Introduction
Human lung malignancies are one of the leading
causes of cancer-associated death and account for ~17% of annual
new tumor diagnoses worldwide. Lung cancers are categorized into
the following subtypes: Lung squamous cell lung carcinoma (L-SCC;
accounting for ~30%) and non-SCCs, which include e.g., lung
adenocarcinomas (L-AC; ~40%) and large cell carcinomas (~10%)
(1,2). Approximately 90% of L-SCCs are
induced by mutagens, particularly toxins inhaled during smoking
(1,3).
Accordingly, clinicians and oncologists are
confronted with high numbers of lung cancer patients seeking
treatment and cure. There are numerous therapeutic options
available, including cytotoxic chemotherapy with carboplatin or
cisplatin, or erlotinib or cetuximab to target epidermal growth
factor, ipilimumab or nivolumab (inhibitors of the immune system)
and ramucirumab (antiangiogenetic mechanism of action) (1). Ongoing research aims to target and
interfere with additional metabolic pathways in cancer cells to
increase sensitivity and response to therapies in lung cancer
patients; numerous studies were performed using murine lung cancer
cell lines (4-6).
The majority of commonly used murine cancer cell
lines were established decades ago. More uniform in shape than
human chromosomes, murine chromosomes are difficult to characterize
by banding cytogenetics, which resulted in a lack of detailed
genetic characterization despite the widespread use of these cell
lines (7,8). The urethane-induced murine lung
cancer cell line LA-4 (also known as LA4 or LA 4), utilized in ~70
studies in the literature, exemplifies the cytogenomic
under-characterization of murine tumor cell lines. LA-4 was
established in 1975(9) from a lung
adenoma induced in a (most likely female) A/He mouse. Cells from a
primary tumor were cultured and 50 cells were cytogenetically
analyzed; on average, 114 (range, 54-133) chromosomes per metaphase
were counted. Stoner et al (9) reported that the subclones LA-1 to
LA-6 were established from the primary tumor; however, further
details were only reported for LA-4 and at present, the other 5
cell lines are no longer available. Reports of LA-4 documented
epithelial morphology with slower growth compared to the primary
tumor. In addition, LA-4 cells did not have any tumorigenic
potential in nude mice. Solid-stain cytogenetics revealed 116
(range, 87-132; 80% of cells within the range of 106-122)
chromosomes (9). According to the
American Type Culture Collection (ATCC) webpage (accessed from
Germany, but cell stocks are the same worldwide if purchased from
ATCC, https://www.lgcstandards-atcc.org/Products/All/CCL-196.aspx?geo_country=de#characteristics),
LA-4 cells have 38-256 chromosomes per cell, with rearranged
chromosomes in ~12% of the metaphases.
As comprehensive cytogenomic characterization of
murine tumor cell lines may be performed using murine multicolor
banding (mcb) combined with molecular karyotyping (8), the present study provided the first
karyotype (including ploidy level) for the LA-4 cell line, with an
overview of chromosomal imbalances and in silico translation
to regions of homology within the human genome; thus, it was
possible to determine for which human tumor type this cell line may
be used as model.
Materials and methods
Cell line
The murine LA-4 cell line (no. CCL-196™; ATCC), were
grown via adherent culture as per the supplier's protocol in Ham's
F-12K medium, supplemented with fetal bovine serum and dimethyl
sulfoxide (all from Thermo Fisher Scientific, Inc.). Subsequently,
tandem cytogenetic analysis and whole genomic DNA extraction were
performed (10) and analyses were
performed as outlined below.
Molecular cytogenetics and
karyotyping
Fluorescence in situ hybridization (FISH) was
performed as previously described using whole-chromosome paints
(‘SkyPaint™ DNA kit M-10 for Mouse Chromosomes’; Applied Spectral
Imaging) for multicolor-FISH and murine chromosome-specific mcb
probe mixes for FISH-banding (10). At least 30 metaphases were analyzed
for each probe set (Zeiss Axioplan microscopy; Zeiss AG), equipped
with ISIS software v2.86 (MetaSystems). Array comparative genomic
hybridization (aCGH) was performed via standard procedures using
the ‘SurePrint G3 Mouse CGH Microarray, 4x180K’ (Agilent
Technologies, Inc.) (10).
Data analysis
Imbalances and breakpoints of LA-4 were determined
from the mcb and aCGH data and aligned to human homologous regions
using Ensembl (https://www.ensembl.org/info/website/tutorials/grch37.html)
and the University of California Santa Cruz (UCSC) Genome Browser
(http://genome-euro.ucsc.edu/cgi-bin/hgGateway?hgsid=95241316&clade=vertebrate&org=Human&db=hg18&redirect=manual&source=genome.ucsc.edu;
GRCh37/hg19), as previously described (10). The data were compared to genetic
changes associated with human cancers (2,11,12).
Results
FISH and aCGH
The LA-4 cell line had a hyper-tetraploid karyotype
with several numerical and structural aberrations. The following
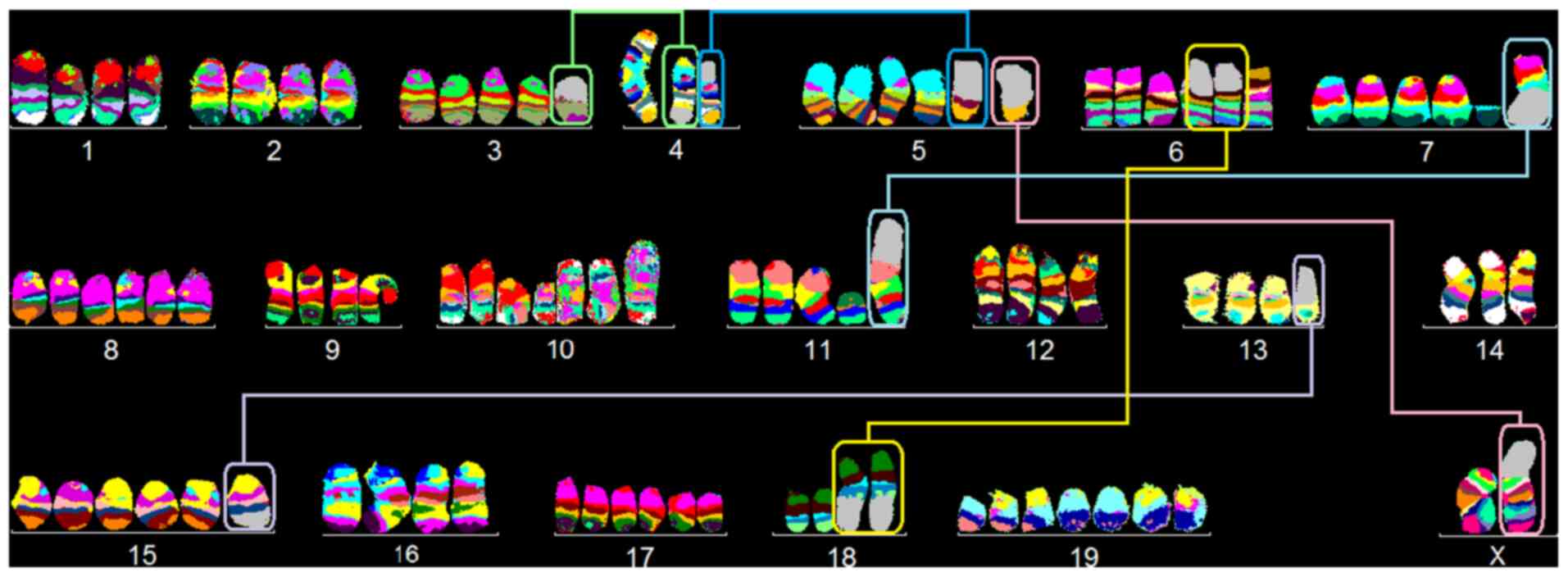
karyotype was observed in 88% of cells (Fig. 1):
85~93<4n>,X,dic(X;5)(Xqter->XA1::5G2->5qter),
-X,-X,-4,-4,der(4)t(3;4)(G;E2),der(5)t(4;5)(A4;C3),del(5)(G2),inv(6)(A1C),+neo(7)(:F3->qter),+dic(7;11)(7A1->7F3::11A2->11qter),+8,+8,del(10)(B5),+idic(10)(A1;A1)x3,neo(11)(:B3->qter),-13,-14,+15,+der(15)t(13;15)(C2;D3),+17,+17,der(18)t(6;18)(B1;E3)x2,+19,+19,+19.
In 12% of LA-4 cells, identical structural
aberrations, but random loss of single chromosomes was observed;
chromosome numbers ranged from 67 to 72. Preferentially lost
chromosomes were 1, 2, 6, 9, 15, 18 and 19.
Overall, the FISH results agreed with the
aCGH-results, as summarized in Fig.
2A. In silico translation to the human genome (only
imbalances >3.5 mega base pairs were included) identified the
corresponding homologous regions (Fig.
2B). Details of aCGH and translation of data are summarized in
Table SI; the original data of
the aCGH experiment are provided in Table SII.
Comparison with the literature
The corresponding translated homologous copy number
variations (CNVs) for LA-4 (Fig.
2B) were compared with common imbalances in related human
cancers (2,11,12)
(Table I). In human L-AC, only
16/36 regions (44%) were affected by CNVs, similar to the cell line
LA-4. Of note, 18/36 (50%) and 21/36 (58%) of CNVs were concordant
with human L-SCCs and head and neck SCC (HNSCC).
 | Table ICopy number changes associated with
molecular subtypes of human L-AC, L-SCC and HNSCC (2,11,12)
compared to those in LA-4 (translated to human). |
Table I
Copy number changes associated with
molecular subtypes of human L-AC, L-SCC and HNSCC (2,11,12)
compared to those in LA-4 (translated to human).
| Chromosomal
region | LA-4 | Human L-AC | Human L-SCC | Human HNSCC |
|---|
| 1p31.3-p22.3 | Gain | Gain | Loss | Loss |
| 2p23~2q12 | Gain | (Gain) | Gain | Gain |
| 3pter~p24.3 | Gain | - | Loss | Loss |
| 3p21.2-3p12 | Loss | - | Loss | Loss |
| ~4 | Gain | Gain | Loss | - |
| ~5p | Gain | Gain | Gain | Gain |
| 5q14q14 | Loss | Gain | Loss | Loss |
| 5q15~q22 | Gain | Gain | Loss | Loss |
| 6pter-p22.1 | Loss | Gain | Gain | Gain |
| 6p22.1-6q12 | Gain | Gain | Gain | Gain |
| 6q15-q15 | Loss | Loss | Gain | Loss |
| 6q16.3-qter | Gain | (Loss) | Gain | Gain |
| ~7pter-p14 | Gain | Gain | Gain | Gain |
| 7p14-p10 | Loss | Gain | Gain | Gain |
| ~7q | Gain | Gain | Gain | Gain |
| ~8p | Gain | - | Loss | - |
| ~8q12-q22.1 | Loss | Gain | Gain | Gain |
| 8q22.1-qter | Gain | Gain | Gain | Gain |
| 9q21.2-q22.3 | Loss | - | Gain | Gain |
| ~10q | Gain | - | Loss | Gain |
| ~11pter-q14.3 | Gain | Gain | - | Gain |
| 12pter-q23 | Gain | Gain | Gain | Gain |
| 12q23-qter | Loss | Gain | - | Gain |
| ~13 | Loss | Gain | Loss | Gain |
| 14q10-q12 | Loss | Gain | Gain | Gain |
| 14q22-q23 | Loss | Gain | - | Gain |
| 15q25-qter | Gain | - | Gain | Gain |
| ~16 | Gain | Gain | Gain | Gain |
| ~17p | Gain | Loss | Loss | Gain |
| ~17q | Gain | Gain | Gain | Gain |
| 18pter-p11.2 | Gain | - | Gain | Gain |
| 18q22-qter | Loss | Loss | Loss | Gain |
| ~19 | Gain | - | Gain | Gain |
| 21q22.3-qter | Gain | - | - | Gain |
| ~22 | Gain | - | Gain | Gain |
| Overall | | 16/36 | 18/36 | 21/36 |
Discussion
The LA-4 cell line is derived from a mutagen
(urethane)-induced primary tumor. In 1975, LA-4 cells were
demonstrated to have a karyotype with 87-132 chromosomes and 80% of
the cells had chromosome numbers ranging from 106-122; no obvious
structural aberrations were observed at that time (9). The only other cytogenetic analyses
performed revealed structural aberrations in 12% of the cells with
chromosome numbers ranging from 38 to 256 (https://www.lgcstandards-atcc.org/Products/All/CCL-196.aspx?geo_country=de#characteristics).
In the present study, the majority of LA-4 cells had
85 to 93 chromosomes, i.e., a hyper-tetraploid karyotype. There
were three distinct (pseudo-)dicentric derivatives, two
neocentrics, four unbalanced translocations, two chromosomes with
terminal deletions and one chromosome with a balanced inversion.
Thus, it was not possible to confirm the original observation that
LA-4 harbored no structural rearrangements (9). Also, in contrast to the information
from ATCC, these derivative chromosomes were present in 100% and
not limited to only 12% of the cells, a result confirmed by the
aCGH results (https://www.lgcstandards-atcc.org/Products/All/CCL-196.aspx?geo_country=de#characteristics).
However, as observed by Stoner et al (9), there was a small subpopulation with
only 67-72 chromosomes per cell. The observed dicentric and
neocentric chromosomes are unusual and only rarely reported as
stable derivatives in tumor cell lines; of note, they have also
been observed in human cancer cell lines (13). In previous studies of 23 murine
tumor cell lines, these phenomena were only observed in the
colorectal cancer cell line CMT-93 [for an overview see (14)].
These results indicate that LA-4 underwent a
karyotype evolution during the past 46 years of cell culture.
However, the LA-4 cells examined in the present study had an
overall stable karyotype, with a tendency towards chromosomal loss
in ~12% of cells. Of note, tetraploidization was an early event
that was present in the original tumor (9). Previously documented cell line
evolution, e.g., for HeLa (15),
suggests that variant clones of LA-4 may be present in different
laboratories. Thus, it is highly recommended that an aCGH or
cytogenetic study of the locally available and used LA-4 cells is
performed to compare their chromosomal/genetic constitution with
that reported in the present study prior to further
experimentation.
LA-4 was established from a benign lung cancer, an
adenoma. As reported in the present study, these genetic data
establish that LA-4 cells have evolved in vitro, through the
acquisition of compounding genetic changes, which were not
initially present in 1975(9).
However, the comparison in Table
I, which aligned LA-4 with specific human lung tumor types, was
rather inconclusive at first observation. Aberrations typical for
human L-ACs were ~44% concordant and human L-SCCs were ~50%
concordant. Of note, LA-4 cells exhibited ~58% concordance with
imbalances observed in human HNSCCs. Thus, it may be cautiously
proposed that LA-4 cells demonstrate characteristic (genetic)
features most consistent with human SCC cells. As their lung origin
is indubitable and they are mutagen-induced, similar to the
majority of human L-SCCs, LA-4 may be considered a well-suited
model for non-metastatic human L-SCC.
Supplementary Material
Regions of copy number gain and loss,
and breakpoints of balanced rearrangements observed in LA-4, with
the corresponding homologous regions in humans listed by cytoband
and position (GRCh37/hg19).
Original data of array comparative
genomic hybridization, also including data of imbalances too small
to be included in this evaluation.
Acknowledgements
The technical support from Dr Nadezda Kosyakova
(Jena University Hospital, Friedrich Schiller University, Institute
of Human Genetics, Jena, Germany) and the help of Dr Heather E.
Williams (Columbia University Irving Medical Center, Department of
Pathology & Cell Biology, New York, USA) in revising the
English language of the manuscript are gratefully acknowledged.
Funding
Funding: The present study was supported by the Wilhelm Sander
Stiftung (grant no. 2013.032.1).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article and in Tables SI and SII.
Authors' contributions
TL conceived the study and obtained funding. SA, MB,
FK and SK performed the FISH analysis. MR performed the aCGH study
and pre-evaluation. SA performed the overall data interpretation.
TL and SA checked and approved the authenticity of the raw data and
drafted and edited the manuscript. All authors read and agreed to
the final draft of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Derman BA, Mileham KF, Bonomi PD, Batus M
and Fidler MJ: Treatment of advanced squamous cell carcinoma of the
lung: A review. Transl Lung Cancer Res. 4:524–532. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Yakut T, Schulten HJ, Demir A, Frank D,
Danner B, Egeli U, Gebitekin C, Kahler E, Gunawan B, Urer N, et al:
Assessment of molecular events in squamous and non-squamous cell
lung carcinoma. Lung Cancer. 54:293–301. 2006.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Barbone F, Bovenzi M, Cavallieri F and
Stanta G: Cigarette smoking and histologic type of lung cancer in
men. Chest. 112:1474–1479. 1997.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Nurwidya F, Andarini S, Takahashi F,
Syahruddin E and Takahashi K: Implications of insulin like growth
factor 1 receptor activation in lung cancer. Malays J Med Sci.
23:9–21. 2016.PubMed/NCBI
|
|
5
|
Hashemi-Sadraei N and Hanna N: Targeting
FGFR in squamous cell carcinoma of the lung. Target Oncol.
12:741–755. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tan AC: Targeting the PI3K/Akt/mTOR
pathway in non-small cell lung cancer (NSCLC). Thorac Cancer.
11:511–518. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Leibiger C, Kosyakova N, Mkrtchyan H, Glei
M, Trifonov V and Liehr T: First molecular cytogenetic high
resolution characterization of the NIH 3T3 cell line by murine
multicolor banding. J Histochem Cytochem. 61:306–312.
2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Azawi S, Liehr T, Rincic M and Manferrari
M: Molecular cytogenomic characterization of the murine breast
cancer cell lines C 127I, EMT6/P and TA3 Hauschka. Int J Mol Sci.
21(4716)2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Stoner GD, Kikkawa Y, Kniazeff AJ, Miyai K
and Wagner RM: Clonal isolation of epithelial cells from mouse lung
adenoma. Cancer Res. 35:2177–2185. 1975.PubMed/NCBI
|
|
10
|
Kubicova E, Trifonov V, Borovecki F, Liehr
T, Rincic M, Kosyakova N and Hussein SS: First molecular
cytogenetic characterization of murine malignant mesothelioma cell
line AE17 and in silico translation to the human genome. Curr
Bioinform. 12:11–18. 2017.
|
|
11
|
Wolff E, Girod S, Liehr T, Vorderwülbecke
U, Ries J, Steininger H and Gebhart E: Oral squamous cell
carcinomas are characterized by a rather uniform pattern of genomic
imbalances detected by comparative genomic hybridisation. Oral
Oncol. 34:186–190. 1998.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sy SM, Wong N, Lee TW, Tse G, Mok TS, Fan
B, Pang E, Johnson PJ and Yim A: Distinct patterns of genetic
alterations in adenocarcinoma and squamous cell carcinoma of the
lung. Eur J Cancer. 40:1082–1094. 2004.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Camps J, Mrasek K, Prat E, Weise A, Starke
H, Egozcue J, Miró R and Liehr T: Molecular cytogenetic
characterisation of the colorectal cancer cell line SW480. Oncol
Rep. 11:1215–1218. 2004.PubMed/NCBI
|
|
14
|
Rhode H, Liehr T, Kosyakova N, Rinčic M
and Azawi SSH: Molecular cytogenetic characterization of two murine
colorectal cancer cell lines. OBM Genet. 2(037)2018.
|
|
15
|
Frattini A, Fabbri M, Valli R, De Paoli E,
Montalbano G, Gribaldo L, Pasquali F and Maserati E: High
variability of genomic instability and gene expression profiling in
different HeLa clones. Sci Rep. 5(15377)2015.PubMed/NCBI View Article : Google Scholar
|