Introduction
Colorectal cancer (CRC) is one of the most common
malignancies worldwide. Although most localized cases of the
disease are treated surgically, a considerable number of patients
experience disease recurrence. Adjuvant chemotherapy after curative
surgery has been shown to reduce the recurrence rate and therefore
all CRC patients with clinical stage III disease are recommended to
receive adjuvant chemotherapy (1),
even though only a proportion of these derive a benefit. Patients
with clinical stage II disease are mostly treated with surgery
alone, however some may benefit from adjuvant therapy because of a
high risk of recurrence (2,3). To maximize the efficacy of adjuvant
chemotherapy, accurate predictive markers are needed to select
patients who will benefit most from treatment.
5-FU-based chemotherapy is the current standard of
care for adjuvant therapy following surgical treatment of CRC
(4). Thymidylate synthase (TS) is a
target enzyme for 5-FU (5), leading
to extensive studies of TS mRNA expression (6), TS protein expression (7,8) and TS
gene polymorphisms (9,10) as potential predictive factors for
the efficacy of 5-FU-based adjuvant chemotherapy. Currently
however, no information regarding TS status is recommended for
routine clinical use in the selection of patients to receive
5-FU-based chemotherapy (11).
TS shows unique genetic variants comprising a
variable number of tandem repeat (VNTR) and a single nucleotide
polymorphism (SNP) in its 5' untranslated region (5'UTR) (12-14).
These may be predictive markers for 5-FU efficacy and for adverse
events from this treatment. We previously reported that VNTR and
SNP can give rise to four TS allele types: 2G, 2C, 3G and 3C. These
may affect the translational activity of TS mRNA, thus influencing
TS protein expression and therefore constitute a marker for the
efficacy of 5-FU-based adjuvant chemotherapy (14-16).
In addition to these four allele types, other rarer alleles
comprising more than three repeats and novel SNPs in the 2R allele
have also been reported (17), thus
giving rise to a larger number of allele types. Furthermore, we
observed that frequent loss of heterozygosity (LOH) of the TS locus
in tumors can affect the genotype, thereby indirectly influencing
the TS expression level in tumors (18,19).
This potential change in genotype status due to LOH should be
considered in studies of the TS genotype as a predictive marker.
The status of VNTR, SNPs in both 2R and 3R, and LOH must all be
evaluated before TS genotype information can be introduced into the
clinical setting.
In this study, we analyzed the TS VNTR, the SNPs in
both the 2R and 3R alleles, as well as the LOH status of the TS
locus in order to explore their potential significance as
prognostic and predictive markers of 5-FU-based adjuvant
chemotherapy in CRC.
Materials and methods
Patient cohort and DNA isolation
Matched tumor and normal tissue samples were
obtained following surgical resection for primary colorectal
adenocarcinoma in 246 patients. The patients were all Japanese and
comprised 146 males and 100 females, ranging in age from 33 to 93
years (mean age 66.0 years). The resected tissues were fixed in
formalin and embedded in paraffin followed by H&E staining and
histological diagnosis. Tumor tissue was dissected manually from 10
µm sections of formalin-fixed, paraffin-embeded tissue blocks.
After deparaffinization using xylene and ethanol, genomic DNA was
isolated using a QIAamp DNA FFPE Tissue Kit (QIAGEN, Hilden,
Germany) following the protocol provided by the manufacturer.
Approval for this project was obtained from the Kanazawa University
Genome/Gene Analysis Research Ethics Committee.
Genotyping of TS VNTR and SNP
TS genotypes for the VNTR and the SNP in the 3R
allele were determined by PCR and PCR-restriction fragment length
polymorphism (RFLP) using the forward primer TS25:
5'-AGGCGCGCGGAAGGGGTCCT-3' and reverse primer TS18:
5'-TCCGAGCCGGCCACAGGCAT-3' as described previously (14) with a modification of PCR conditions.
PCR with the genomic DNA template was performed in reaction
mixtures containing 1X TaKaRa HS Taq buffer (TaKaRa Bio, Otsu,
Japan), 200 µM deoxyribonucleoside triphosphates, 500 nM of each
primer, 0.5 unit of TaKaRa HS Taq DNA polymerase (TaKaRa Bio) and
100 ng of genomic DNA. The cycling conditions were: one cycle at
95˚C for 3 min, 35 cycles at 98˚C for 10 sec and 68˚C for 60 sec,
with a final extension at 72˚C for 5 min. Aliquots of amplified
fragments were separated on 3% agarose gels to determine the TS
VNTR genotype.
Samples showing the 2R/3R or 3R/3R genotypes were
analyzed further for the G/C polymorphism in the 3R allele by using
the RFLP method. HaeIII digestion of the 3R fragment
produced 66-, 37-, 28- and 10-bp bands for the 3G allele, and 94-,
37- and 10-bp bands for the 3C allele after separation on 3%
agarose gels. For samples with 2R/2R or 2R/3R genotypes, the G/C
polymorphism in the 2R allele was determined by PCR-PERFLP method,
consisting of PCR followed by primer extension (PE) and RFLP
analysis with HaeIII digestion. The PCR reaction was
performed using reverse primer TS21: 5'-CAGCTCCGAGCCGGCCACAG-3'
instead of TS18. Five microliters of PCR product was mixed with
extension primer TS105: 5'-TCCGAGCCAGCCACAGGCAT-3' labeled with
fluorescein 5'-isothiocyanate to a total volume of 7.5 µl. The
mixture was denatured for 5 min at 98˚C, annealed for 10 min at
room temperature and then combined with 2.5 µl of PE reaction
mixture containing 0.5 unit of Vent (exo-) DNA polymerase (New
England BioLabs, Ipswich, MA), 200 µM deoxyribonucleoside
triphosphates, 1X ThermoPol Reaction Buffer provided by the
manufacturer, followed by incubation for 10 min at 72˚C. The
product of primer extension was digested with HaeIII,
separated on 3% agarose gels and visualized with the Typhoon
fluoroimager. The 2G allele produced a 48 bp fragment and the 2C
allele a 76 bp fragment with the fluorescein 5'-isothiocyanate
label. The TS genotype was thus classified into 2G/2G, 2G/2C,
2C/2C, 2G/3G, 2G/3C, 2C/3G, 2C/3C, 3G/3G, 3G/3C, or 3C/3C by
comprehensive genotyping of the VNTR and SNP in the TS 5'UTR.
Analyses were performed at least twice to confirm the genotype.
LOH analysis
LOH of the TS locus was determined in three distinct
ways depending on the TS genotype observed in the normal tissue.
The G/C SNP in the 2R allele was not taken into consideration for
LOH analyses. Samples that were 2R/3G or 2R/3C were analyzed by PCR
followed by separation on Spreadex gel (Elchrom Scientific, Cham,
Switzerland). Samples that were 2R/2R, 3G/3G or 3C/3C were
evaluated for LOH using the microsatellite marker D18S59, as
described previously (18). Samples
that were 3G/3C were analyzed using the PCR-PERFLP method, as
described above for the SNP genotyping method with the 2R allele.
PCR-PERFLP avoids interference due to heteroduplex formation,
thereby allowing the exact allele ratio to be determined. The 3G
allele produced a 76 bp fragment and the 3C allele a 104 bp
fragment with PCR-PERFLP, as visualized by Typhoon fluoroimaging.
The image was analyzed using ImageQuant software and the relative
ratio between 3G and 3C alleles in tumor DNA was normalized using
the ratio measured in the corresponding normal tissue DNA sample.
LOH was defined as either the complete absence of one allele, or a
decrease in intensity of one allele by at least 50%. LOH of 18q was
analyzed using the microsatellite markers D18S58, D18S61 and
D18S64. Forward primers were labeled with fluorescein
5'-isothiocyanate and the same method as for microsatellite marker
D18S59 was used.
Statistical analysis
Relationships between variables were analyzed by
Chi-square analysis or the Scheffe post-hoc test used following
ANOVA. The cumulative survival rate was estimated using the
Kaplan-Meier method and statistical significance was assessed by
the log-rank test. Cox regression modelling was used for
multivariate analysis. P-values less than 0.05 were considered
significant.
Results
Genotype analysis in normal tissue
samples
We previously investigated and compared the TS
genotypes between matched normal and tumor tissues from 151
patients with colorectal cancer. The results suggest that frequent
LOH of the TS locus detected in the tumors affect the functional TS
genotyping (18). Therefore, in the
present study, TS genotyping was carried out on DNA from normal
tissue rather than from tumor samples in order to avoid possible
artifacts from LOH. The VNTR genotype distribution amongst the 246
cases was: 2R/2R (n=9), 2R/3R (n=70), 3R/3R (n=162) and 3R/5R
(n=5). Because the 5R allele is rare and analysis of the G/C SNP in
the repeat component is difficult, the 5 cases with 3R/5R genotype
were excluded. The remaining 241 cases of 2R/2R, 2R/3R and 3R/3R
VNTR genotype cases were then screened for the G/C SNP in both the
2R and 3R alleles (Fig. 1). The
combined VNTR and SNP genotype frequencies were: 2G/2C (n=1), 2C/2C
(n=8), 2G/3G (n=3), 2G/3C (n=3), 2C/3G (n=25), 2C/3C (n=39), 3G/3G
(n=48), 3G/3C (n=81) and 3C/3C (n=33). The previously reported 2R
allele with a G→C SNP located in the first tandem repeat (17) was not found in our subjects. This
should have resulted in a 121 bp fragment following PCR-PERFLP and
gel electrophoresis (Fig. 1B). The
2G allele was quite rare in our population and showed no
significant associations with any clinicopathological variable or
with clinical course (data not shown). Therefore, SNP information
for the 2R allele was not considered in further analysis. The
distribution of clinicopathological features according to TS
VNTR/SNP genotype are shown in Table
I. As reported previously (14), the 3G allele was less frequent in
females (P=0.04, chi-square test) (male G=115, female G=62, male
C=79, female C=68). No significant associations were apparent
between the normal tissue TS genotype and any other
clinicopathological feature.
 | Figure 1TS VNTR and SNP analysis. (A) VNTR
analysis using PCR amplification and separation of products on a 3%
agarose gel. (B) SNP analysis in the 2R allele using PERFLP
followed by separation on a 3% agarose gel and scanning with a
fluoroimager. (C) SNP analysis in the 3R allele by RFLP and
separation on a 3% agarose gel. The DNA fragments stained using
ethidium bromide are displayed as white pixels on a black
background, whereas those labeled by fluoresein are displayed as
black pixels on a white background. The numbers on these panels
indicate the same samples being analyzed. The genotypes are as
follows: 1, 2G/2C; 2, 2C/2C; 3, 2G/3C; 4, 2C/3G; 5, 2C/3C; 6,
3G/3G; 7, 3G/3C; and 8, 3C/3C. M indicates the size marker (50,
100, 200 and 300 bp) and NC indicates that there was no template
control. TS, thymidylate synthase gene; VNTR, variable number
tandem repeat; SNP, single nucleotide polymorphisml RFLP,
restriction fragment length polymorphism. |
 | Table IThymidylate synthase genotype and
clinicopathological features. |
Table I
Thymidylate synthase genotype and
clinicopathological features.
| | 2R/3R | 3R/3R |
|---|
| Parameter | 2R/3R | 2R/3G | 2R/3C | 3G/3G | 3G/3C | 3C/3C |
|---|
| Total | 9 | 28 | 42 | 48 | 81 | 33 |
| Sex | | | | | | |
|
Male | 5 | 18 | 23 | 37 | 41 | 19 |
|
Female | 4 | 10 | 19 | 11 | 40 | 14 |
| Age (years) | | | | | | |
|
Mean | 63.3 | 70.4 | 63.7 | 66.3 | 65.5 | 66.3 |
|
SD | 15.8 | 11.1 | 11.2 | 13.2 | 11.1 | 13.5 |
| Stage | | | | | | |
|
I | 1 | 4 | 6 | 3 | 7 | 3 |
|
II | 5 | 5 | 17 | 18 | 32 | 10 |
|
III | 3 | 13 | 17 | 14 | 27 | 13 |
|
IV | 0 | 6 | 2 | 13 | 15 | 7 |
| Tumor site | | | | | | |
|
Proximal | 2 | 9 | 12 | 19 | 32 | 9 |
|
Distal | 7 | 19 | 30 | 29 | 49 | 24 |
| Pathology | | | | | | |
|
Tub1 | 7 | 8 | 17 | 21 | 39 | 13 |
|
Tub2 | 1 | 17 | 21 | 23 | 36 | 16 |
|
Muc | 1 | 0 | 1 | 2 | 1 | 2 |
|
Por | 0 | 3 | 3 | 2 | 5 | 2 |
LOH and the residual TS allele in
colorectal cancer
The appropriate method for LOH analysis was selected
according to the genotype found in the normal tissue, as shown in
Fig. 2. The genotype frequency
according to LOH status is also shown in Fig. 2. A novel method for LOH analysis of
the 3G/3C genotype was employed in this study and involved the use
of PCR-PERFLP to avoid interference with heteroduplex product
(Fig. 3). Fifteen cases could not
be evaluated for LOH status because both the TS genotype and the
D18S59 microsatellite marker were homozygous. Therefore, complete
TS VNTR/SNP genotype information together with tumor LOH status was
available for 226 patients. The frequencies were: 2R/2R (n=3),
2R/3G (n=11), 2R/3C (n=20), 3G/3G (n=13), 3G/3C (n=34), 3C/3C
(n=14), 2R/loss (n=26), 3G/loss (n=58) and 3C/loss (n=47). The
overall frequency of LOH for the TS locus was 58.0% (131/226).
Lossed alleles were evenly distributed between 2R (n=23), 3G (n=58)
and 3C (n=50). Associations between LOH status and
clinicopathological features are shown in Table II. The absence of LOH for TS was
significantly associated with proximal tumor location and with
mucinous histology. No other statistically significant associations
were observed.
 | Table IILoss of heterozygosity of the
thymidylate synthase locus and clinicopathological features. |
Table II
Loss of heterozygosity of the
thymidylate synthase locus and clinicopathological features.
| Parameter | No LOH | LOH | P-value |
|---|
| Total | 95 | 131 | |
| Sex | | | 0.63 |
|
Male | 55 | 80 | |
|
Female | 40 | 51 | |
| Age (years) | | | 0.16 |
|
Mean | 64.6 | 66.9 | |
|
SD | 12.9 | 11.3 | |
| Stage | | | 0.61 |
|
I | 11 | 12 | |
|
II | 30 | 52 | |
|
III | 39 | 46 | |
|
IV | 15 | 21 | |
| Tumor site | | | <0.0001 |
|
Proximal | 48 | 30 | |
|
Distal | 47 | 101 | |
| Pathology | | | 0.038 |
|
Tub1 | 42 | 55 | |
|
Tub2 | 39 | 69 | |
|
Muc | 6 | 1 | |
|
Por | 8 | 6 | |
Patient prognosis and TS LOH
status
A number of studies have reported that LOH at a
given gene locus is associated with poor prognosis. We therefore
analyzed the prognostic significance of TS LOH status before
examining the prognostic role of the TS genotype. This was
performed for 153 patients with clinical stage II or III disease
who underwent curative surgery and where clinical information
including long term follow-up and use of adjuvant therapy was
available. Of these patients, 90 (59%) showed TS LOH and had
significantly shorter survival (P=0.0005) compared to patients with
no LOH (n=63; Fig. 4). We also
compared the frequency of LOH between the tumors at clinical stage
II and III by Chi-square test. We found no statistical difference
in frequency of LOH between the two groups of tumors (P=0.22).
Multivariate analysis with the parameters listed in Table III demonstrated that TS LOH
status, receipt of adjuvant chemotherapy and clinical stage were
independent prognostic factors in this patient cohort.
 | Table IIIMultivariate analysis of prognostic
factors. |
Table III
Multivariate analysis of prognostic
factors.
| Variable | Hazard ratio | 95% CI | P-value |
|---|
| TS LOH | | | |
|
Yes vs.
no | 3.01 | 1.36-6.64 | 0.0065 |
| Adjuvant
chemotherapy | | | |
|
Yes vs.
no | 0.53 | 0.28-0.98 | 0.045 |
| Sex | | | |
|
Male vs.
female | 1.22 | 0.66-2.26 | 0.53 |
| Age | | | |
|
≥66 vs.
<66 | 1.15 | 0.62-2.16 | 0.66 |
| Stage | | | |
|
II vs.
III | 0.53 | 0.29-0.96 | 0.039 |
| Tumor site | | | |
|
Proximal vs.
distal | 0.63 | 0.28-1.43 | 0.27 |
| Pathology | | | |
|
Tub2 vs.
Tub1 | 1.27 | 0.368-2.38 | 0.45 |
|
Muc vs.
Tub1 | 2.42 | 0.50-11.7 | 0.27 |
|
Por vs.
Tub1 | 0.53 | 0.068-4.21 | 0.55 |
TS LOH is often accompanied by 18q
LOH
TS is located on 18p11.32. Earlier studies reported
that 18q LOH was associated with poor prognosis in CRC (20,21).
Thus, we analyzed the relation between TS LOH and 18q LOH in 41
randomly selected samples comprising 15 cases with no TS LOH and 26
cases with TS LOH. Three microsatellite markers (D18S58, D18S61,
D18S64) located at 18q22.3, 18q22.2 and 18q21.32, respectively,
were used to determine 18q LOH. Fig.
5 shows the TS LOH status as well as that of each 18q
microsatellite marker. Whenever LOH was observed at the TS gene
locus, it was also consistently present at other 18q loci. On the
other hand, three tumors (from patients no. 4, 13 and 16 shown in
Fig. 5) showed LOH at one or more
18q microsatellite markers in the absence of LOH at TS. In these
tumors, LOH was not observed for all three markers, indicating the
chromosomal loss occurred in a relatively small area of 18q. These
results suggest that LOH at TS and 18q are simultaneous events in
most CRC, although a few tumors have small areas of LOH at 18q
without allelic loss at the TS locus.
TS genotype as a prognostic and
predictive factor in tumors with LOH
Since TS LOH was a strong prognostic factor
(Fig. 4) and also influences the TS
genotype observed in tumors, we explored the role of TS genotype
separately in patient groups stratified according to their LOH
status. In patients without TS LOH (n=63), the tumor genotype is
identical to that found in normal tissue. These patients were
classified into 6 groups: 2R/2R (n=2), 2R/3G (n=6), 2R/3C (n=13),
3G/3G (n=10), 3G/3C (n=23) and 3C/3C (n=9). The relatively small
number of cases for each genotype prevented analysis of the
prognostic value of these groups. When the genotypes were grouped
into L-type (2R/2R, 2R/3C, 3C/3C; n=24) and H-type (2R/3G, 3G/3C,
3G/3G; n=39) according to criteria from our previous report
(14), no prognostic significance
was observed in the overall patient group, in patients treated by
surgery alone, or in patients treated with adjuvant chemotherapy
(data not shown).
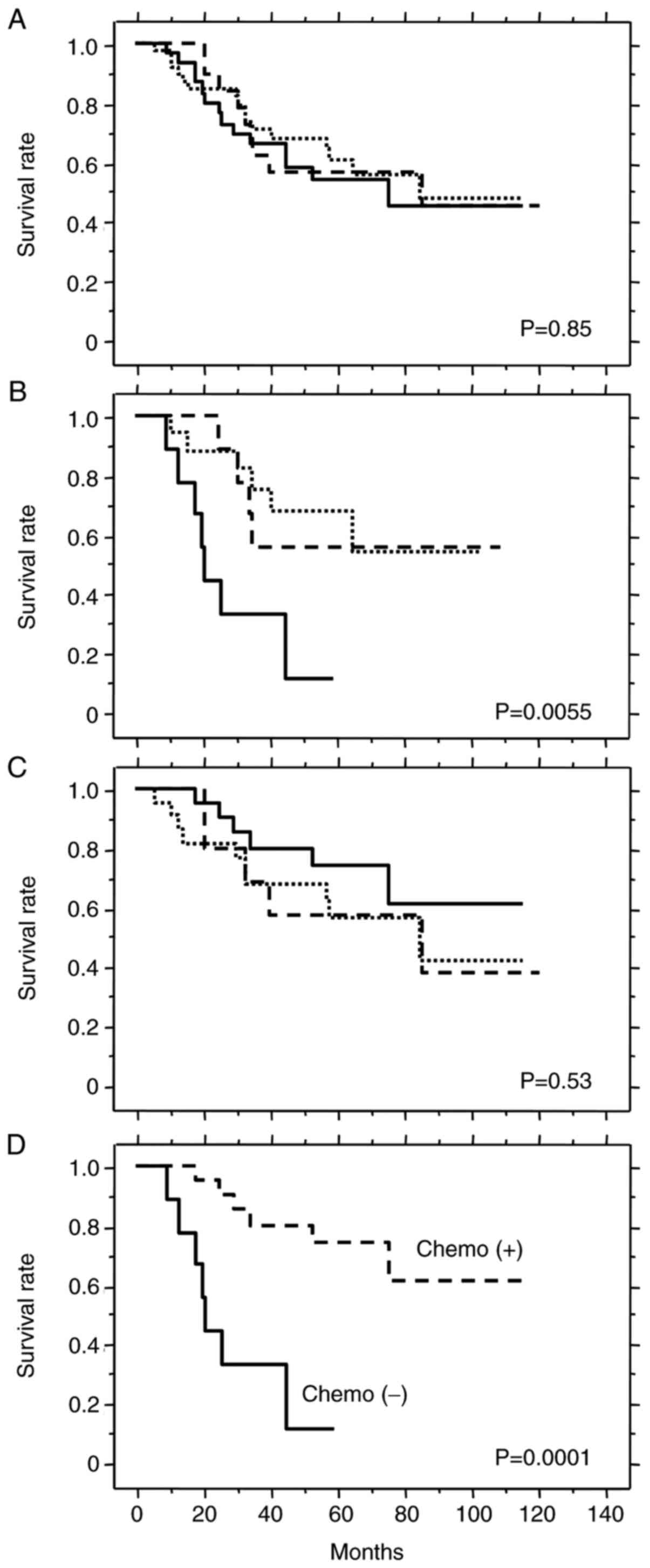
In patients with TS LOH (n=90), the tumor TS
genotypes were: 2R/loss (n=20), 3G/loss (n=39) and 3C/loss (n=31).
No prognostic significance was observed for these genotypes in the
overall group of patients with TS LOH (Fig. 6A) or in patients who received
adjuvant chemotherapy (Fig. 6C).
However, the 3C/loss genotype was associated with significantly
shorter survival in patients treated by surgery alone (Fig. 6B). These results suggest that
patients with the 3C/loss genotype have poor prognosis when treated
by surgery alone, but may benefit from chemotherapy as observed by
the similar survival rate to patients with other genotypes. Indeed,
patients with the 3C/loss genotype who received adjuvant
chemotherapy survived significantly longer than those treated by
surgery alone (Fig. 6D). Thus, the
3C/loss genotype appears to be a prognostic marker for poor
outcome, as well as a predictive marker for good response to
5-FU-based chemotherapy.
Discussion
In this report we genotyped TS for VNTR status and
for SNPs located within the 2R and 3R alleles. We have evaluated
the TS locus for LOH in CRC. In agreement with our previous
observations, LOH was quite frequent regardless of the TS genotype
(18,19). TS LOH was a prognostic factor for
poor survival (Fig. 4),
independently of clinical stage and other clinicopathological
features. Because of the high frequency of TS LOH (58%) and also
its significant prognostic impact, the TS genotype cannot be
combined with LOH status to give one simple prognostic indicator
similar for example to the ‘3G-containing type’ we used previously
(14). The 3G/3G and 3G/loss
genotypes are identical in that both have only the 3G allele,
however their prognostic significance is quite different due to the
presence of LOH in the latter. Important prognostic information
derived from the LOH status would be lost if the 3G/3G and 3G/loss
genotypes were combined into a simple ‘3G type’.
In exploring the predictive value of a given factor
for adjuvant chemotherapy, it is also important to consider its
prognostic value in the absence of such treatment. The TS LOH
status is essential for the correct use of TS genotype as a
prognostic and predictive factor in 5-FU-based adjuvant
chemotherapy. The different number of TS genotypes in tumor DNA is
another reason to stratify patients according to their TS LOH
status. In cases with no LOH, 6 major TS genotypes are observed
(2R/2R, 2R/3G, 2R/3C, 3G/3G, 3G/3C, 3C/3C) whereas three groups are
seen in cases with LOH (2R/loss, 3G/loss, 3C/loss). Future
investigations into the role of TS genotype in the clinical setting
will require large patient cohorts so that the LOH status of TS can
also be taken into account.
In the current study we classified patients
according to their TS LOH status and then followed by investigating
the prognostic and predictive significance of TS genotype. In cases
with no LOH, the overall patient group showed relatively good
prognosis (Fig. 4) and there were 6
major genotype groups, making it difficult to obtain statistically
meaningful results because of the relatively small patient numbers.
Moreover, no prognostic or predictive significance was observed
when these 6 genotypes were classified into just two groups (H and
L) according to our previous results (14). This indicates that TS genotype is
not a useful marker in patients without TS LOH, although study of a
larger number of patients is required to confirm this
observation.
Cases with TS LOH showed poor prognosis (Fig. 4). These were further classified into
three simple TS genotype groups (2R/loss, 3G/loss, 3C/loss) in
order to explore their prognostic and predictive values (Fig. 6). The 3C/loss genotype was a marker
for poor prognosis in patients treated by surgery alone (Fig. 6B). Furthermore, the 3C/loss genotype
also predicted good response to 5-FU-based adjuvant chemotherapy
(Fig. 6D). Despite the relatively
small number of patients (n=31), this result reached a high level
of statistical significance (P=0.0001). Using an in vitro
reporter assay, we previously showed the 3C allele was associated
with lower translational activity compared to the 3G allele
(14). Low expression of TS mRNA
(6) and of the TS protein (8) in CRC have both been associated with
good response to 5-FU-based chemotherapy. The current result
showing the TS 3C/loss genotype is a marker for good response to
5-FU-based adjuvant chemotherapy (Fig.
6D) is therefore consistent with our previous in vitro
observations and with the results of Salonga et al (6) and Soong et al (8). Although we cannot explain why this
genotype was associated with poor prognosis (Fig. 6B), the result concurs with a
previous study showing that low TS expression is a marker of worse
prognosis in patients treated by surgery alone (8).
Due to the retrospective nature of this study and
the potential for biases, further analyses are required to validate
the results, particularly for the TS genotype groups in cases with
LOH. The 2G allele was rare and no 2R allele with the G→C SNP in
the first tandem repeat was found in our patient cohort, suggesting
that SNP typing of the 2R allele can be omitted in further studies
of the Japanese population. However, there is considerable ethnic
variation in the allele frequency for 2R and the incidence is
higher in Western populations (22). Therefore, additional analysis of the
SNP in the 2R allele may be required for Caucasian populations.
The LOH status of TS was closely associated with 18q
LOH status, with the latter being reported previously as a
prognostic factor in CRC (23,24).
The mechanism by which 18q LOH is linked to poor prognosis of CRC
patients is not known, although the loss of several tumor
suppressor genes in this region including DCC, SMAD4
and SMAD22 has been implicated. The current study sheds
light on loss of the whole of chromosome 18 as a prognostic factor.
LOH of 18p and 18q should be analyzed simultaneously to investigate
whether the minimally lost regions on 18q or the whole allelic loss
of chromosome 18 have stronger prognostic significance.
TS LOH is a significant prognostic factor in CRC.
Furthermore, the 3C/loss genotype appears to be prognostic in
patients treated by surgery alone and predictive in patients who
receive 5-FU-based adjuvant chemotherapy. Since TS LOH status
influences the TS genotype of tumors and also has a significant
prognostic role, TS LOH should be incorporated into all future
studies of TS genotype, particularly in relation to its predictive
value. Stratification of CRC patients into subgroups defined by TS
LOH status is therefore essential in obtaining clear evidence for a
clinical role of the TS genotype.
Acknowledgements
The authors would like to thank Dr. Barry Iacopetta
(School of Surgery, University of Western Australia) for the
critical reading of the manuscript and for providing valuable
suggestions.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MKo and KK conceived and designed the current study.
MKo, HB, MKa and KK collected clinical samples. MKo, HB, MKa, HT
and KK performed the experiments and analyzed the data. MKo, TM and
KK drafted the manuscript. MKo and TM confirmed the authenticity of
all the raw data. All authors read and approved the final version
of manuscript.
Ethics approval and consent to
participate
The current study was performed in accordance with
the Declaration of Helsinki. Since tissues used in this study were
obtained from the patients diagnosed between 1999 and 2010, written
informed consent was available for most but not all patients.
However in accordance with Japanese ethical guidelines and law, the
study protocol was reviewed and approved by the Kanazawa University
Human Genome/Gene Analysis Research Ethics Committee (approval nos.
181 and 264). Following instruction by the Ethics Committee at
approval, all patients were publicly provided with an opportunity
to opt-out from registration to the current study. None declined.
All samples were anonymized before analysis was performed to
guarantee the protection of privacy.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
NIH consensus conference. Adjuvant therapy
for patients with colon and rectal cancer. JAMA. 264:1444–1450.
1990.PubMed/NCBI
|
|
2
|
Benson AB III, Schrag D, Somerfield MR,
Cohen AM, Figueredo AT, Flynn PJ, Krzyzanowska MK, Maroun J,
McAllister P, Van Cutsem E, et al: American Society of Clinical
Oncology recommendations on adjuvant chemotherapy for stage II
colon cancer. J Clin Oncol. 22:3408–3419. 2004.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Schmoll HJ, Van Cutsem E, Stein A,
Valentini V, Glimelius B, Haustermans K, Nordlinger B, van de Velde
CJ, Balmana J, Regula J, et al: ESMO Consensus Guidelines for
management of patients with colon and rectal cancer. a personalized
approach to clinical decision making. Ann Oncol. 23:2479–2516.
2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Gill S, Loprinzi CL, Sargent DJ, Thomé SD,
Alberts SR, Haller DG, Benedetti J, Francini G, Shepherd LE,
Francois Seitz J, et al: Pooled analysis of fluorouracil-based
adjuvant therapy for stage II and III colon cancer: Who benefits
and by how much? J Clin Oncol. 22:1797–1806. 2004.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Danenberg PV: Thymidylate synthetase-a
target enzyme in cancer chemotherapy. Biochim Biophys Acta.
473:73–92. 1977.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Salonga D, Danenberg KD, Johnson M,
Metzger R, Groshen S, Tsao-Wei DD, Lenz HJ, Leichman CG, Leichman
L, Diasio RB and Danenberg PV: Colorectal tumors responding to
5-fluorouracil have low gene expression levels of dihydropyrimidine
dehydrogenase, thymidylate synthase, and thymidine phosphorylase.
Clin Cancer Res. 6:1322–1327. 2000.PubMed/NCBI
|
|
7
|
Edler D, Glimelius B, Hallström M,
Jakobsen A, Johnston PG, Magnusson I, Ragnhammar P and Blomgren H:
Thymidylate synthase expression in colorectal cancer: A prognostic
and predictive marker of benefit from adjuvant fluorouracil-based
chemotherapy. J Clin Oncol. 20:1721–1728. 2002.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Soong R, Shah N, Salto-Tellez M, Tai BC,
Soo RA, Han HC, Ng SS, Tan WL, Zeps N, Joseph D, et al: Prognostic
significance of thymidylate synthase, dihydropyrimidine
dehydrogenase and thymidine phosphorylase protein expression in
colorectal cancer patients treated with or without
5-fluorouracil-based chemotherapy. Ann Oncol. 19:915–919.
2008.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Iacopetta B, Grieu F, Joseph D and Elsaleh
H: A polymorphism in the enhancer region of the thymidylate
synthase promoter influences the survival of colorectal cancer
patients treated with 5-fluorouracil. Br J Cancer. 85:827–830.
2001.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Tsuji T, Hidaka S, Sawai T, Nakagoe T,
Yano H, Haseba M, Komatsu H, Shindou H, Fukuoka H, Yoshinaga M, et
al: Polymorphism in the thymidylate synthase promoter enhancer
region is not an efficacious marker for tumor sensitivity to
5-fluorouracil-based oral adjuvant chemotherapy in colorectal
cancer. Clin Cancer Res. 9:3700–3704. 2003.PubMed/NCBI
|
|
11
|
Scartozzi M, Maccaroni E, Giampieri R,
Pistelli M, Bittoni A, Del Prete M, Berardi R and Cascinu S:
5-Fluorouracil pharmacogenomics: Still rocking after all these
years? Pharmacogenomics. 12:251–265. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Horie N, Aiba H, Oguro K, Hojo H and
Takeishi K: Functional analysis and DNA polymorphism of the
tandemly repeated sequences in the 5'-terminal regulatory region of
the human gene for thymidylate synthase. Cell Struct Funct.
20:191–197. 1995.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Mandola MV, Stoehlmacher J, Muller-Weeks
S, Cesarone G, Yu MC, Lenz HJ and Ladner RD: A novel single
nucleotide polymorphism within the 5'tandem repeat polymorphism of
the thymidylate synthase gene abolishes USF-1 binding and alters
transcriptional activity. Cancer Res. 63:2898–2904. 2003.PubMed/NCBI
|
|
14
|
Kawakami K and Watanabe G: Identification
and functional analysis of single nucleotide polymorphism in the
tandem repeat sequence of thymidylate synthase gene. Cancer Res.
63:6004–6007. 2003.PubMed/NCBI
|
|
15
|
Kawakami K, Omura K, Kanehira E and
Watanabe Y: Polymorphic tandem repeats in the thymidylate synthase
gene is associated with its protein expression in human
gastrointestinal cancers. Anticancer Res. 19:3249–3252.
1999.PubMed/NCBI
|
|
16
|
Kawakami K, Salonga D, Park JM, Danenberg
KD, Uetake H, Brabender J, Omura K, Watanabe G and Danenberg PV:
Different lengths of a polymorphic repeat sequence in the
thymidylate synthase gene affect translational efficiency but not
its gene expression. Clin Cancer Res. 7:4096–4101. 2001.PubMed/NCBI
|
|
17
|
Lincz LF, Scorgie FE, Garg MB and Ackland
SP: Identification of a novel single nucleotide polymorphism in the
first tandem repeat sequence of the thymidylate synthase 2R allele.
Int J Cancer. 120:1930–1934. 2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Kawakami K, Ishida Y, Danenberg KD, Omura
K, Watanabe G and Danenberg PV: Functional polymorphism of the
thymidylate synthase gene in colorectal cancer accompanied by
frequent loss of heterozygosity. Jpn J Cancer Res. 93:1221–1229.
2002.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Uchida K, Hayashi K, Kawakami K, Schneider
S, Yochim JM, Kuramochi H, Takasaki K, Danenberg KD and Danenberg
PV: Loss of heterozygosity at the thymidylate synthase (TS) locus
on chromosome 18 affects tumor response and survival in individuals
heterozygous for a 28-bp polymorphism in the TS gene. Clin Cancer
Res. 10:433–439. 2004.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Jen J, Kim H, Piantadosi S, Liu ZF, Levitt
RC, Sistonen P, Kinzler KW, Vogelstein B and Hamilton SR: Allelic
loss of chromosome 18q and prognosis in colorectal cancer. N Engl J
Med. 331:213–221. 1994.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lanza G, Matteuzzi M, Gafá R, Orvieto E,
Maestri I, Santini A and del Senno L: Chromosome 18q allelic loss
and prognosis in stage II and III colon cancer. Int J Cancer.
79:390–395. 1998.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Marsh S, Collie Duguid ES, Li T, Liu X and
McLeod HL: Ethnic variation in the thymidylate synthase enhancer
region polymorphism among Caucasian and Asian populations.
Genomics. 58:310–312. 1999.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ogunbiyi OA, Goodfellow PJ, Herfarth K,
Gagliardi G, Swanson PE, Birnbaum EH, Read TE, Fleshman JW, Kodner
IJ and Moley JF: Confirmation that chromosome 18q allelic loss in
colon cancer is a prognostic indicator. J Clin Oncol. 16:427–433.
1998.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Watanabe T, Wu TT, Catalano PJ, Ueki T,
Satriano R, Haller DG, Benson AB III and Hamilton SR: Molecular
predictors of survival after adjuvant chemotherapy for colon
cancer. N Engl J Med. 344:1196–1206. 2001.PubMed/NCBI View Article : Google Scholar
|