed oligoastrocytomas have neoplastic glial cells with morphological characteristics of astrocytes and oligodendrocytes. Regarding the diagnosis of neuroendocrine tumors, immunohistochemical staining of synaptophysin, an integral membrane glycoprotein produced in the presynaptic vesicles of neurons, can be used as a biomarker for the neuronal differentiation of DGs (18,19).
According to the 2007 WHO classification, DGs are graded as grade II (low grade), grade III (anaplastic) or grade IV (glioblastoma). This grading of DGs is based on histological features such as nuclear atypia, mitotic activity, necrosis, and microvascular proliferation. The presence of necrosis and/or microvascular proliferation leads to a diagnosis of glioblastoma. In this context, since Ki-67 is a biomarker to determine cell proliferation, the Ki-67 labeling index (fraction of Ki-67-positive tumor cells) is often correlated with the clinical course of cancer and increases with the malignancy grade (<5% in low-grade tumors, 5-10% in anaplastic gliomas and >10% in glioblastomas) (2,20). Automated IHC was performed on 5-µm-thick formalin-fixed paraffin-embedded sections with an avidin-biotin-peroxidase complex on Benchmark XT (Ventana Medical System Inc.), using the Ventana Kit including DAB reagent to search for the expression of GFAP (1:50, clone EP672Y, Cell Marque), Ki-67 (1:50 clone 30-9, Ventana Medical Systems), Synaptophysin (1:50, clone MRQ-40, Cell Marque) and IDH1-R132H (1:40, clone H09, Dianova GmbH). Assays were performed according to the manufacturer's instructions. Negative controls were carried out ommitting the primary antibody. The level of Ki-67 expression was defined semiquantitatively according to the fraction of positive nuclear staining and was scored as high (>5% positive nuclear staining) or low (<5% nuclear staining). The semiquantitative evaluation was done by two neuropathologists blinded to all patient's details.
Molecular assays Tumoral DNA extraction
Five 5 µm-thick unstained sections were cut from the paraffin block. The area(s) with the highest density of tumor cells was identified by microscopic examination and marked on the H&E-stained slide. The areas of tumor were required to contain more than 80% of tumor cells. This cut-off value (minimum ratio of tumor cells to normal cells) was chosen based on the allelic ratios observed in specimens with different tumor/normal ratios, and reported by other researchers (21). If necessary, tissue dissection was performed and the H&E-stained slide served as a guide. Sections were deparaffinized by incubations at room temperature in xylene for 5 min, 100% ethanol for 5 min, and dried for 30 min while protected from dust.
DNA was released from the tissue fragment by incubation in a proteinase K solution, prepared from kit components according to the manufacturer's instructions (Roche Diagnostics, Mannheim, Germany), for 8 h at 56˚C followed by 60 min at 90˚C to inactivate proteinase K. DNA was isolated by a DNA Sample Preparation kit (Roche Diagnostics,), according to the manufacturer's instructions. The DNA extract was stored at -20˚C until used in aliquots.
Multiplex ligation-dependent probe amplification (MLPA)
This is a technique by which multiple different sequences corresponding to specific known genomic regions can be targeted in a single, semi-quantitative polymerase chain reaction (PCR)-based experiment (22). We used the MLPA SALSA P088-C2 kit (MRC-Holland) and performed the MLPA analysis according to the manufacturer's recommendations. Briefly, DNA (100 to 200 ng) was denatured and subsequently cooled to 25˚C. After adding the probe mix and hybridization buffer, the sample was denatured, and the probes were allowed to hybridize (16 h at 60˚C). After ligation of both probe pairs (15 min at 54˚C) and inactivation of ligase (5 min at 98˚C), PCR was performed by adding the polymerase master mix, using the Applied Biosystems Veriti 96-well thermal cycler (Applied Biosystems), followed by 35 cycles of denaturation at 95˚C for 30 sec, annealing at 60˚C for 30 sec, and extension at 72˚C for 60 sec, with a final extension of 20 min at 72˚C. For capillary electrophoresis, a mixture of 1 µl product amplification, 0.2 µl weight marker (LIZ 500), and 9 µl of HiDi formamide, denatured for 3 min at 90˚C, was used for each patient on an ABI 310 Genetic Analyzer (Applied Biosystems) using a 36-cm capillary with POP4 resin. Separation and quantification were performed with a 1.6-kV injection voltage and a 15-sec injection time. Data were analyzed using the Coffalyser software, as proposed by MRC-Holland in the MLPA General Protocol version MDP-V007. This analysis comprised three stages: The record of fragments, a comparative analysis and analysis of the Coffalyser score. In each set of MLPA experiments, in addition to the tumor samples to be analyzed, we included at least three normal control DNA samples for data processing.
PCR amplification and direct sequencing
The genomic regions spanning the R132 codon of the IDH1 gene and the R172 codon of the IDH2 gene were amplified and sequenced (23). PCR amplification was performed using 50 ng of genomic DNA, 0.5 µmol of each primer (IDH1.F: 5'-CGGTCTTCAGAGAAGCCATT-3' and IDH1.R: 5'-GCAAAATCACATTATTGCCAAC-3'; IDH2.F: 5'-AGCCCATCATCTGCAAAAAC-3' and IDH2.R: 5'-CTAGGCGAGGAGCTCCAGT-3'), 1X HotStarTaq Plus Master Mix Kit (QIAGEN, Hilden Germany) in a total volume of 20 µl. The reaction mixture was subjected to an initial denaturation of 95˚C for 5 min, followed by 40 cycles of amplification consisting of denaturation at 95˚C for 40 sec, annealing at 55˚C for 40 sec and extension 72˚C for 1 min. The PCR products were purified (NucleoSpin Gen and PCR Clean-up) and sequenced in both sense and antisense directions by using the BigDye Terminator version 1.1 Cycle Sequencing kit (Applied Biosystems) according to the manufacturer's instructions. Sequencing products were purified (ZR DNA Sequencing Clean-up kit, Zymo Research) and analyzed on ABI 310 (Applied Biosystems). The sequence electropherograms were analyzed by using Sequencing Analysis Software 6 (Life Technologies). Each case was classified as either positive or negative for the IDH mutation based on the sequencing results.
Statistical analysis
The principal endpoint was overall survival (OS), defined as the length of time from the glioma diagnosis to the patient's death or last follow up. OS analysis was performed using the Kaplan-Meier estimator and the log-rank test for group comparison. Analyses were conducted using the MedCalc Statistical Software version 19.1 (MedCalc Software bv; https://www.medcalc.org; 2019) (24).
Results
Patient characteristics
Data from 139 patients with operated glial tumors were collected in the period from 2005 to 2017 (Table I). According to the exclusion criteria, 20 cases were ruled out. In this series of 119 patients, 65% were male and 35% female, with a median age of 52 years (22-80 years) and 55 years (23-83 years), respectively. A great majority of cases (91/119, 76%) were 55 years or older.
 |
Table I
Patient characteristics.
|
Table I
Patient characteristics.
| Variable |
Value |
| Factors |
|
| Age, median (range), years |
52 (22-80) |
| Sex (male/female), n (%) |
78/41 (65.0/35.0) |
| Immunostaining and molecular alterations |
|
| Ki67 >5%, n (%) |
105/119 (88.2) |
| IDH-mutant, n (%) |
29/119 (24.4) |
| IDH-mutant (IHC), n |
24/29 |
| IDH-mutant (MLPA), n |
27/29 |
| IDH-mutant (sequence), n |
29/29 |
| 1p/19q codeletion, n (%) |
8/119 (6.7) |
2007 WHO classification for gliomas
According to the 2007 WHO classification, these 119 cases were classified as follows: 17 astrocytomas (grade II: 5; grade III: 9; grade III/IV: 3), 11 unclassified gliomas (grade II: 6; grade III: 1; grade III/IV: 4), 73 glioblastomas, 15 oligodendrogliomas (grade II: 6; grade III: 9), one oligoastrocytoma grade II, and two samples in which gliosis or low grade glioma could not be distinguished (Fig. S1). All samples were suitable for molecular studies.
Molecular assays of gliomas
These studies showed IDH1 mutation in 26 tumors (p.R132H: 25; p.R132C: 1), IDH2 mutation in three tumors (p.R172K: 1, p.R172G: 1, p.R172C: 1) and 1p/19q co-deletion in eight tumors (6.72%).
The p.R132H-IDH1 mutation, occurring in approximately 90% of cases, was detected by all the methods performed (IHC, MLPA and sequencing), the p.R132C-IDH1 and p.R172K-IDH2 mutations were detected by two methods (MLPA and sequencing), and the p.R172G and p.R172C mutations of the IDH2 gene were detected only by sequencing. These results indicate the importance of the accurate choice of methods used for the detection of IDH mutations, especially in young patients (Table I).
2016 WHO classification for gliomas
The molecular reclassification allowed dividing DGs into eight subgroups according to the IDH mutations and 1p/19q co-deletion (Table II). Based on the new updated WHO classification, the 119 cases were reclassified in: Five oligodendrogliomas IDH-mutant, 1p/19q co-deleted (4.20%), three anaplastic oligodendrogliomas IDH-mutant, 1p/19q co-deleted (2.52%), eight diffuse astrocytomas IDH-mutant (6.72%), two diffuse astrocytomas IDH-wild type (1.68%), six anaplastic astrocytomas IDH-mutant (5.04%), 10 anaplastic astrocytomas IDH-wild type (8.40%), seven glioblastomas IDH-mutant (5.88%), and 78 glioblastomas IDH-wild type (65.56%). In 32 cases (27%), diagnosis changed (Fig. 2; Table SI). Regarding tumor histology, 60% of oligodendrogliomas, 35% of astrocytomas and 100% of unclassified gliomas were re-classified, while glioblastomas maintained their initial classification (Table SI). Regarding the tumor grade, 18.5% of samples changed; almost all of them (95%) increased their grade from II to III (Table SI).
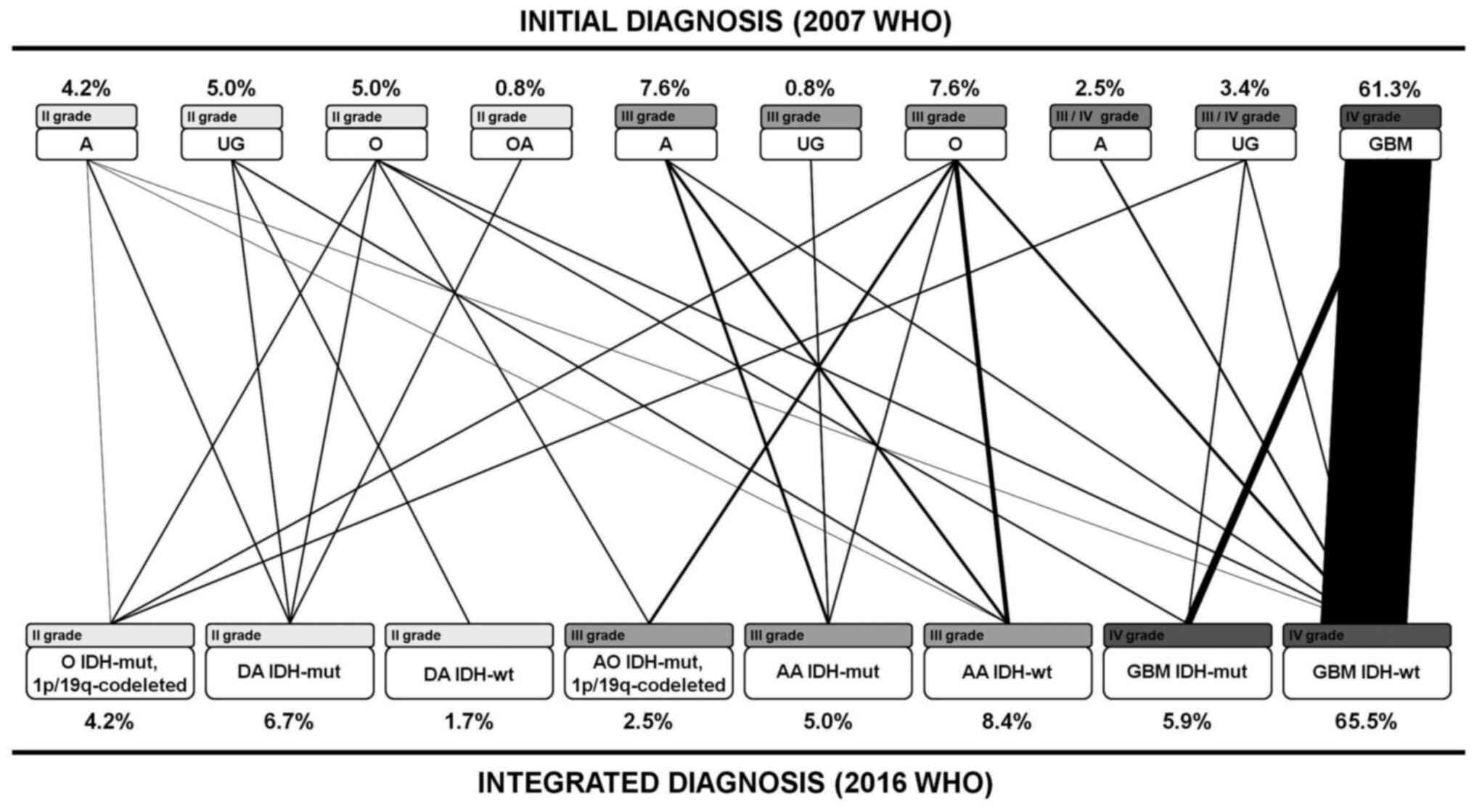 |
Figure 2
Reclassification of 119 tumors according to the updated 2016 WHO classification. A, astrocytoma; UG, unclassified glioma; O, oligodendroglioma; OA, oligoastrocytoma; DA, diffuse astrocytoma; AO, anaplastic oligodendroglioma; AA, anaplastic astrocytoma; GBM, glioblastoma; IDH, isocitrate dehydrogenase; wt, wild-type; mut, mutant; WHO, World Health Organization.
|
 |
Table II
Reclassification of the 119 cases according to the updated 2016 World Health Organization classification.
|
Table II
Reclassification of the 119 cases according to the updated 2016 World Health Organization classification.
| Grade |
No. |
Percentage |
| Grade II |
|
|
| Oligodendroglioma IDH-mutant,1p/19q-codeleted |
5 |
4.20 |
| Diffuse astrocytoma IDH-mutant |
8 |
6.72 |
| Diffuse astrocytoma IDH-wild type |
2 |
1.68 |
| Grade III |
|
|
| Anaplastic oligodendroglioma IDH-mutant, 1p/19q-codeleted |
3 |
2.52 |
| Anaplastic astrocytoma IDH-mutant |
6 |
5.04 |
| Anaplastic astrocytoma IDH-wild type |
10 |
8.40 |
| Grade IV |
|
|
| Glioblastoma IDH-mutant |
7 |
5.88 |
| Glioblastoma IDH-wild type |
78 |
65.56 |
Prognostic value of the histological and tumor grade
We also evaluated the prognostic value of the histological grade (Fig. 3). Both the 2007 and 2016 WHO classifications of gliomas had a prognostic value for the main OS in all groups. Diffuse astrocytomas IDH-wild type were not included due to the low number of cases. The histological subgroup associated with longer OS was grade II glioma (OS-2007 WHO: 35.6 months and OS-2016 WHO: 46.8 months). In contrast, grade IV glioma (glioblastoma) was the subgroup associated with the poorer outcome (OS-2007 WHO: 10.4 months and OS-2016 WHO: 11.1 months).
 |
Figure 3
Survival probability according to the (A) previous World Health Organization classification and (B) updated classification. The mean survival time is reported with 95% CI. 95% CI, 95% confidence interval; DF, degrees of freedom.
|
We next evaluated the OS of tumor grade subgroups with respect to their IDH status (Fig. 4A). For all subgroups, IDH-mutant (IDH-mut) tumors were associated with a better prognosis than IDH-wild-type (IDH-wt) tumors (grade II: OS-IDH-mut: 46.8 months vs. OS-IDH-wt: 22.0 months; grade III: Subgroups OS-IDH-mut: 41.3 months vs. OS-IDH-wt: 12.8 months; grade IV: OS-IDH-mut: 17.4 months vs. OS-IDH-wt: 10.2 months). These results validated the size and representativeness of each glioma molecular subgroup for future studies. Based on these OS results, we were able to reclassify the tumor grades according to the OS as: Grade IV (OS: 10.5 months) for anaplastic astrocytoma IDH-wt and glioblastoma IDH-wt, grade III (OS: 17.4 months) for glioblastoma IDH-mut, and grade II (OS: 45.8 months) for oligodendroglioma IDH-mut, 1p/19q co-deletion, diffuse astrocytoma IDH-mut, anaplastic oligodendroglioma IDH-mut, 1p/19q co-deletion, and anaplastic astrocytoma IDH-mut (Fig. 4B).
 |
Figure 4
Survival probability of tumor subgroups according to the 2016 World Health Organization classification. (A) Tumor grade-based Ki-67 labeling index and IDH status. (B) Classification of tumor grade based on the patient's overall survival. The mean survival time is reported with 95% CI. 95% CI, 95% confidence interval; DF, degrees of freedom; IDH, isocitrate dehydrogenase; wt, wild-type; mut, mutant.
|
The accuracy introduced by the molecular biomarkers (IDH status and 1p/19q co-deletion) allowed a better classification of the diagnosis and prognosis of patients with gliomas.
Discussion
In the present study, we evaluated the impact of the new 2016 WHO classification of the CNS tumors in a cohort of 119 DGs from Argentine patients, previously classified according to the 2007 WHO classification based only on histological characteristics. Integrated diagnosis allowed defining tumor lineage in all cases.
Our results showed that 18.5% of the samples were re-classified. This change in glioma subgroups was associated with the introduction of the 1p/19q co-deletion, which was decisive in the subdivision of astrocytomas and oligodendrogliomas, as well as in the disappearance of the oligoastrocytoma subgroup. According to the new WHO classification, diagnosis of oligoastrocytomas should be extremely rare, since they can be re-classified as astrocytoma or oligodendroglioma by incorporating molecular and cytogenetic data (25). These results correlate with other studies, which have reported that the main changes in the 2016 WHO classification were in the astrocytoma and oligodendroglioma subgroups (26,27).
Although the low number of oligodendroglioma cases in our cohort is a limitation, the variability between different studies demonstrates the difficulty of correct diagnosis of this tumor subgroup without the introduction of molecular biomarkers (26,27).
Recently, the Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy- Not Official WHO (cIMPACT-NOW) recommended diagnostic criteria for ‘Diffuse astrocytic glioma, IDH-wild type, with molecular features of glioblastoma, WHO grade IV’ (28). Different studies have demonstrated the importance of identifying IDH-wild type diffuse or anaplastic astrocytomas that would behave most aggressively, similarly to glioblastomas, and thus with a clinical course corresponding to WHO grade IV (29-31). Epidermal growth factor receptor amplification, combined with whole chromosome 7 gain and whole chromosome 10 loss (+ 7/- 10) and telomerase reverse transcriptase promoter mutation, will soon be used to test our cases and results will be published.
The most frequent IDH mutation in DGs was p.R132H of the IDH1 gene, which can be reliably detected with IDH1-R132H IHC staining. However, four IDH mutations (p.R132C of the IDH1 gene and p.R172G, p.R172C and p.R172K of the IDH2 gene) could not be detected by this method, causing a misdiagnosis of these tumors. IDH-mutant tumors with immunostaining zones negative for IDH1-R132H have been reported, probably due to the loss of the IDH1-mutant allele in those tumor portions. In these cases, a confirmatory genetic analysis may be needed (32). In IDH-mutant gliomas, the 1p/19q co-deletion is pathognomonic for oligodendrogliomas (23).
Glioblastoma was the most frequent type of glioma (71.4%) diagnosed at our cohort and was subclassified according to the IDH status. Glioblastoma IDH-mutant accounted for 8.2% of all glioblastomas. These results were similar to those previously reported for other cohorts (23,26,27). All gliomas classified as glioblastoma with the 2007 WHO classification remained the same after the revision of the cases and molecular tests performed, indicating that the morphological criteria for glioblastoma were unequivocal.
Astrocytoma IDH-wt and IDH-mut showed frequencies (10.1 and 11.7% respectively) similar to those previously reported (23,26,27). These differences in astrocytoma diagnosis could be related to their diagnostic re-classification, overall, to glioblastoma. This could be explained by the fact that, currently, the classification between astrocytoma and glioblastoma depends only on the tumor histological characteristics.
During a median follow-up of 2 years (range 0.0-15.0 years), 95 out of the 119 patients had died. We evaluated the survival data according to the histological grade by univariate analysis (Fig. 3). Both the 2007 and the 2016 WHO classifications of gliomas were prognostic for OS (P=0.001 and P<0.001, respectively). We found that the 2016 WHO classification was highly accurate in predicting OS, confirming the value of adding molecular characteristics to diagnosis, especially the 1p/19q co-deletion to define tumor lineage. Patients with grade II gliomas had longer OS than those with grade III and IV gliomas in both classifications.
We next analyzed the impact of the IDH status on the tumor grade (Fig. 4A). IDH-mut gliomas were the molecular subgroup associated with the longest OS in all cases, compared to IDH-wt tumors (grade II: 46.8 vs. 22.0 months; grade III: 41.3 vs. 12.8 months; grade IV: 17.4 vs. 10.2 months). These results suggest that the IDH status is a strong biomarker of prognosis. These mutations change the enzymatic activity, allowing α-ketoglutarate to be reduced to 2-hydroxyglutarate in a NADPH-dependent reaction (33). This leads to an accumulation of high levels of 2-hydroxyglutarate in glioma tissues, which can inhibit several enzymes, causing epigenetic alterations in DNA and histones, and thus altering gene expression and promoting oncogenic transformation (34). Thus, the IDH status allows identifying a subgroup of glial tumors that have a better survival prognosis (14). In addition, these data are in accordance with previous studies indicating that IDH-mut gliomas are associated with a better prognosis than IDH-wt tumors (26,35,36). In our study, we were able to re-classify tumor grades based on OS and not using the Ki-67 labeling index (Fig. 4B). Previous studies in astrocytic tumors have reported that this index could not correctly determine the WHO grade or estimate the OS of patients (37). At this point, the histological evaluation of the H&E-stained tissues and the detection of the IDH status would be more suitable to determine the tumor grade.
The main limitation of this study was the biased distribution of tumors, which included many glioblastomas IDH-wt, and a small number of tumors of the other categories. Despite this, we found that the incorporation of molecular biomarkers in the new WHO classification improves tumor characterization and prognostic value of the subgroups. The impact was greater for low-grade gliomas, mainly oligodendrogliomas. This would allow better treatment options for the patients.
In summary, our study demonstrates the importance of the prognostic value of the histological classification and genetic profile based on the IDH status and 1p/19q co-deletion. Finally, the differentiation of subgroups based on molecular biomarkers provides prognostic information complementary to that provided by histological classification.
Supplementary Material
Histopathological characteristics of gliomas. (A) Representative images of H&E-stained sections (magnification, x10) of astrocytoma and oligodendroglioma according to World Health Organization grades. (B) Immunohistochemistry staining (magnification, x10) of glioblastoma IDH-mut. H&E, hematoxylin and eosin stain; IDH, isocitrate dehydrogenase; IDH-mut, IDH-mutant; GFAP, glial fibrillary acidic protein.
Change of classification of the 119 cases.
Acknowledgements
The authors would like to thank Mr. Pablo Casal (Department of Microbiology, Faculty of Biochemical and Pharmaceutical Sciences, National University of Rosario) for the review of the draft of this manuscript and Miss Paula Rocha [Center for Pathological Diagnosis SRL (Gamma Group)] for her commitment and dedication to this project.
Funding
This research was supported by the Agencia Santafecina de Ciencia, Tecnología e Innovación para el Apoyo al desarrollo de soluciones tecnológicas en empresas (grant no. IP‑2015‑00003), Santa Fe, Argentina.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
MFR and GRP designed and conceived the study. MFR, LCB, MVG and GRP acquired the data. MFR, MVG and ARG made contributions to analysis and interpretation of data obtained from anatomopathological studies. LCB, MT and GRP made contributions to analysis and interpretation of data obtained from molecular studies. GRP performed statistical analysis. MFR and GRP prepared the manuscript. MT and ARG revised the manuscript. ARG and MT contributed to the critical revision of the article for important intellectual content and gave final approval of the version to be published. The authenticity of all the raw data was confirmed by MFR and GRP. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Regional Ethics Committee (Santa Fe, Argentina) and respected the basic ethical principles and standards set forth in the Helsinki Declaration. The requirement for informed consent was waived in view of the retrospective design of the study. However, patients were given the opportunity to decline participation via the opt-out route.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Chen R, Smith-Cohn M, Cohen AL and Colman H: Glioma subclassifications and their clinical significance. Neurotherapeutics. 14:284–297. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wesseling P, Kros JM and Jeuken JWM: The pathological diagnosis of diffuse gliomas: Towards a smart synthesis of microscopic and molecular information in a multidisciplinary context. Diagnostic Histopathology. 17:486–494. 2011.
|
|
3
|
Kristensen BW, Priesterbach-Ackley LP, Petersen JK and Wesseling P: Molecular pathology of tumors of the central nervous system. Ann Oncol. 30:1265–1278. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Brandner S and von Deimling A: Diagnostic, prognostic and predictive relevance of molecular markers in gliomas. Neuropathol Appl Neurobiol. 41:694–720. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Diwanji TP, Engelman A, Snider JW and Mohindra P: Epidemiology, diagnosis, and optimal management of glioma in adolescents and young adults. Adolesc Health Med Ther. 8:99–113. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 114:97–109. 2007.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Fuller GN and Scheithauer BW: The 2007 revised world health organization (WHO) classification of tumours of the central nervous system: Newly codified entities. Brain Pathol. 17:304–307. 2007.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P and Ellison DW: The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 131:803–820. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, von Deimling A, Aldape K, Brat D, Collins VP, Eberhart C, et al: International Society of Neuropathology-Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol. 24:429–435. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Iwamoto FM, Nicolardi L, Demopoulos A, Barbashina V, Salazar P, Rosenblum M and Hormigo A: Clinical relevance of 1p and 19q deletion for patients with WHO grade 2 and 3 gliomas. J Neurooncol. 88:293–298. 2008.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Louis DN, Wesseling P, Paulus W, Giannini C, Batchelor TT, Cairncross JG, Capper D, Figarella-Branger D, Lopes MB, Wick W and van den Bent M: cIMPACT-NOW update 1: Not otherwise specified (NOS) and not elsewhere classified (NEC). Acta Neuropathol. 135:481–484. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C and von Deimling A: Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 116:597–602. 2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al: IDH1 and IDH2 mutations in gliomas. N Engl J Med. 360:765–773. 2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, El Hallani S, Boisselier B, Mokhtari K, Hoang-Xuan K and Delattre JY: Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol. 27:4150–4154. 2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kouwenhoven MC, Kros JM, French PJ, Biemond-ter Stege EM, Graveland WJ, Taphoorn MJ, Brandes AA and van den Bent MJ: 1p/19q loss within oligodendroglioma is predictive for response to first line temozolomide but not to salvage treatment. Eur J Cancer. 42:2499–2503. 2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Boots-Sprenger SH, Sijben A, Rijntjes J, Tops BB, Idema AJ, Rivera AL, Bleeker FE, Gijtenbeek AM, Diefes K, Heathcock L, et al: Significance of complete 1p/19q co-deletion, IDH1 mutation and MGMT promoter methylation in gliomas: Use with caution. Mod Pathol. 26:922–929. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Garman RH: Histology of the central nervous system. Toxicol Pathol. 39:22–35. 2011.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wiedenmann B, Franke WW, Kuhn C, Moll R and Gould VE: Synaptophysin: A marker protein for neuroendocrine cells and neoplasms. Proc Natl Acad Sci USA. 83:3500–3504. 1986.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Choi BH and Khang SK: Immunohistochemical Expression of Synaptophysin in Brain Tumors. The Korean Journal of Pathology, 35: 433-439.
|
|
20
|
van den Bent MJ: Interobserver variation of the histopathological diagnosis in clinical trials on glioma: A clinician's perspective. Acta Neuropathol. 120:297–304. 2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Skotheim RI, Diep CB, Kraggerud SM, Jakobsen KS and Lothe RA: Evaluation of loss of heterozygosity/allelic imbalance scoring in tumor DNA. Cancer Genet Cytogenet. 127:64–70. 2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Torres-Martin M, Pena-Granero C, Carceller F, Gutiérrez M, Burbano RR, Pinto GR, Castresana JS, Melendez B and Rey JA: Homozygous deletion of TNFRSF4, TP73, PPAP2B and DPYD at 1p and PDCD5 at 19q identified by multiplex ligation-dependent probe amplification (MLPA) analysis in pediatric anaplastic glioma with questionable oligodendroglial component. Mol Cytogenet. 7(1)2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, et al: Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1,010 diffuse gliomas. Acta Neuropathol. 118:469–474. 2009.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Schoonjans F, Zalata A, Depuydt CE and Comhaire FH: MedCalc: A new computer program for medical statistics. Comput Methods Programs Biomed. 48:257–262. 1995.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sahm F, Reuss D, Koelsche C, Capper D, Schittenhelm J, Heim S, Jones DT, Pfister SM, Herold-Mende C, Wick W, et al: Farewell to oligoastrocytoma: In situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta Neuropathol. 128:551–559. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tabouret E, Nguyen AT, Dehais C, Carpentier C, Ducray F, Idbaih A, Mokhtari K, Jouvet A, Uro-Coste E, Colin C, et al: Prognostic impact of the 2016 WHO classification of diffuse gliomas in the French POLA cohort. Acta Neuropathol. 132:625–634. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Iuchi T, Sugiyama T, Ohira M, Kageyama H, Yokoi S, Sakaida T, Hasegawa Y, Setoguchi T and Itami M: Clinical significance of the 2016 WHO classification in Japanese patients with gliomas. Brain Tumor Pathol. 35:71–80. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Brat DJ, Aldape K, Colman H, Holland EC, Louis DN, Jenkins RB, Kleinschmidt-DeMasters BK, Perry A, Reifenberger G, Stupp R, et al: cIMPACT-NOW update 3: Recommended diagnostic criteria for ‘Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV.’. Acta Neuropathol. 136:805–810. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML, Smirnov IV, et al: Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 372:2499–2508. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Weller M, Weber RG, Willscher E, Riehmer V, Hentschel B, Kreuz M, Felsberg J, Beyer U, Löffler-Wirth H, Kaulich K, et al: Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol. 129:679–693. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Wijnenga MMJ, Dubbink HJ, French PJ, Synhaeve NE, Dinjens WNM, Atmodimedjo PN, Kros JM, Dirven CMF, Vincent AJPE and van den Bent MJ: Molecular and clinical heterogeneity of adult diffuse low-grade IDH wild-type gliomas: Assessment of TERT promoter mutation and chromosome 7 and 10 copy number status allows superior prognostic stratification. Acta Neuropathol. 134:957–959. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Preusser M, Wohrer A, Stary S, Hoftberger R, Streubel B and Hainfellner JA: Value and limitations of immunohistochemistry and gene sequencing for detection of the IDH1-R132H mutation in diffuse glioma biopsy specimens. J Neuropathol Exp Neurol. 70:715–723. 2011.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Losman JA and Kaelin WG Jr: What a difference a hydroxyl makes: Mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 27:836–852. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ye D, Ma S, Xiong Y and Guan KL: R-2-hydroxyglutarate as the key effector of IDH mutations promoting oncogenesis. Cancer Cell. 23:274–276. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Bush NA and Butowski N: The effect of molecular diagnostics on the treatment of glioma. Curr Oncol Rep. 19(26)2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lipp ES and McLendon RE: Tissue is the issue: Biomarkers of prognosis and classification in adult gliomas. Semin Oncol Nurs. 34:430–442. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Stoyanov GS, Dzhenkov DL, Kitanova M, Donev IS and Ghenev P: Correlation between Ki-67 index, World health organization grade and patient survival in glial tumors with astrocytic differentiation. Cureus. 9(e1396)2017.PubMed/NCBI View Article : Google Scholar
|


















