Introduction
Colorectal cancer (CRC) is a serious threat to
public health, as it ranks second in terms of mortality and third
in terms of incidence. Over 1.8 million new colorectal cancer cases
and 881,000 cancer-related deaths occurred in 2018, accounting for
approximately 1 in 10 cancer cases and deaths (1). Although progress has been made in the
early diagnosis of CRC and the systemic therapy, the prognosis for
advanced CRC remains poor. Therefore, it is of utmost importance to
identify new sensitive and specific biomarkers for the prognosis of
patients with CRC.
The Rho GTPase-activating protein 25 (ARHGAP25) gene
is located on the human chromosome 2p13 and encodes a protein of
639 amino acids with a potential Rho/Rac GAP domain (151-340 aa)
(2). The ARHGAP25 protein is a
member of the ARHGAP family and plays an important role in
regulating B cell chemotaxis and the germinal center reaction
through the CXCL12/CXCR4 pathway (3). ARHGAP25 is present in leukocytes
where it works as a negative regulator of the phagocytosis of
neutrophils, and transendothelial migration of leukocytes (4,5). Our
previous work demonstrated that ARHGAP25 overexpression
significantly inhibited CRC cell growth, suppressed cell migration
and invasion, and reduced the expression of MMPs, EMT-associated
factors and β-catenin. The expression of ARHGAP25 in colorectal
cancer tissues was markedly lower than that in the normal adjacent
tissue. In addition, the data of ARHGAP25 expression collected from
both the GEO database and TCGA database indicated that ARHGAP25
mRNA expression was downregulated in patients with CRC when
compared with the corresponding healthy controls (6). In the present study, the expression
of ARHGAP25 in tumor tissues of patients with CRC was detected, and
its association with the clinical characteristics and prognosis of
patients was analyzed. Therefore, ARHGAP25 was used as a predictive
target for prognosis, which is innovative and prospective.
Materials and methods
Patients
Patients who underwent colorectal tumor resection at
the Longhua Hospital of Shanghai University of Traditional Chinese
Medicine from February 24, 2009 to June 1, 2017, were included in
this study. The main inclusion criteria were as follows: i)
Patients who underwent resection of a large intestine tumor; ii)
assessment of histologically proven colorectal cancer, staging and
resectable specimens in accordance with the International Union
against Cancer, 2017 (UICC) guidelines; and iii) patients who were
well informed about the long-term follow-up. The main exclusion
criteria were the following: i) Presence of another cancer; ii)
presence of serious heart, liver or kidney complications; and iii)
pregnant or lactating women, as well as children and individuals
suffering from mental illness.
Ethical statement
The present study was approved by the Institutional
Review Board of Longhua Hospital Shanghai University of Traditional
Chinese Medicine (Approval no. 2018LCSY004). The study was
performed in full agreement with the national ethical and
regulatory guidelines, and with the 1964 Declaration of Helsinki
and its later amendments or comparable ethical standards. Informed
consent was obtained from all participants included in the present
study. All patients signed a written informed consent to
participate in this study.
Tissue sample collection
A sample of fresh tissue (including surgical and
bioptic tissue) was selected; the collected CRC tissue sample
included the malignant tumor tissue and the adjacent tissue
surgically removed or obtained from the biopsy. After the surgery,
the tissue samples were collected under the guidance of clinical
tumor pathologists to avoid misdiagnosis, with the following
citeria: a) Sampling sites included colorectal cancer tissue and
adjacent tissue (the latter 1-3 cm away from the cancerous tissue);
b) clear clinical diagnosis with the removal of the specimens
without radiotherapy or chemotherapy performed under the guidance
of pathologists; c) the survival of the cancerous tissue was
ensured; d) the surgically removed tissue samples were quickly
submerged into liquid nitrogen and then stored in a liquid nitrogen
tank or at -80˚C within 30 min from the collection of the surgical
specimen; e) images of the samples were obtained using a digital
camera; then, they were cut into small pieces but not too small
(size 0.5x0.5 cm, thickness <0.5 cm), placed into the frozen
storage tube, and into the specimen box. The label was attached on
the lid to record the specimen number, collection time and specimen
type. A total of 3-5 cryopreservation tubes containing the
specimens were stored for each patient, and each tissue weighed no
less than 2 g.
Expression of ARHGAP25
Immunohistochemistry (IHC) was performed to assess
ARHGAP25 expression. Specific steps were carried out according to
the instructions of the immunohistochemistry kit. In brief, antigen
repair was performed using 0.01 M sodium citrate buffer at high
pressure for 15 min, followed by cooling for 5 min and washing with
PBS buffer 3 times for 3 min. Subsequently, 10% goat serum (diluted
20 times with PBS) (cat. no. SL038; Beijing Solarbio Science &
Technology Co., Ltd.) was directly added to the 4-mm thick tissue
sections and incubated at 37˚C for 10 min. They were then incubated
overnight with the primary antibody against ARHGAP25 (diluted
1:200; rabbit IgG1; product code ab192020; Abcam) at 4˚C. Next,
they were incubated with the secondary antibody (diluted 1:1,000;
product no. D-3004-100; Shanghai Long Island Biotech Co., Ltd.) at
25˚C for 30 min. Finally, the slides were incubated with DAB
staining solution at room temperature for 15 min (product no.
FL-6001-03; Shanghai Long Island Biotech Co., Ltd.) until the
required staining levels were reached and counterstained with
hematoxylin for 3 min (product code BA4097; BASO), followed by
differentiation with 1% alcohol hydrochloric acid and light
microscopic observation to control the degree of staining. The
sections were then rinsed with running water for 10 min and placed
in an oven at 65˚C to remove all moisture. The slides were observed
under an Olympus inverted light microscope at a magnification of
x200. The expression of ARHGAP25 was mainly observed in the
cytoplasm of the tissue cells as brownish yellow particles. The
expression of ARHGAP25 was subjected to log2
transformation before the statistical analysis. The median method
was used to classify colorectal cancer as high and low expression
of ARHGAP25, and the median value was 3.07. All samples were
divided into high and low ARHGAP25 groups, with the median ARHGAP25
value as the cutoff point: The ARHGAP25-high expression group was
the one with an expression higher than the median value, the
ARHGAPDH25-low expression group was the one with an expression
lower than the median value.
Semi-quantification of IHC
A simple method of automated digital IHC image
analysis algorithm was used for an unbiased, quantitative
assessment of the intensity of the antibody staining in the tissue
sections. The IHC Profiler, an open source plugin for the
quantitative evaluation and automated scoring of IHC images of
human tissue samples was used to calculate the positive area of CRC
tissue and adjacent tissue. The IHC Profiler, which is compatible
with the ImageJ software version 1.53 (National Institutes of
Health), performs IHC analysis using color deconvolution and
computerized pixel profiling leading to the assignment of an
automated score to the respective image. This method used in the
present study has been thoroughly validated using high volume IHC
digital datasets representing multiple protein markers expressed in
the cytoplasm (7,8).
Main outcome
The primary outcome was the overall survival (OS),
defined as the the date of surgical resection of the colorectal
tumor until death from any cause. The follow-up was based on
appointments, scheduled questionnaires, and telephone surveys, and
was carried out until the end of the study on May 31, 2020, which
served as the establishment of an outcome and endpoints.
Variables
The medical records and electronic records of the
patients were analyzed, and the following parameters were
collected: Demographic factors (age and sex), tumor-related factors
(degree of invasion, lymph node metastasis, distant metastasis, TNM
stage, nerve invasion, vascular invasion, resection edge, tumor
deposition, tumor location, histological type and histological
grade), expression of the tumor-associated protein ARHGAP25 and
postoperative treatment radiotherapy, chemotherapy, targeted
therapy and traditional Chinese medicine (TCM).
Statistical analysis
A total of 153 patients with colorectal cancer were
included in this study, and among these, 124 had complete data and
no missing values. The median follow-up time was 54.9 months
[interquartile range (IQR): 27.30, 60.33]. Among the 153 eligible
patients, 42 (27.5%) succumbed to CRC, 77 (50.3%) survived, and the
remaining 34 were lost to follow-up.
The scatter diagram was plotted using the GraphPad
Prism 8.0.2 software (GraphPad Software, Inc.), and the statistical
significance was analyzed by paired t-test. The OS curve was
plotted using the Kaplan Meier method, and the statistical
significance was calculated by log-rank test and Gehan Breslow
Wilcoxon test. The aforementioned data were analyzed by GraphPad
Prism 8.0.2 software.
Cox proportional hazards regression analysis was
used for univariate analysis, multivariate analysis and
hierarchical analysis. Five models based on variable adjustment
were constructed in multivariate analysis: M0, no variable; M1,
age, sex; M2, degree of invasion, lymph node metastasis, distant
metastasis, TNM stage; M3, tumor location, histological type,
histological grade, tumor deposition; M4, chemotherapy, TCM
treatment; M5, age, sex, degree of invasion, lymph node metastasis,
distant metastasis, TNM stage, tumor location, histological type,
histological grade, tumor deposition, chemotherapy, TCM treatment.
The risk ratio (HR) with 95% confidence interval (CI) and two-sided
P-values were calculated. Variables containing >20% missing
values and zero variance were deleted. The present study used three
methods to overcome the missing values (<20%): Complete case
analysis, multiple interpolation and missing index method (9-11).
Values of P<0.05 were considered to indicate a statistically
significant difference. The aforementioned data were analyzed by
the software R programming language, R version 3.5.3 (https://www.r-project.org/).
Results
ARHGAP25 expression in CRC and tumor
adjacent tissues
The results of IHC on 26 pairs of CRC tissues and
corresponding adjacent tissues revealed that the expression of
ARHGAP25 in the adjacent tissues was higher than that in the CRC
tissues (P=0.0016). The statistical difference was analyzed
by paired t-test (Fig. 1A).
Moreover, the sample size was increased by the addition of 127 CRC
tissue samples, for a total of 153 CRC tissues that were subjected
to IHC: a) Adjacent tissues (ARHGAP25), the ARHGAP25-positive area
was 2,785; b) CRC tissue (low ARHGAP25), the ARHGAP25-positive area
was 520; c) CRC tissue (high ARHGAP25), the ARHGAP25-positive area
was 1,001; d) adjacent tissues (ARHGAP25), ARHGAP25-positive area
was 3,448; e) CRC tissue (low ARHGAP25), the ARHGAP25-positive area
was 280; and f) CRC tissue (high ARHGAP25), the ARHGAP25-positive
area was 1,071. The magnification was x200 (Fig. 1B).
 | Figure 1(A) IHC on 26 pairs of CRC tissues and
corresponding adjacent tissues showing that the expression of
ARHGAP25 in the adjacent tissues was higher than that in the CRC
tissues (P=0.0016). The statistical difference was evaluated by
paired t-test. (B) The expression of ARHGAP25 in CRC and tumor
adjacent tissues was detected with IHC, dividing the patients into
low and high ARHGAP25 expression. (a) adjacent tissues (ARHGAP25),
the ARHGAP25-positive area was 2,785; (b) CRC tissue (low
ARHGAP25), the ARHGAP25-positive area was 520; (c) CRC tissue (high
ARHGAP25), the ARHGAP25-positive area was 1,001; (d) adjacent
tissues (ARHGAP25), the ARHGAP25-positive area was 3,448; (e) CRC
tissue (low ARHGAP25), the ARHGAP25-positive area was 280; and (f)
CRC tissue (high ARHGAP25), the ARHGAP25-positive area was 1,071.
Original magnification, x200. IHC, immunohistochemistry; ARHGAP25,
Rho GTPase-activating protein 25; CRC, colorectal cancer; IQR,
interquartile range. |
Patient characteristics
A total of 153 patients suffering from colorectal
cancer were enrolled in the present study, and among them, 124 had
complete data with statistical analysis and no missing values. The
details of the baseline characteristics are listed in Table I.
 | Table IBaseline characteristics of patients
with colorectal cancer. |
Table I
Baseline characteristics of patients
with colorectal cancer.
| Variables | Overall | Missing |
|---|
| Age, median
(IQR) | 69.00 (61.00,
79.00) | 0 |
| ARHGAP25, median
(IQR) | 3.07 (2.86,
3.21) | 0 |
| OS, median (IQR) | 45.27 (27.30,
60.33) | 0 |
| Sex (%) | | 0 |
|
Male | 93 (60.78) | |
|
Female | 60 (39.22) | |
| Invasion degree
(%) | | 1 (0.65) |
|
T1 | 6 (3.92) | |
|
T2 | 36 (23.53) | |
|
T3 | 65 (42.49) | |
|
T4 | 45 (29.41) | |
| Lymph node
metastasis (%) | | 0 |
|
No | 87 (56.86) | |
|
Yes | 66 (43.14) | |
| Tumor deposits
(%) | | 0 |
|
No | 138 (90.20) | |
|
Yes | 15 (9.80) | |
| Distant metastasis
(%) | | 1 (0.65) |
|
No | 142 (92.81) | |
|
Yes | 10 (6.54) | |
| TNM stage (%) | | 0 |
|
I | 39 (25.49) | |
|
II | 42 (27.45) | |
|
III | 61 (39.87) | |
|
IV | 11 (7.19) | |
| Tumor location
(%) | | 0 |
|
Right
hemicolon | 40 (26.14) | |
|
Left
hemicolon | 46 (30.07) | |
|
Rectum | 67 (43.79) | |
| Histological
subtype (%) | | 0 |
|
Adenocarcinoma | 115 (75.16) | |
|
Mucinous
adenocarcinoma | 30 (19.61) | |
|
Signet-ring
cell carcinoma | 3 (1.96) | |
|
Other | 5 (3.27) | |
| Histological grade
(%) | | 4 (2.61) |
|
Well
differentiated | 1 (0.65) | |
|
Moderately
differentiated | 130 (84.97) | |
|
Poorly
differentiated | 17 (11.11) | |
|
Undifferentiated | 1 (0.65) | |
| Chemotherapy
(%) | | 27 (17.65) |
|
No | 42 (27.45) | |
|
Yes | 84 (54.90) | |
| TCM treatment
(%) | | 27 (17.65) |
|
No | 51 (33.33) | |
|
Yes | 75 (49.02) | |
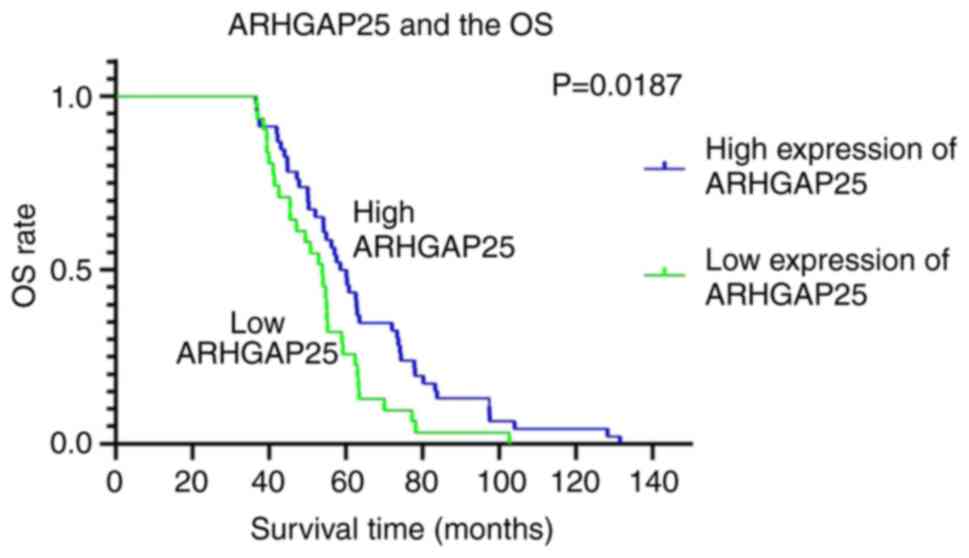
Correlation between ARHGAP25
expression and OS
The total 153 patients with CRC were divided into
two subgroups according to the low and high ARHGAP25 expression,
which accounted for 49.67% (76/153) and 50.33% (77/153),
respectively. The OS of patients with CRC with high expression of
ARHGAP25 was significantly higher than that of the patients with
low expression of ARHGAP25. The median survival of the patients
with high expression of ARHGAP25 was 60.13 months, and the median
survival with those with low expression of ARHGAP25 was 53.87
months (P<0.05) (Fig.
2). A high expression of ARHGAP25 indicated a favorable
prognosis of patients with CRC. The OS curve of ARHGAP25 revealed
that the survival time of patients with high expression of ARHGAP25
was significantly longer than that of patients with low expression
of ARHGAP25 (Fig. 2).
Univariate analysis
Univariate Cox analysis was used to evaluate the
prognostic value of ARHGAP25 expression and other factors affecting
the survival of patients with CRC using complete data. The results
revealed that the expression of ARHGAP25 was associated with the
improvement of OS in patients with CRC (P=0.011; HR, 0.213; 95% CI,
0.065-0.701). In addition, age (P=0.012; HR, 1.051; 95% CI,
1.011-1.093), lymph node metastasis (P<0.001; HR, 5.312; 95% CI,
2.191-12.879), distant metastasis (P=0.007; HR, 5.529; 95% CI,
1.584-19.295), TNM stage (P=0.0205; HR, 4.356; 95% CI,
1.254-15.13), signet ring cell carcinoma (P=0.001; HR, 13.625; 95%
CI, 3.025-61.366), and tumor deposition (P=0.011; HR, 3.566; 95%
CI, 1.333-9.538) were associated with adverse OS in patients with
CRC (Table II).
 | Table IIUnivariate analysis of the prognostic
value of ARHGAP25 expression and other prognostic factors for
predicting overall survival. |
Table II
Univariate analysis of the prognostic
value of ARHGAP25 expression and other prognostic factors for
predicting overall survival.
| Variables | No. of
patients | HR | 95% CI | P-value |
|---|
| ARHGAP25 | 124 | 0.213 | 0.065-0.701 | 0.011 |
| Age | 124 | 1.051 | 1.011-1.093 | 0.012 |
| Sex | | | | |
|
Male | 75 | NA | NA | NA |
|
Female | 49 | 1.542 | 0.702-3.387 | 0.281 |
| Invasion
degree | | | | |
|
T1 | 6 | NA | NA | NA |
|
T2 | 30 | 0.966 | 0-Inf | 0.999 |
|
T3 | 58 | 4.63E+08 | 0-Inf | 0.999 |
|
T4 | 30 | 2.22E+08 | 0-Inf | 0.999 |
| Lymph node
metastasis | | | | |
|
No | 76 | NA | NA | NA |
|
Yes | 48 | 5.312 | 2.191-12.879 | <0.001 |
| Tumor deposits | | | | |
|
No | 115 | NA | NA | NA |
|
Yes | 9 | 3.566 | 1.333-9.538 | 0.011 |
| Distant
metastasis | | | | |
|
No | 117 | NA | NA | NA |
|
Yes | 7 | 5.529 | 1.584-19.295 | 0.007 |
| TNM stage | | | | |
|
Non-IV | 116 | NA | NA | NA |
|
IV | 8 | 4.356 | 1.254-15.13 | 0.0205 |
| Tumor location | | | | |
|
Right
hemicolon | 33 | NA | NA | NA |
|
Left
hemicolon | 41 | 0.387 | 0.14-1.071 | 0.068 |
|
Rectum | 50 | 0.459 | 0.186-1.133 | 0.091 |
| Histological
type | | | | |
|
Adenocarcinoma | 92 | NA | NA | NA |
|
Mucinous
adenocarcinoma | 27 | 1.865 | 0.751-4.63 | 0.179 |
|
Signet ring
cell carcinoma | 2 | 13.625 | 3.025-61.366 | 0.001 |
|
Other | 3 | 19.727 | 3.907-99.592 | <0.001 |
| Histological
grade | | | | |
|
Well | 1 | NA | NA | NA |
|
Moderate | 107 | 2443062 | 0-Inf | 0.997 |
|
Poor | 15 | 8535588 | 0-Inf | 0.997 |
|
Undifferentiated | 1 | 1.9E+08 | 0-Inf | 0.997 |
| Chemotherapy | | | | |
|
No | 42 | NA | NA | NA |
|
Yes | 82 | 1.105 | 0.474-2.577 | 0.817 |
| TCM treatment | | | | |
|
No | 51 | NA | NA | NA |
|
Yes | 73 | 0.661 | 0.299-1.463 | 0.307 |
Multivariate analysis
Multivariate analysis was performed to determine
whether the expression of ARHGAP25 was still associated with OS
after adjusting for other variables. The complete case analysis
revealed the following results: M0 (adjusted P=0.011; adjusted HR,
0.213; 95% CI, 0.065-0.701), M1 (adjusted P=0.017; adjusted HR,
0.238; 95% CI, 0.073-0.777), M2 (adjusted P=0.024; adjusted HR,
0.168; 95% CI, 0.036-0.789), M3 (adjusted P=0.034; adjusted HR,
0.220; 95% CI, 0.055-0.889), M4 (adjusted P=0.012; adjusted HR,
0.221; 95% CI, 0.068-0.721) and M5 (adjusted P=0.003; adjusted HR,
0.096; 95% CI, 0.021-0.449) (Fig.
3).
 | Figure 3Forest plot of the multivariate
analysis data of the prognostic value of ARHGAP25 expression for
predicting overall survival in patients with colorectal cancer. M0,
no variables; M1, age and sex; M2, invasion degree, lymph node
metastasis, distant metastasis and TNM stage; M3, tumor location,
histological type, histological grade and tumor deposits; M4,
chemotherapy and TCM treatment; M5, age, sex, invasion degree,
lymph node metastasis, distant metastasis, TNM stage, tumor
location, histological type, histological grade, tumor deposits,
chemotherapy and TCM treatment. ARHGAP25, Rho GTPase-activating
protein 25; TCM, traditional Chinese medicine. |
Multiple imputation analysis revealed the following
results: M0 (adjusted P=0.009; adjusted HR, 0.280; 95% CI,
0.108-0.724); M1 (adjusted P=0.008; adjusted HR, 0.280; 95% CI,
0.110-0.716); M2 (adjusted P=0.012; adjusted HR, 0.244; 95% CI,
0.081-0.735); M3 (adjusted P=0.007; adjusted HR, 0.246; 95% CI,
0.089-0.680); M4 (adjusted P=0.009; adjusted HR, 0.283; 95% CI,
0.110-0.728); M5 (adjusted P<0.001; adjusted HR, 0.106; 95% CI,
0.030-0.369) (Fig. 3).
The analysis of missing index cases resulted in the
following results: M0 (adjusted P=0.008; adjusted HR, 0.280; 95%
CI, 0.109-0.722); M1 (adjusted P=0.008; adjusted HR, 0.283; 95% CI,
0.111-0.722); M2 (adjusted P=0.012; adjusted HR, 0.244; 95% CI,
0.081-0.736); M3 (adjusted P=0.010; adjusted HR, 0.267; 95% CI,
0.098-0.727); M4 (adjusted P=0.746; adjusted HR, 0.280; 95% CI,
0.108-0.728); M5 (adjusted P=0.005; adjusted HR, 0.220; 95% CI,
0.077-0.632) (Fig. 3). The
addition of different grouping variables still revealed that
ARHGAP25 expression was associated to the improvement of OS,
indicating that ARHGAP25 is a protective factor to prevent poor
prognosis in patients with CRC.
Stratified analysis
The analysis with complete cases revealed that the
predictive value of ARHGAP25 for OS in patients with CRC was
stronger in males, elderly patients (>70 years old), T3 stage,
lymph node metastasis, TNM stage III, right hemicolon tumor
location and poor differentiation (P<0.05). In addition,
ARHGAP25 expression had a significant effect on patients with CRC
without distant metastasis, without tumor deposits and without
having received TCM treatment (P<0.05).
The analysis with missing-indicator cases produced
results similar to those of the analysis with complete cases. In
fact, it was revealed that ARHGAP25 expression was relevant to OS
in elderly patients with CRC (>70 years old), males, T3-4 stage,
lymph node metastasis, TNM stage III and poor differentiation
(P<0.05). Additionally, ARHGAP25 expression was a significant
predictor of OS in patients without distant metastasis, without
tumor deposits and without having received TCM treatment
(P<0.05).
The analysis with multiple imputation cases also
revealed that ARHGAP25 expression was associated to OS in patients
with CRC >70 years and male (P<0.05). Moreover, ARHGAP25
expression had a significant effect on patients with CRC with T3-4
stage, lymph node metastasis (adjusted P=0.007), TNM stage III and
poorly differentiated adenocarcinoma (P<0.05). Additionally,
ARHGAP25 expression was a significant predictor of OS in patients
without distant metastasis, without tumor deposits, who received
chemotherapy and without having received TCM treatment (P<0.05)
(Table III).
 | Table IIIStratified analysis of the
correlation between ARHGAP25 expression and the prognosis of
patients with colorectal cancer. |
Table III
Stratified analysis of the
correlation between ARHGAP25 expression and the prognosis of
patients with colorectal cancer.
| | Complete | | Missing
indicator | | Multiple
imputation |
|---|
| Variables | Total | HR | 95% CI | P-value | Total | HR | 95% CI | P-value | Total | HR | 95% CI | P-value |
|---|
| Age | | | | | | | | | | | | |
|
≤70 | 69 | 0.517 | (0.044-6.098) | 0.600 | 83 | 0.323 | (0.061-1.715) | 0.185 | 83 | 0.323 | (0.061-1.715) | 0.185 |
|
>70 | 55 | 0.160 | (0.040-0.639) | 0.010 | 70 | 0.234 | (0.074-0.735) | 0.013 | 70 | 0.234 | (0.074-0.735) | 0.013 |
| Sex | | | | | | | | | | | | |
|
Male | 75 | 0.084 | (0.016-0.429) | 0.003 | 93 | 0.117 | (0.035-0.397) | 0.001 | 93 | 0.117 | (0.035-0.397) | 0.001 |
|
Female | 49 | 0.633 | (0.106-3.793) | 0.616 | 60 | 0.962 | (0.203-4.562) | 0.961 | 60 | 0.962 | (0.203-4.563) | 0.961 |
| Invasion
degree | | | | | | | | | | | | |
|
T1 | 6 | NA | NA | NA | 6 | NA | NA | NA | 6 | NA | NA | NA |
|
T2 | 30 | NA | NA | NA | 36 | 0.708 | (0.019-26.436) | 0.852 | 37 | 0.708 | (0.019-26.438) | 0.852 |
|
T3 | 58 | 0.130 | (0.028-0.598) | 0.009 | 65 | 0.165 | (0.041-0.660) | 0.011 | 65 | 0.165 | (0.041-0.661) | 0.011 |
|
T4 | 30 | 0.029 | (0.000-8.144) | 0.218 | 45 | 0.126 | (0.021-0.757) | 0.024 | 45 | 0.126 | (0.021-0.757) | 0.024 |
|
Missing | | | | | 1 | NA | NA | NA | | | | |
| Lymph node
metastasis | | | | | | | | | | | | |
|
No | 76 | 0.258 | (0.034-1.945) | 0.189 | 87 | 0.374 | (0.070-2.013) | 0.252 | 87 | 0.374 | (0.070-2.013) | 0.252 |
|
Yes | 48 | 0.106 | (0.017-0.649) | 0.015 | 66 | 0.175 | (0.050-0.616) | 0.007 | 66 | 0.175 | (0.050-0.616) | 0.007 |
| Distant
metastasis | | | | | | | | | | | | |
|
No | 117 | 0.252 | (0.070-0.909) | 0.035 | 142 | 0.312 | (0.111-0.877) | 0.027 | 143 | 0.313 | (0.111-0.878) | 0.027 |
|
Yes | 7 | 0.083 | (0.000-24.291) | 0.391 | 10 | 0.080 | (0.002-3.206) | 0.180 | 10 | 0.080 | (0.002-3.254) | 0.181 |
|
Missing | | | | | 1 | NA | NA | NA | | | | |
| TNM stage | | | | | | | | | | | | |
|
I | 32 | NA | NA | NA | 39 | 0.900 | (0.022-37.516) | 0.956 | 39 | 0.900 | (0.022-37.519) | 0.956 |
|
II | 39 | 0.073 | (0.005-1.099) | 0.059 | 42 | 0.077 | (0.006-0.100) | 0.050 | 42 | 0.077 | (0.006-0.100) | 0.050 |
|
III | 45 | 0.134 | (0.018-0.978) | 0.047 | 61 | 0.215 | (0.054-0.852) | 0.029 | 61 | 0.215 | (0.054-0.852) | 0.029 |
|
IV | 8 | 0.052 | (0.000-8.258) | 0.252 | 11 | 0.058 | (0.002-1.836) | 0.106 | 11 | 0.058 | (0.002-1.836) | 0.106 |
| Tumor location | | | | | | | | | | | | |
|
Right
hemicolon | 33 | 0.125 | (0.017-0.920) | 0.041 | 40 | 0.215 | (0.044-1.049) | 0.057 | 40 | 0.215 | (0.044-1.049) | 0.057 |
|
Left
hemicolon | 41 | 1.287 | (0.026-63.899) | 0.899 | 46 | 0.541 | (0.026-11.280) | 0.692 | 46 | 0.541 | (0.026-11.281) | 0.692 |
|
Rectum | 50 | 0.195 | (0.031-1.218) | 0.080 | 67 | 0.321 | (0.088-1.171) | 0.085 | 67 | 0.321 | (0.088-1.171) | 0.085 |
| Histological
type | | | | | | | | | | | | |
|
Adenocarcinoma | 92 | 0.339 | (0.060-1.920) | 0.221 | 115 | 0.358 | (0.111-1.159) | 0.087 | 115 | 0.358 | (0.111-1.159) | 0.087 |
|
Mucinous
adenocarcinoma | 27 | 0.466 | (0.041-5.275) | 0.538 | 30 | 0.549 | (0.058-5.211) | 0.602 | 30 | 0.549 | (0.058-5.212) | 0.602 |
|
Signet ring
cell carcinoma | 2 | NA | NA | NA | 3 | NA | NA | NA | 3 | 0.549 | (0.058-5.212) | 0.602 |
|
Other | 3 | NA | NA | NA | 5 | <0.001 |
(0.000-12336.405) | 0.367 | 5 | <0.001 |
(0.000-12340.361) | 0.367 |
| Histological
grade | | | | | | | | | | | | |
|
Well | 1 | NA | NA | NA | 1 | NA | NA | NA | 1 | NA | NA | NA |
|
Moderate | 107 | 0.373 | (0.087-1.591) | 0.182 | 130 | 0.445 | (0.148-1.338) | 0.150 | 131 | 0.446 | (0.148-1.344) | 0.151 |
|
Poor | 15 | 0.002 | (0.000-0.350) | 0.019 | 17 | 0.007 | (0.000-0.224) | 0.005 | 18 | 0.006 | (0.000-0.214) | 0.005 |
|
Undifferentiated | 1 | NA | NA | NA | 1 | NA | NA | NA | 3 | 0.003 | (0-Inf) | 0.999 |
|
Missing | | | | | 4 | NA | NA | NA | | | | |
| Tumor deposits | | | | | | | | | | | | |
|
No | 115 | 0.217 | (0.059-0.798) | 0.021 | 138 | 0.295 | (0.101-0.861) | 0.026 | 138 | 0.295 | (0.101-0.861) | 0.026 |
|
Yes | 9 | 0.036 | (0.000-5.606) | 0.196 | 15 | 0.039 | (0.001-1.077) | 0.055 | 15 | 0.039 | (0.001-1.077) | 0.055 |
| Chemotherapy | | | | | | | | | | | | |
|
No | 42 | 0.141 | (0.018-1.117) | 0.064 | 42 | 0.141 | (0.018-1.117) | 0.064 | 49 | 0.280 | (0.049-1.587) | 0.150 |
|
Yes | 82 | 0.264 | (0.059-1.175) | 0.080 | 84 | 0.260 | (0.058-1.166) | 0.079 | 104 | 0.282 | (0.086-0.921) | 0.036 |
|
Missing | | | | | 27 | 0.406 | (0.077-2.153) | 0.290 | | | | |
| TCM treatment | | | | | | | | | | | | |
|
No | 51 | 0.122 | (0.027-0.554) | 0.006 | 51 | 0.122 | (0.027-0.554) | 0.006 | 66 | 0.209 | (0.061-0.712) | 0.012 |
|
Yes | 73 | 0.824 | (0.104-6.518) | 0.854 | 75 | 0.830 | (0.102-6.740) | 0.861 | 87 | 0.618 | (0.114-3.360) | 0.578 |
|
Missing | | | | | 27 | 0.406 | (0.077-2.153) | 0.290 | | | | |
Discussion
CRC is a huge global burden in terms of
complications, mortality, side effects after the treatment, use of
health care services, and medical costs (12). Therefore, the use of clinical
samples to identify more new prognostic and predictive biomarkers
is of great significance.
Rac/Rho-like GTPases are important regulators of
several different cell functions, including cell polarity control,
membrane transport, transcription regulation, survival, adhesion,
and proliferation. GTPase-activating proteins (GAP) are negative
regulators of Rac/Rho-like GTPase because they convert thes active
GTP binding state to the inactive GDP binding state (13,14).
According to the homologous catalytic domain, approximately 70
proteins act as Rac/Rho GAP in different human tissues (15,16).
ARHGAP25 protein is a Rac-specific GAP mainly expressed in
hematopoietic cells. The RhoE/ROCK/ARHGAP25 signaling pathway has
been revealed to control the invasion potential of alveolar
rhabdomyosarcoma cells (17). In
addition, ARHGAP25 was revealed to be downregulated in lung cancer
and inhibit its growth, migration and invasion through its effect
on the Wnt/β-catenin signaling pathway (18).
Our previous study (6) showed that the downregulation of
ARHGAP25 in CRC was associated to the progression of CRC, and the
upregulation of ARHGAP25 reduced CRC metastasis in vitro and
in vivo. In addition, ARHGAP25 inhibited the migration and
invasion of CRC cells by inhibiting the Wnt/β-catenin pathway,
reduced the expression of MMPs and inhibited EMT. Moreover,
ARHGAP25 was downregulated in the colon of patients with CRC
compared with normal adjacent tissues (6). Despite this evidence, more studies
are required to further determine the prognostic value of ARHGAP25
expression. Therefore, the present study aimed to evaluate the
importance of ARHGAP25 expression in predicting the outcome of
patients with CRC.
The present study showed that ARHGAP25 expression
was significantly associated with a favorable prognostic value when
analyzed as a continuous variable. Our results indicated that older
age, lymph node metastasis, distant metastasis, signet ring cell
carcinoma, and tumor deposition were particularly associated with
poor OS, which was also consistent with previous studies (19-21).
Multivariate analysis showed that ARHGAP25 was
associated with the improvement of the prognosis of patients with
CRC after adjusting data, suggesting that ARHGAP25 is an
independent prognostic factor for patients with CRC.
Further stratified analysis of the complete case
analysis revealed that ARHGAP25 expression significantly predicted
OS, especially in the elderly and male patients. A previous study
revealed that elderly patients (>75 years) had worse survival
outcomes than younger patients, because younger patients can
receive surgery, radiotherapy, and chemotherapy, while older
patients usually receive palliative care (22). A population-based study conducted
in Germany confirmed the survival advantage of women compared with
men with CRC (23). The present
study similar to other studies also showed that ARHGAP25 expression
had a significant predictive effect on OS in patients with T3
stage, lymph node metastasis, TNM stage III, right-sided colon
cancer, and poorly differentiated patients. T3-4 staging is a risk
factor for the prognosis of CRC (24-26),
and lymph node metastasis is an independent prognostic factor for
patients with CRC (27-29).
An analysis of patients based on disease stages showed that
patients with early tumors have a higher survival rate than
patients with a tumor in an advanced stage (30). Recently, evidence revealed
differences in molecular, pathological and clinical features
between right-sided colon cancer and left-sided colon cancer
(31). In the wild-type RAS
population of CRYSTAL and FIRE-3, the prognosis of patients with
left-sided colon tumors was significantly better than that of
patients with right-sided colon tumors (30). In addition, patients with poor
differentiation have significantly worse OS (32,33).
Our study revealed that ARHGAP25 expression was an important
predictor of OS, especially in patients without tumor deposits.
However, previous studies have revealed that the presence of tumor
deposits is an independent risk factor in the prognosis of patients
with CRC (34,35). This bias in the data may be caused
by the use of a small number of cases with tumor deposits in our
study. In addition, our analysis showed that ARHGAP25 expression
had a prognostic significance in the patients who did not receive
TCM treatment. Our previous study revealed that TCM treatment is a
protective factor for OS (36).
Similarly, the missing index method and multiple
imputation data analysis revealed that ARHGAP25 expression
significantly predicted OS, especially in the elderly, men, T3
stage, lymph node metastasis, or TNM stage III. In addition, our
findings indicated that ARHGAP25 expression significantly predicted
OS in patients with TNM stage IV. Overall, ARHGAP25 exhibited
certain advantages in extending OS.
However there are some limitations in the present
study. The association between serum CEA level, R0 surgical
resection, surgical type (open, laparoscopic, robotic),
postoperative complications and OS were not included in this study.
The detailed reasons are the following: i) Clinically, 5 ng/ml is
generally regarded as the cut-off value of CEA expression. The
association between CEA expression and OS was analyzed by the
log-rank test of survival analysis, and the result was
statistically significant, the lower the CEA expression, the longer
the OS (37). In addition,
considering that the influence of surgery and other therapeutic
means on CEA, dynamically change this value, CEA was not included
in this study. ii) In our study, 153 patients mainly underwent
surgical R0 resection. Only 4 cases underwent R1 resection and 3
cases underwent R2 resection, thus the remaining patients underwent
R0 resection. As is well known, the OS of patients following R0
resection is longer than other surgical resection methods (38,39),
and our study results also reached the same conclusion. Cox
regression of the survival analysis was used to compare the effects
of the other two surgical types (open and laparoscopic) on OS, and
the results were not statistically significant. Thus, they were not
included in this study. iii) The number of postoperative
complications in the 153 patients was extremely low; 5 cases in
total including postoperative bleeding, postoperative wound
infection, vaginal fistula, wound infection, and peritoneal
adhesions. Therefore, they were not included in the comparison.
In conclusion, the present study demonstrated the
association between ARHGAP25 expression and prognosis. Therefore,
ARHGP25 could be considered as a potential indicator for a
favorable prognosis of CRC, particularly in patients with poor
prognostic factors.
Acknowledgements
Not applicable.
Funding
Funding: This research was supported by the National Natural
Science Foundation of China (grant nos. 81603548 and 82174450).
Availability of data and materials
The raw data supporting the findings of this article
are available from the authors, without undue reservation.
Authors' contributions
YuZ and YL acquired, analyzed, and interpreted the
data, drafted the article, and critically revised the study for
important intellectual content. In addition, they contributed
equally to this work. YiZ collected and analyzed the data. XZ
performed the pathological and immunohistochemical analysis. LT
conceived and designed the study, helped in the manuscript
preparation and English language editing and critically revised it
for important intellectual content. MY analyzed and interpreted the
data. YuZ and LT confirm the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of Longhua Hospital Shanghai University of Traditional
Chinese Medicine (Approval no. 2018LCSY004). The study was
performed in full agreement with the national ethical and
regulatory guidelines, and with the 1964 Declaration of Helsinki
and its later amendments or comparable ethical standards. All
patients signed a written informed consent to participate in this
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA, Torre LA and Jemal A: Global cancer statistics 2018:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 68:394–424.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Katoh M and Katoh M: Identification and
characterization of ARHGAP24 and ARHGAP25 genes in silico. Int J
Mol Med. 14:333–338. 2004.PubMed/NCBI
|
|
3
|
Lindner SE, Egelston CA, Huard SM, Lee PP
and Wang LD: Arhgap25 deficiency leads to decreased numbers of
peripheral blood B cells and defective germinal center reactions.
Immunohorizons. 4:274–281. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Csépányi-Kömi R, Sirokmány G, Geiszt M and
Ligeti E: ARHGAP25, a novel Rac GTPase-activating protein,
regulates phagocytosis in human neutrophilic granulocytes. Blood.
119:573–582. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Csépányi-Kömi R, Wisniewski É, Bartos B,
Lévai P, Németh T, Balázs B, Kurz AR, Bierschenk S, Sperandio M and
Ligeti E: Rac GTPase activating protein ARHGAP25 regulates
leukocyte transendothelial migration in mice. J Immunol.
197:2807–2815. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tao L, Zhu Y, Gu Y, Zheng J and Yang J:
ARHGAP25: A negative regulator of colorectal cancer (CRC)
metastasis via the Wnt/β-catenin pathway. Eur J Pharmacol.
858(172476)2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Jensen EC: Quantitative analysis of
histological staining and fluorescence using ImageJ. Anat Rec
(Hoboken). 296:378–381. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Varghese F, Bukhari AB, Malhotra R and De
A: IHC profiler: An open source plugin for the quantitative
evaluation and automated scoring of immunohistochemistry images of
human tissue samples. PLoS One. 9(e96801)2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Choi J, Dekkers OM and le Cessie S: A
comparison of different methods to handle missing data in the
context of propensity score analysis. Eur J Epidemiol. 34:23–36.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Pedersen AB, Mikkelsen EM, Cronin-Fenton
D, Kristensen NR, Pham TM, Pedersen L and Petersen I: Missing data
and multiple imputation in clinical epidemiological research. Clin
Epidemiol. 9:157–165. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
White IR and Carlin JB: Bias and
efficiency of multiple imputation compared with complete-case
analysis for missing covariate values. Stat Med. 29:2920–2931.
2010.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Wong CK, Lam CL, Poon JT, McGhee SM, Law
WL, Kwong DL, Tsang J and Chan P: Direct medical costs of care for
Chinese patients with colorectal neoplasia: A health care service
provider perspective. J Eval Clin Pract. 18:1203–1210.
2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Vega FM and Ridley AJ: Rho GTPases in
cancer cell biology. FEBS Lett. 582:2093–2101. 2008.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Etienne-Manneville S and Hall A: Rho
GTPases in cell biology. Nature. 420:629–635. 2002.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Bernards A: GAPs galore! A survey of
putative Ras superfamily GTPase activating proteins in man and
Drosophila. Biochim Biophys Acta. 1603:47–82. 2003.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Tcherkezian J and Lamarche-Vane N: Current
knowledge of the large RhoGAP family of proteins. Biol Cell.
99:67–86. 2007.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Thuault S, Comunale F, Hasna J, Fortier M,
Planchon D, Elarouci N, De Reynies A, Bodin S, Blangy A and
Gauthier-Rouvière C: The RhoE/ROCK/ARHGAP25 signaling pathway
controls cell invasion by inhibition of Rac activity. Mol Biol Cell
Cell. 27:2653–2661. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Xu K, Liu B and Ma Y: The tumor
suppressive roles of ARHGAP25 in lung cancer cells. Onco Targets
Ther. 12:6699–6710. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Derwinger K, Carlsson G and Gustavsson B:
A study of lymph node ratio as a prognostic marker in colon cancer.
Eur J Surg Oncol. 34:771–775. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Mehrkhani F, Nasiri S, Donboli K, Meysamie
A and Hedayat A: Prognostic factors in survival of colorectal
cancer patients after surgery. Colorectal Dis. 11:157–161.
2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Nitsche U, Friess H, Agha A, Angele M,
Eckel R, Heitland W, Jauch KW, Krenz D, Nüssler NC, Rau HG, et al:
Prognosis of mucinous and signet-ring cell colorectal cancer in a
population-based cohort. J Cancer Res Clin Oncol. 142:2357–2366.
2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Serra-Rexach JA, Jimenez AB,
Garcia-Alhambra MA, Pla R, Vidán M, Rodriguez P, Ortiz J,
García-Alfonso P and Martín M: Differences in the therapeutic
approach to colorectal cancer in young and elderly patients.
Oncologist. 17:1277–1285. 2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Majek O, Gondos A, Jansen L, Emrich K,
Holleczek B, Katalinic A, Nennecke A, Eberle A and Brenner H: GEKID
Cancer Survival Working Group. Sex differences in colorectal cancer
survival: Population-based analysis of 164,996 colorectal cancer
patients in Germany. PLoS One. 8(e68077)2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Compton C, Fenoglio-Preiser CM, Pettigrew
N and Fielding LP: American joint committee on cancer prognostic
factors consensus conference: Colorectal working group. Cancer.
88:1739–1757. 2000.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Newland RC, Dent OF, Lyttle MN, Chapuis PH
and Bokey EL: Pathologic determinants of survival associated with
colorectal cancer with lymph node metastases. A multivariate
analysis of 579 patients. Cancer. 73:2076–2082. 1994.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Shepherd NA, Baxter KJ and Love SB: The
prognostic importance of peritoneal involvement in colonic cancer:
A prospective evaluation. Gastroenterology. 112:1096–1102.
1997.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Park YJ, Park KJ, Park JG, Lee KU, Choe KJ
and Kim JP: Prognostic factors in 2230 Korean colorectal cancer
patients: Analysis of consecutively operated cases. World J Surg.
23:721–726. 1999.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Vasile L, Olaru A, Munteanu M, Pleşea IE,
Surlin V and Tudoraşcu C: Prognosis of colorectal cancer: Clinical,
pathological and therapeutic correlation. Rom J Morphol Embryol.
53:383–391. 2012.PubMed/NCBI
|
|
29
|
Missiaglia E, Jacobs B, D'Ario G, Di Narzo
AF, Soneson C, Budinska E, Popovici V, Vecchione L, Gerster S, Yan
P, et al: Distal and proximal colon cancers differ in terms of
molecular, pathological, and clinical features. Ann Oncol.
25:1995–2001. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Tejpar S, Stintzing S, Ciardiello F,
Tabernero J, Van Cutsem E, Beier F, Esser R, Lenz HJ and Heinemann
V: Prognostic and predictive relevance of primary tumor location in
patients With RAS wild-type metastatic colorectal cancer:
Retrospective analyses of the CRYSTAL and FIRE-3 trials. JAMA
Oncol. 3:194–201. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Derwinger K, Kodeda K, Bexe-Lindskog E and
Taflin H: Tumour differentiation grade is associated with TNM
staging and the risk of node metastasis in colorectal cancer. Acta
Oncol. 49:57–62. 2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kanazawa T, Watanabe T, Kazama S, Tada T,
Koketsu S and Nagawa H: Poorly differentiated adenocarcinoma and
mucinous carcinoma of the colon and rectum show higher rates of
loss of heterozygosity and loss of E-cadherin expression due to
methylation of promoter region. Int J Cancer. 102:225–229.
2002.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Mirkin KA, Kulaylat AS, Hollenbeak CS and
Messaris E: Prognostic significance of tumor deposits in stage III
colon cancer. Ann Surg Oncol. 25:3179–3184. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Nagayoshi K, Ueki T, Nishioka Y, Manabe T,
Mizuuchi Y, Hirahashi M, Oda Y and Tanaka M: Tumor deposit is a
poor prognostic indicator for patients who have stage II and III
colorectal cancer with fewer than 4 lymph node metastases but not
for those with 4 or more. Dis Colon Rectum. 57:467–474.
2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Liu X, Xiu LJ, Jiao JP, Zhao J, Zhao Y, Lu
Y, Shi J, Li YJ, Ye M, Gu YF, et al: Traditional Chinese medicine
integrated with chemotherapy for stage IV non-surgical gastric
cancer: A retrospective clinical analysis. J Integr Med.
15:469–475. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yang X, Hao J, Zhu CH, Niu YY, Ding XL,
Liu C and Wu XZ: Survival benefits of western and traditional
Chinese medicine treatment for patients with pancreatic cancer.
Medicine (Baltimore). 94(e1008)2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Tsouma A, Aggeli C, Lembessis P, Zografos
GN, Korkolis DP, Pectasides D, Skondra M, Pissimissis N, Tzonou A
and Koutsilieris M: Multiplex RT-PCR-based detections of CEA, CK20
and EGFR in colorectal cancer patients. World J Gastroenterol.
16:5965–5974. 2010.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Diaconescu A, Alexandrescu S, Ionel Z,
Zlate C, Grigorie R, Brasoveanu V, Hrehoret D, Ciurea S, Botea F,
Tomescu D, et al: Resection of concomitant hepatic and extrahepatic
metastases from colorectal cancer-a worthwhile operation? Chirurgia
(Bucur). 112:673–682. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Luo M, Chen SL, Chen J, Yan H, Qiu Z, Chen
G, Lu L and Zhang F: Resection vs ablation for lesions
characterized as resectable-ablative within the colorectal liver
oligometastases criteria: A propensity score matching from
retrospective study. PeerJ. 8(e8398)2020.PubMed/NCBI View Article : Google Scholar
|