Introduction
Colon cancer (CC) is among the five most common
cancers in Vietnam and globally. In 2020, 1,148,515 new cases were
reported and 576,858 deaths were attributed to CC worldwide
(1). CC is the fourth deadliest
cancer in Vietnam (both sexes combined), with an increasing
tendency of new cases (2,3). Biomarker discoveries have provided
the possibility for early diagnosis and partly supported mortality
reduction of CC (4). Among
important discovered oncogenes, KRAS proto-oncogene, a
homolog of Kirsten Ras sarcoma virus (KRAS) oncogene in
humans, has been showing the highest detected mutation rate in CC
with >35% of patients (5-7).
Most of the mutations at these sites continuously trigger
mitogen-activated protein kinase (MAPK) pathways and subsequently,
the proliferation of cancer cells (8-12).
The presence of a KRAS mutation is a predictive factor for
resistance to anti-epidermal growth factor receptor (EGFR)
treatment. Therefore, the mutated gene is considered a worse
prognosis in advanced colon cancer (13-17).
However, the impact of KRAS mutation is
controversial in colon cancer stage I-III. The worse prognosis for
survival has been indicated in some research, particularly in some
subgroups determined by disease stage, tumor site, sex, BRAF
wild-type, and microsatellite instability (MSI) status (18-21),
while other studies have not shown these associations (22-24).
KRAS status has been described in colonic polyps and
KRAS mutations were listed as a potential molecular factor for the
risk of developing advanced neoplasia (25,26).
Currently, the progression from polyp to cancer has been
identified. The development of normal epithelial cells to
adenocarcinoma generally follows a progression of histological and
concurrent epigenetic and genetic changes (27). Colon cancer without the occurrence
of polyps has different pathogeneses compared with polyposis
syndrome colon cancer (28,29).
Non-polyp status was examined by colonoscopy and/or confirmed by
observing the dissected tissues after surgery. By gathering the
results of this group, patients with certain polyposis syndrome
were excluded from the present research.
Due to the promotion of KRAS gene mutations to the
tumor invasion and metastatic processes (30), numerous studies have referred to
the KRAS gene as a worse prognostic factor in survival, worldwide.
However, few studies on KRAS mutations in Vietnamese
patients with CC have been reported and the KRAS status for
colon cancer without the occurrence of polyps has not been
described (31,32). In addition, the associations of the
KRAS status with the survival of the Vietnamese patients
with NPCC are also under establishment. Therefore, the present
study aimed to determine KRAS status and possible
associations of KRAS status with clinicopathological features and
survival in non-polyp colon cancer stage II-III in patients from
Vietnam.
Materials and methods
Data in the present study were collected from 194
patients (males, 53.1% and females, 46.9%; median age, 58 years)
with non-polyp colon cancers at stages II or III from January 2016
to August 2020 at The Nuclear Medicine and Oncology Center of Bach
Mai Hospital, The Oncology Department of Viet Duc Hospital, and
National Cancer Hospital (Hanoi, Vietnam). The inclusion criteria
were as follows: i) Stages II-III colon cancer according to the 8th
edition of the American Joint Committee on Cancer (AJCC) staging
system; ii) no occurrence of polyps; iii) radical surgery and
adjuvant therapy with FOLFOX-4 or XELOX; iv) testing for the
KRAS status; and v) access to the medical records of
patients. The exclusion criteria were as follows: i) Diagnosed with
a second cancer; ii) medical history indicating the removal of
colorectal polyps; iii) patients with the appearance of colon
polyps in any numbers at any time; and iv) inability to answer the
research questions due to illness. The present study was approved
(approval no. NCS28/HMU-IRB) by the Ethics Committee of Hanoi
Medical University (Hanoi, Vietnam). Written informed consent was
obtained from all participants.
All patients underwent radical treatment and
followed-up with a scheduled exam at hospitals. Studied variables
included clinical and subclinical features such as age, sex, tumor
site (33), histopathology
(34), cancer stage, and
KRAS status.
DNA isolation and mutation
analysis
KRAS mutations were investigated with 10%
formalin-fixed (carried out at room temperature for ~12 h),
paraffin-embedded (FFPE) tissue. A total of five of 10-µm-thick
tissue slides from each FFPE tissue block (194 samples) were used
for the isolation of genomic DNA. Paraffin in FFPE tissue slides
was removed using an FFPE deparaffinization solution (Merck KGaA).
The DNA was then extracted from tissue samples with the aid of
PureLink™ Genomic DNA Mini Kit according to the manufacturer's
instructions (Invitrogen; Thermo Fisher Scientific, Inc.).
KRAS mutations were detected with KRAS XL
StripAssay (ViennaLab Diagnostics GmbH) in a five-step procedure
including amplification, hybridization, stringent wash and color
development. Briefly, prepared DNA samples were mixed with
Taq-polymerase (ViennaLab Diagnostics GmbH) and an amplification
mixture from the manufacturer. The specific DNA sequences were
amplified in 35 cycles of polymerase chain reactions (PCR). Pre-PCR
was performed at 37˚C for 10 min and 94˚C for 2 min. The
thermocycling conditions were as follows: 94˚C for 1 min, 70˚C for
50 sec, 56˚C for 50 sec, 60˚C for 1 min (35 cycles) and a final
extension at 60˚C for 3 min. The amplification products were stored
on ice or at 2-8˚C until further use. The PCR products were then
denatured and hybridized with probes on strips in the hybridization
buffer. After hybridization ended, solutions were removed, and the
strips were washed. The conjugated solution was then added to the
strips, followed by a second washing step. Lastly, the color
developer was added to the strips in the dark for the appearance of
purple positive bands, followed by a third washing step. The strips
were analyzed using the KRAS collector sheet included in the kit
box or StripAssay Evaluator software (version 2.12.2018.212) to
determine KRAS mutation at codons 12, 13, 59, 60, 61, 117, and 146.
Both the collector sheet and the software were developed and
provided by ViennaLab Diagnostics GmbH, Austria (35,36).
Statistical analysis
Statistical analysis was performed using SPSS 21.0
(IBM Corp.). Non-normal variables (age) were reported as a median.
Nonparametric tests were used to compare the median of two groups
of non-normal distribution. Differences between groups were
assessed using the Chi-square tests. All tests were two-sided, with
a significance level of P<0.05. Multivariate analysis was
estimated using binary logistic regression models. The Kaplan-Meier
method was used to calculate the survival rate, and the log-rank
test was performed to compare survival rates. Cox's regression
model with a 95% confidence interval (CI) was used for multivariate
survival analysis. A P<0.05 was considered to indicate a
statistically significant difference.
Results
During the course of the study, a total of 194
patients who satisfied the selected conditions were included in the
analysis. Among them, 91 (46.9%) were females, of whom 39 (20.1%)
carried the wild-type while 52 (26.8%) had the mutated KRAS.
On the other hand, 103 (53.1%) were males including 62 (32.0%)
wild-type and 41 (21.1%) mutated KRAS. The age of the
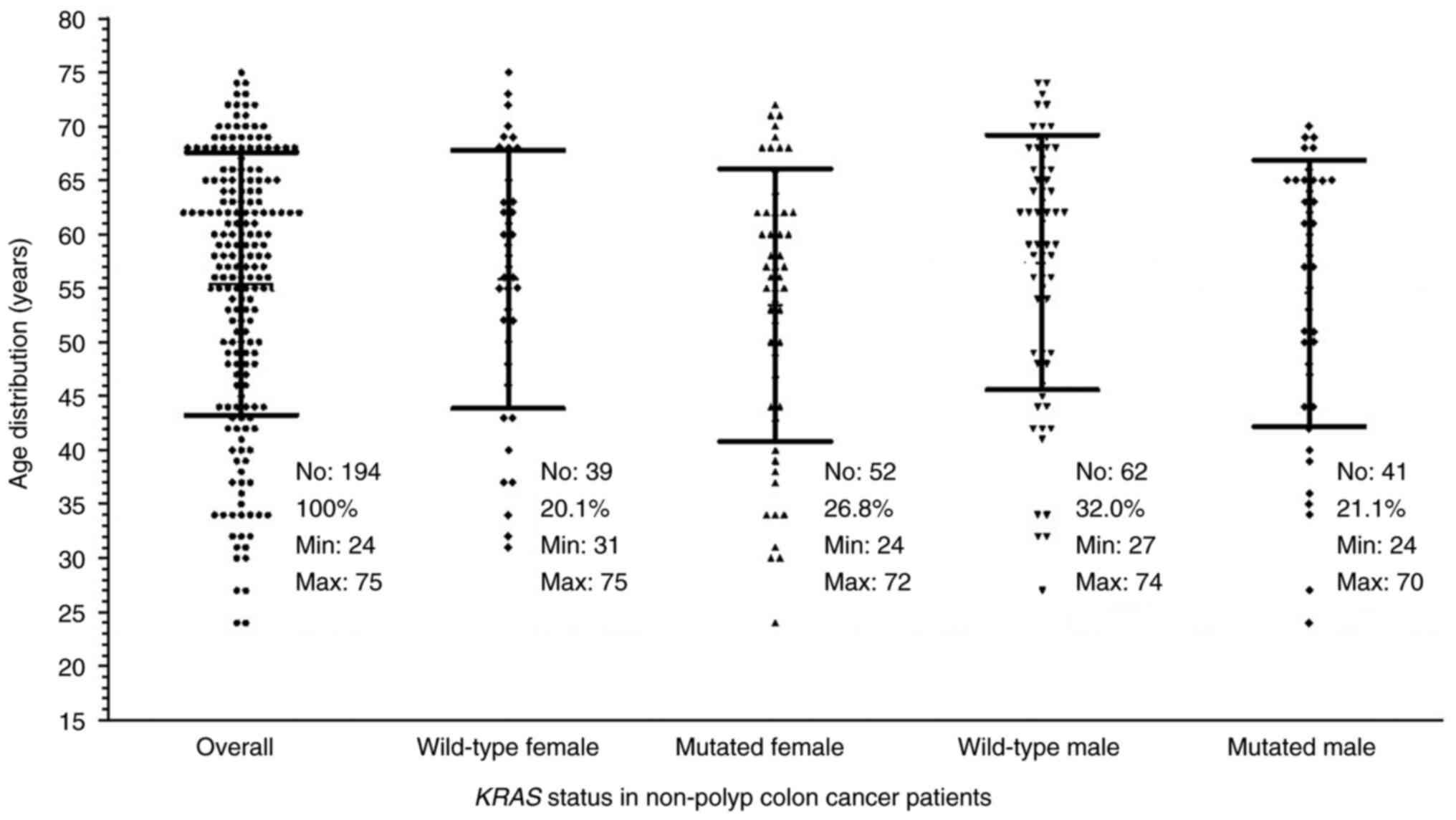
patients ranged from 24 to 75 with a median age of 58-years-old. As
summarized in Fig. 1, the minimum
age in the mutated KRAS-carrying males and females was lower
than those of the patients with the wild-type KRAS. The
median age of the mutated KRAS patients was 57 (58 for
males; 56 for females) which was also lower than the 59 for the
wild-type KRAS group (60 for males; 57 for females).
Furthermore, pair-wise comparisons using nonparametric tests showed
no significant differences between the mutated and the wild-type
groups, even in subgroups by sex.
Mutations of the KRAS gene in
Vietnamese patients with NPCC at stages II and III
Point mutations of the KRAS gene in exon 2
(codons 12 and 13), 3 (codons 59 and 61) and 4 (codons 117 and 146)
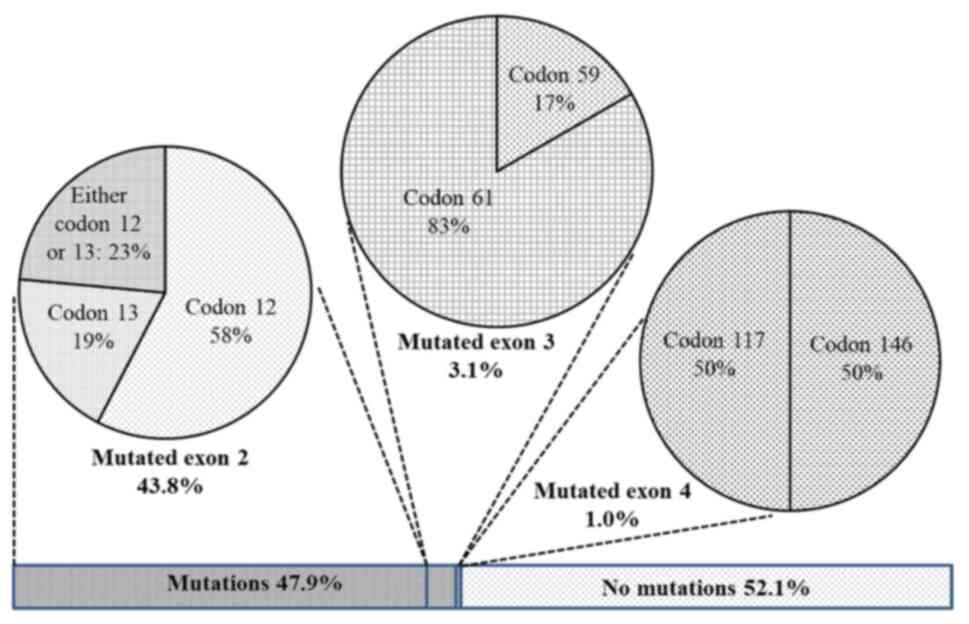
were determined. Overall, the KRAS mutation was identified
in 47.9% of the patients (Fig. 2).
Of which, mutations in exon 2 were dominant with 85 cases
accounting for 43.8% of recruited patients (or 91.4% of the total
detected mutations). Mutations in exon 3 including 1 case at codon
59 and 5 cases at codon 61 resulted in 3.1%. Only two cases of
mutated exon 4 were detected (1%), one carried mutation at codon
117 and the other at codon 146 (data not shown). None of the
patients had more than one mutation.
Associations of KRAS status and
clinical features
A total of seven clinical symptoms were recorded
before surgery and displayed in Fig.
3. Abdominal pain was predominant among the symptoms with 157
cases, accounting for 80.9% of patients. Diarrhea was the second
most common symptom that was reported in 51 cases (26.3%). Anemia
and weight loss were lower with 42 (21.6%) and 39 (20.1%) cases,
respectively. Other notable symptoms were bleeding with 36 cases
(18.6%), blood in stool with 33 cases (17%) and constipation with
36 cases (18.6%).
A pair-wise comparison between the wild-type and
mutated KRAS groups for each symptom was conducted to
identify the possible association of a symptom with the KRAS
status. Most of the observed symptoms were unlikely to be
correlated with the KRAS mutations (Fig. 3). Abdominal pains were highly
recorded in both groups with 84.2 and 77.4% in the wild-type group
and mutation groups, respectively. Other symptoms which did not
exhibit considerable changes between the two groups included
bleeding (14.9 vs. 22.6%), blood in the stool (21.8 vs. 11.8%),
diarrhea (25.7 vs. 26.9%), constipation (19.8 vs. 17.2%) and weight
loss (19.8 vs. 20.4%), respectively. However, the analysis revealed
a significant increase in cases of anemia among the mutated group
with P=0.041. Particularly, 28.0% of mutated KRAS patients
exhibited anemia symptoms while only 15.8% of the wild-type group
did (Fig. 3), indicating an
association of anemia with KRAS mutation in NPCC.
Association of KRAS mutation with
clinicopathological features in patients with NPCC
The possible associations of clinicopathological
features with KRAS status in patients with NPCC is revealed
in Table I. Notably, the
KRAS mutation rate in females was statistically higher than
in males. With 52 females, the KRAS mutation rate was
identified in 55.9% of the cases, far higher than 44.1% identified
in the males. The mutations were also detected at a higher rate in
right-sided cancers (RC) compared with left-sided cancers (LC)
(54.8% for the RC vs. 45.2% for the LC; P=0.047). By contrast,
KRAS mutation rates were relatively compatible among the
younger and older patients.
 | Table IClinicopathological features and
KRAS status. |
Table I
Clinicopathological features and
KRAS status.
| KRAS
mutation status |
|---|
| Characteristic | Total (n=194) | Wild type (n=101,
52.1%) | Mutated (n=93,
47.9%) | P-value |
|---|
| Age | | | | 0.714 |
|
<50 | 56 | 28 (27.7%) | 28 (30.1%) | |
|
≥50 | 138 | 73 (72.3%) | 65 (69.9%) | |
| Sex | | | | 0.016a |
|
Male | 103 | 62 (61.4%) | 41 (44.1%) | |
|
Female | 91 | 39 (38.6%) | 52 (55.9%) | |
| Tumour sites | | | | 0.047a |
|
Right | 92 | 41 (40.6%) | 51 (54.8%) | |
|
Left | 102 | 60 (59.4%) | 42 (45.2%) | |
| Histological
types | | | | 0.583 |
|
Adenocarcinoma | 164 | 84 (83.2%) | 80 (86.0%) | |
|
Others | 30 | 17 (16.8%) | 13 (14.0%) | |
| Invasion depth | | | | 0.333 |
|
T3 | 81 | 47 (46.5%) | 34 (36.6%) | |
|
T4a | 92 | 43 (42.6%) | 49 (52.7%) | |
|
T4b | 21 | 11 (10.9%) | 10 (10.8%) | |
| Lymphnode
status | | | | 0.190 |
|
N0 | 93 | 52 (51.5%) | 41 (44.1%) | |
|
N1 | 71 | 32 (30.7%) | 40 (43.0%) | |
|
N2 | 30 | 18 (17.8%) | 12 (12.9%) | |
| pTNM stage | | | | 0.303 |
|
II | 93 | 52 (51.5%) | 41 (44.1%) | |
|
III | 101 | 49 (48.5%) | 52 (55.9%) | |
A high rate of adenocarcinoma (AC) was revealed in
164 cases (84.5% of patients) while other types were identified in
30 cases (15.5% of patients) (Table
I). The number of NPCC AC cases in mutated vs. wild-type
KRAS was almost identical, 80 and 84 cases, respectively,
indicating that the KRAS mutation unlikely caused AC. The
KRAS mutation rate among the AC patients with NPCC was also
not different from that of the other types.
The invasion depth of tumors in the patients with
NPCC could possibly be linked to the KRAS mutation. The T4
stage (including T4a and b) was confirmed in 59 cases of the
mutated group, accounting for 63.5%. This stage was confirmed in
only 54 wild-type patients (53.5% of the wild-type group).
Similarly, stage III tumors were determined with a higher rate
among the mutated patients (52 out of 93 cases, or 55.9%) compared
with the same stage among the wild-type patients (49 out of 101
cases, or 48.5%). However, none of the associations between the
staging and KRAS mutation were statistically significant,
including lymph node status (Table
I).
Multivariate regression analysis:
Correlations of KRAS mutations with other prognostic markers
The association of KRAS mutations in females
among Vietnamese patients with NPCC was confirmed by multivariate
logistic regression. The KRAS mutation rate was
significantly higher in females with an odds ratio (OR)=2.144 (95%
CI: 1.184-3.882; P=0.012]. The analysis further confirmed the
correlation of KRAS mutations with the right NPCC with
P=0.048. Therein, the OR of left-sided colon cancer (LCC) to
right-sided colon cancer (RCC) was 0.551 (95% CI: 0.305-0.996). In
addition to the differences aforementioned, there was no evidence
of an association between tumor staging and histologic types with
KRAS mutation among Vietnamese patients with NPCC (Table II).
 | Table IIAssociations between KRAS
mutation and prognostic markers. |
Table II
Associations between KRAS
mutation and prognostic markers.
| | Multivariate
logistic regression |
|---|
| KRAS
mutation | | OR | 95% CI | P-value |
|---|
| Age | ≥50/<50 | 0.821 | 0.426-1.582 | 0.555 |
| Sex | Female/Male | 2.144 | 1.184-3.882 | 0.012a |
| Tumour
location | Left/Right | 0.551 | 0.305-0.996 | 0.048a |
| Invasion depth | T4/T3 | 1.421 | 0.776-2.600 | 0.255 |
| Nodal status | N (+)/(-) | 1.239 | 0.681-2.253 | 0.483 |
| Histology |
Adenocarcinoma/Others | 1.726 | 0.740-4.026 | 0.206 |
Treatment outcomes
During this study, one patient discontinued the
follow-up examinations at the three aforementioned hospitals.
Therefore, the treatment outcomes of 193 patients including 100
(51.8%) wild-type and 93 (48.2%) mutated KRAS cases were
analyzed. The mean follow-up duration was 38.8±13.2 months (min, 12
months; max, 78 months). The averages of disease-free survival
(DFS) and overall survival (OS) were 48.9±2.3 and 56.1±2.2 months,
respectively. For patients with a 3-year follow-up, the DFS was
53.8% and the OS was 73.0% (data not shown).
Association between KRAS status and
survival
Sorting patients into two groups by only their
KRAS status revealed that mutated KRAS resulted in a
worse trend for survival than the wild-type. The 3-year DFS of the
mutated KRAS group was 48.8% while for that of the wild-type
group it was 58.3%. However, the difference was not statistically
significant (P=0.205). Similarly, the 4-year OS of the mutated
KRAS group and wild-type groups were 63.0 and 67.0%,
respectively (P=0.525) (data not shown).
Different and notable statistical data was observed
after sorting patients into smaller groups with different criteria.
As could be observed from Table
III, KRAS mutation was associated with lower ratios of
3-year DFS and 4-year OS in patients with NPCC stage II. Among
stage II patients, 53.2% of the mutated KRAS group had
3-year DFS, notably lower than that of the wild-type group (79.8%)
with P=0.008. Similarly, the 4-year OS ratios of mutated
KRAS and wild-type groups were 66.0 and 94.1%, respectively
(P=0.021).
 | Table IIIAssociations between KRAS
status and survival in specified groups. |
Table III
Associations between KRAS
status and survival in specified groups.
| | 3-year DFS | 4-year OS |
|---|
| Specified groups
(n) | n | KRAS wt
(%) | Mutated KRAS
(%) | P-value | KRAS wt
(%) | Mutated KRAS
(%) | P-value |
|---|
| Age <50 | 56 | 60.3 | 46.2 | 0.364 | 67.6 | 63.7 | 0.254 |
| Age ≥50 | 137 | 57.7 | 50.0 | 0.376 | 63.7 | 63.4 | 0.937 |
| Male | 102 | 55.0 | 51.2 | 0.658 | 66.9 | 61.2 | 0.455 |
| Female | 91 | 64.1 | 46.6 | 0.146 | 67.3 | 65.7 | 0.798 |
| Right-sided | 91 | 62.3 | 62.4 | 0.795 | 56.6 | 73.9 | 0.429 |
| Left-sided | 102 | 55.9 | 32.2 | 0.012a | 72.7 | 52.8 | 0.070a |
| Adenocarcinoma | 163 | 63.2 | 51.0 | 0.124 | 72.5 | 66.6 | 0.318 |
| pT3 | 81 | 71.4 | 73.4 | 0.946 | 86.8 | 85.3 | 0.671 |
| pT4 | 112 | 46.7 | 34.7 | 0.301 | 50.0 | 52.4 | 0.877 |
| Stage II | 92 | 79.8 | 53.2 | 0.008a | 94.1 | 66.0 | 0.021a |
| Stage III | 101 | 35.7 | 45.3 | 0.379 | 36.8 | 60.3 | 0.184 |
Significant changes were also observed in groups of
patients with LCC. The 3-year DFS) of the mutated KRAS LCC
group was 32.2% compared with 55.9% of the wild-type KRAS
LCC group (P=0.012). The 4-year OS rates of the mutated and
wild-type KRAS LCC were 52.8 and 72.7%, respectively
(P=0.070). In addition, the effect of KRAS mutations on the
survival of patients did not significantly differ according to age
groups, sexes, RCC, histopathology of AC, and tumor stages
(Table III).
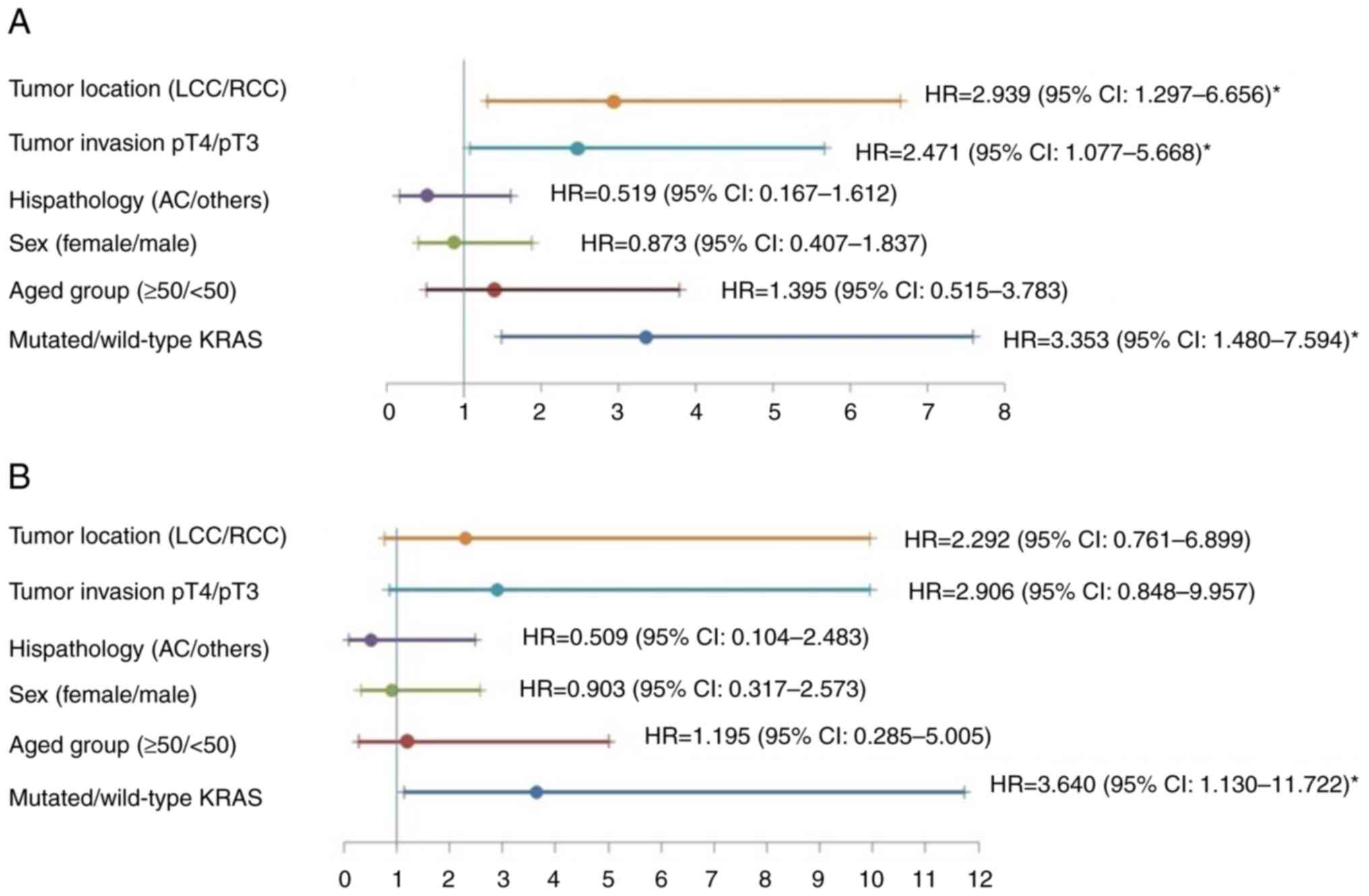
Further analyses were performed for stage II
patients using the Cox's model. The KRAS mutations were
independently related to worse DFS and OS. The hazard rate (HR) of
DFS was 3.353 (95% CI: 1.480-7.594; P=0.004; Fig. 4A), and that of OS was 3.640 (95%
CI: 1.130-11.722, P=0.030; Fig.
4B).
Discussion
The association of mutations in the KRAS gene
with poor DFS of colon cancer has been well-studied worldwide
(37). More than 3000-point
mutations of the KRAS gene in colorectal cancer are reported
in the literature. Most (~82%) reported mutations were in codon 12.
Mutations at codons 13 and 61 accounted for ~17 and ~1%,
respectively (18,38,39).
A meta-analysis, by Roth et al, of four large clinical
trials in CC stage II-III patients who underwent radical treatment
indicated the KRAS mutation rate was 37.0% (36.0% in stage
II, 37.5% in stage III) (22).
Another study on 228 patients with CC showed that the rate of
mutated KRAS was 39.9%, of which 26.0% was in codon 12, 6.6%
in codon 13, and 3.5% in codon 61(21). The rate of mutated KRAS in
Vietnamese patients with CC was 41.0, and >80% of mutations were
detected at codon 12 or 13 in Exon 2 (56/64), while a minority (8
cases) was identified at codon 61 in Exon 3(31).
However, the KRAS mutation rate in NPCC has
rarely been reported in Vietnam. According to the results of the
present study in which point mutations of KRAS in exons 2
(codons 12 and 13), 3 (codons 59 and 61) and 4 (codons 117 and 146)
were determined, the overall KRAS mutation rate was
identified to be 47.9% in patients with NPCC (Fig. 2), notably a high rate in the
country. This detected rate was surprisingly higher than previous
studies reporting 36-38% KRAS mutation rates (5,21,22).
The mutations mostly occurred in exon 2 of the gene. Particularly,
the mutation rate in KRAS exon 2 was 43.8% of the recruited
patients, markedly higher than a previous study on CC in Vietnam
(37.1%) and China (42.2%) (32,40).
The mutation rate in KRAS exon 3 (3.1%) was relatively
similar to that reported in a study by Guo et al (40). While the mutations in exon 4 were
rare in Vietnamese patients with NPCC with only 2 cases, or 1%
reported in the present study.
Clinicopathological properties such as abdominal
pain, anemia and various symptoms were identified in >83% of
patients. Of those, diarrhea and anemia were likely correlated to
the tumors in the right colon (41). Logically, an association between
diarrhea and anemia with KRAS mutations is suspected.
However, the current data has confirmed significantly higher anemia
cases but not diarrhea cases among KRAS-mutated patients
with NPCC. Mutated KRAS had a higher frequency in the right-side
colon compared with the left-side colon.
Another impressive finding in the present study was
the association of KRAS mutations with female patients. The
mutations were detected in 57.1% of female patients (52 out of 91
females) and only 39.8% of male patients (41 out of 103 males).
Females accounted for 55.9% of patients with mutated KRAS
genes (Table I) while males
accounted for 44.1%. These sex ratios among patients with mutated
KRAS genes were contrary to those in a previous study for CC
in Vietnam, which reported 48.2 and 51.8% for female and male
patients with mutated KRAS genes, respectively (32). In addition, the age of KRAS
mutation-carrying patients in each sex was lower than those of the
wild-type groups. However, the correlation of KRAS mutation
with the age of patients was unclear. By applying multivariate
logistic regression, the KRAS mutation rate was
significantly higher in females, and the analysis further confirmed
the prevalence of KRAS mutations in the right-side colon
(Table II).
Several studies have revealed that the KRAS
mutation rate was likely higher in the RCC than in the LCC
(5,20,42).
In the present study, the association of the KRAS mutation
rate with the RCC was clarified. The KRAS mutation rate in
the RCC was 55.4% (51 mutants out of 92), while the rate in the LCC
was only 41.2% (42 mutants out of 102).
The effect of KRAS mutation status on
survival has been reported in previous studies; however, the
results were controversial. Some authors indicated that KRAS
mutations led to a worse prognosis of DFS in patients with CC at
stages II-III (6,18,20,21).
Conversely, an opposite conclusion concerning the prognostic value
of the KRAS gene has been reported (22-24).
By analyzing some specified groups, the prognostic
values of KRAS mutations in the distinct group of patients
with NPCC stage II-III, were clarified. Most previous studies have
reported that RCC was associated with a higher recurrence and lower
survival rate than LCC. Some clinical features were considered to
explain this issue, as in the earlier stage, fewer nodal invasions,
and fewer gene mutations such as BRAF, PIK3CA, CTNNB1 and PTEN
(33,43,44)
were diagnosed. Among LCC patients, mutated KRAS was a
significant predictive factor for poor DFS (32.2% of mutated
KRAS vs. 55.9% of wild-type; P=0.012) but not for OS
(Table III). Some other studies
supported this result. Xie et al indicated mutated-KRAS was
a worse prognosis for OS in LCC (HR: 1.21; 95% CI: 1.08-1.36;
P<0.01) (42). In LCC stage IV,
mutated KRAS was significantly related to a higher risk of
mortality (HR: 1.18; 95% CI: 1.05-1.33) (45).
Although no considered difference in survival time
was revealed in overall NPCC stage II-III patients according to
KRAS mutation status, poor DFS and OS were observed in stage
II patients. The Cox's regression analysis revealed that
KRAS mutations were an independent prognostic factor for
stage II patients (HR of DFS: 3.353; P=0.004; HR of OS: 3.640;
P=0.030; Fig. 4). This data
differed from that reported in Chinese patients with CC (40). A study by Natsume et al also
reported survival in colon cancer stage II, with a 5-year DFS of
mutated KRAS vs. KRAS wild-type: 75.9 vs. 76.7%; and a 5-year OS of
84.3 vs. 87.1% (P>0.05) (46).
A study in early-stage colorectal cancer (stage I-II), with an
average of 72 months of follow-up, revealed that DFS was worse in
patients with KRAS codon 13 mutation (stage I: P=0.015; stage II:
P<0.001) (47). The worse
survival of KRAS-mutated colon cancer stage II could be explained
by the role of the KRAS-mutated gene in colon cancer. KRAS gene
mutations that occur in the earlier stage of colon cancer can cause
earlier recurrence.
The limitations of the present study were the impact
on survival, analyzed only by KRAS status, not in association with
other gene mutations such as BRAF, NRAS and MMR, and with recurrent
patients, the treatment after diagnosis was not standardized.
In conclusion, the KRAS mutation rate in NPCC
stage II-III in Vietnam was very high (47.9%). Of which, mutations
in exon 2 were dominant and detected in 43.8% of the recruited
patients (or 91.4% of the total detected mutations). The patients
who carried the mutated-KRAS further potentially experienced
anemia. Moreover, the KRAS mutations occurred in Vietnamese
females with NPCC and RCC patients. Furthermore, the mutated
KRAS was an independent predictor for poor DFS and OS in
stage II patients and DFS in LCC. These findings provide essential
scientific background for treating and managing NPCC.
Acknowledgements
The authors express their profound gratitude to Dr
Le Thanh Do (Institute for Global Health Innovations, Duy Tan
University, Da Nang, Vietnam) for his advice on the first
draft.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HMC, VHT and NMD wrote the manuscript. HMC and VHT
participated in design of the study. HMC, VHT and NTL were involved
in acquisition of data. HMC, VHT and BTTH participated in analysis
and interpretation of data. HMC, VHT, BTTH and NMD confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no.
NCS28/HMU-IRB) by the Ethics Committee of Hanoi Medical University
(Hanoi, Vietnam). Written informed consent from all participants
was obtained.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Pham T, Bui L, Kim G, Hoang D, Tran T and
Hoang M: Cancers in vietnam-burden and control efforts: A narrative
scoping review. Cancer Control. 26(1073274819863802)2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Pham DX, Phung AHT, Nguyen HD, Bui TD, Mai
LD, Tran BNH, Tran TS, Nguyen TV and Ho-Pham LT: Trends in
colorectal cancer incidence in Ho Chi Minh City, Vietnam
(1996-2015): Joinpoint regression and age-period-cohort analyses.
Cancer Epidemiol. 77(102113)2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Alves Martins BA, de Bulhões GF,
Cavalcanti IN, Martins MM, de Oliveira PG and Martins AMA:
Biomarkers in colorectal cancer: The role of translational
proteomics research. Front Oncol. 9(1284)2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tran CG, Goffredo P, Mott SL, Hart A, You
YN, Vauthey JN, Weigel RJ and Hassan I: The impact of KRAS
mutation, microsatellite instability, and tumor laterality on the
prognosis of nonmetastatic colon cancer. Surgery. 171:657–665.
2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Lee DW, Kim KJ, Han SW, Lee HJ, Rhee YY,
Bae JM, Cho NY, Lee KH, Kim TY, Oh DY, et al: KRAS mutation is
associated with worse prognosis in stage III or high-risk stage II
colon cancer patients treated with adjuvant FOLFOX. Ann Surg Oncol.
22:187–194. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Jones RP, Sutton PA, Evans JP, Clifford R,
McAvoy A, Lewis J, Rousseau A, Mountford R, McWhirter D and Malik
HZ: Specific mutations in KRAS codon 12 are associated with worse
overall survival in patients with advanced and recurrent colorectal
cancer. Br J Cancer. 116:923–929. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Roberts PJ and Der CJ: Targeting the
Raf-MEK-ERK mitogen-activated protein kinase cascade for the
treatment of cancer. Oncogene. 26:3291–3310. 2007.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Krasinskas AM: EGFR Signaling in
Colorectal Carcinoma. Patholog Res Int. 2011(932932)2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Morrison DK: MAP kinase pathways. Cold
Spring Harb Perspect Biol. 4(a011254)2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Avruch J: MAP kinase pathways: The first
twenty years. Biochim Biophys Acta. 1773:1150–1160. 2007.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang W and Liu HT: MAPK signal pathways
in the regulation of cell proliferation in mammalian cells. Cell
Res. 12:9–18. 2002.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Therkildsen C, Bergmann TK,
Henrichsen-Schnack T, Ladelund S and Nilbert M: The predictive
value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment
in metastatic colorectal cancer: A systematic review and
meta-analysis. Acta Oncol. 53:852–864. 2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Misale S, Yaeger R, Hobor S, Scala E,
Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M,
Siravegna G, et al: Emergence of KRAS mutations and acquired
resistance to anti-EGFR therapy in colorectal cancer. Nature.
486:532–536. 2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Siddiqui AD and Piperdi B: KRAS mutation
in colon cancer: A marker of resistance to EGFR-I therapy. Ann Surg
Oncol. 17:1168–1176. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ozer M and Goksu SY: Age-dependent
prognostic value of KRAS mutation in metastatic colorectal cancer.
Future Oncol. 17:4883–4893. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Saltz LB, Lenz HJ, Kindler HL, Hochster
HS, Wadler S, Hoff PM, Kemeny NE, Hollywood EM, Gonen M, Quinones
M, et al: Randomized phase II trial of cetuximab, bevacizumab, and
irinotecan compared with cetuximab and bevacizumab alone in
irinotecan-refractory colorectal cancer: The BOND-2 study. J Clin
Oncol. 25:4557–4561. 2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Yoon HH, Tougeron D, Shi Q, Alberts SR,
Mahoney MR, Nelson GD, Nair SG, Thibodeau SN, Goldberg RM, Sargent
DJ, et al: KRAS codon 12 and 13 mutations in relation to
disease-free survival in BRAF-wild-type stage III colon cancers
from an adjuvant chemotherapy trial (N0147 alliance). Clin Cancer
Res. 20:3033–3043. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sinicrope FA, Mahoney MR, Yoon HH, Smyrk
TC, Thibodeau SN, Goldberg RM, Nelson GD, Sargent DJ and Alberts
SR: Alliance for Clinical Trials in Oncology. Analysis of molecular
markers by anatomic tumor site in stage III colon carcinomas from
adjuvant chemotherapy trial NCCTG N0147 (Alliance). Clin Cancer
Res. 21:5294–5304. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Scott A, Goffredo P, Ginader T, Hrabe J,
Gribovskaja-Rupp I, Kapadia MR, Weigel RJ and Hassan I: The Impact
of KRAS mutation on the presentation and prognosis of
non-metastatic colon cancer: An Analysis from the National Cancer
Database. J Gastrointest Surg. 24:1402–1410. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Shen Y, Han X, Wang J, Wang S, Yang H, Lu
SH and Shi Y: Prognostic impact of mutation profiling in patients
with stage II and III colon cancer. Sci Rep.
6(24310)2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Roth AD, Tejpar S, Delorenzi M, Yan P,
Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C,
et al: Prognostic role of KRAS and BRAF in stage II and III
resected colon cancer: Results of the translational study on the
PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 28:466–474.
2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gavin PG, Colangelo LH, Fumagalli D,
Tanaka N, Remillard MY, Yothers G, Kim C, Taniyama Y, Kim SI, Choi
HJ, et al: Mutation profiling and microsatellite instability in
stage II and III colon cancer: An assessment of their prognostic
and oxaliplatin predictive value. Clin Cancer Res. 18:6531–6541.
2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ogino S, Meyerhardt JA, Irahara N,
Niedzwiecki D, Hollis D, Saltz LB, Mayer RJ, Schaefer P, Whittom R,
Hantel A, et al: KRAS mutation in stage III colon cancer and
clinical outcome following intergroup trial CALGB 89803. Clin
Cancer Res. 15:7322–7329. 2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Juárez M, Egoavil C, Rodríguez-Soler M,
Hernández-Illán E, Guarinos C, García-Martínez A, Alenda C,
Giner-Calabuig M, Murcia O, Mangas C, et al: KRAS and BRAF somatic
mutations in colonic polyps and the risk of metachronous neoplasia.
PLoS One. 12(e0184937)2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chan TL, Zhao W, Leung SY and Yuen ST:
Cancer Genome Project. BRAF and KRAS mutations in colorectal
hyperplastic polyps and serrated adenomas. Cancer Res.
63:4878–4881. 2003.PubMed/NCBI
|
|
27
|
Kuipers EJ, Grady WM, Lieberman D,
Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ and Watanabe T:
Colorectal cancer. Nat Rev Dis Primers. 1(15065)2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Shussman N and Wexner SD: Colorectal
polyps and polyposis syndromes. Gastroenterol Rep (Oxf). 2:1–15.
2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Bond JH: Colon polyps and cancer.
Endoscopy. 35:27–35. 2003.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Boutin AT, Liao WT, Wang M, Hwang SS,
Karpinets TV, Cheung H, Chu GC, Jiang S, Hu J, Chang K, et al:
Oncogenic Kras drives invasion and maintains metastases in
colorectal cancer. Genes Dev. 31:370–382. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ta TV, Nguyen QN, Chu HH, Truong VL and
Vuong LD: RAS/RAF mutations and their associations with epigenetic
alterations for distinct pathways in Vietnamese colorectal cancer.
Pathol Res Pract. 216(152898)2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Nguyen HT, Le DT, Duong QH, Tatipamula VB
and Van Nguyen B: High frequency of microsatellite instability and
its substantial co-existence with KRAS and BRAF mutations in
Vietnamese patients with colorectal cancer. Oncol Lett.
21(41)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sakin A, Arici S, Secmeler S, Can O,
Geredeli C, Yasar N, Demir C, Demir OG and Cihan S: Prognostic
significance of primary tumor localization in stage II and III
colon cancer. World J Gastrointest Oncol. 10:410–420.
2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Nagtegaal ID, Odze RD, Klimstra D, Paradis
V, Rugge M, Schirmacher P, Washington KM, Carneiro F and Cree IA:
WHO Classification of Tumours Editorial Board. The 2019 WHO
classification of tumours of the digestive system. Histopathology.
76:182–188. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
GmbH VD. KRAS XL StripAssay: 2020
[updated; cited]. Available from: https://www.goffinmoleculartechnologies.com/wp-content/uploads/2021/04/5680A-InstructionsForUse-2020-01.pdf.
|
|
36
|
Abd El Kader Y, Emera G, Safwat E, Kassem
HA and Kassem NM: The KRAS StripAssay for detection of KRAS
mutation in Egyptian patients with colorectal cancer (CRC): A pilot
study. J Egypt Natl Canc Inst. 25:37–41. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Formica V and Sera F: KRAS and BRAF
Mutations in Stage II and III colon cancer: A systematic review and
meta-analysis. J Natl Cancer Inst. 114:517–527. 2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Aoki Y, Niihori T, Narumi Y, Kure S and
Matsubara Y: The RAS/MAPK syndromes: Novel roles of the RAS pathway
in human genetic disorders. Hum Mutat. 29:992–1006. 2008.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Jin CE, Yeom SS, Koo B, Lee TY, Lee JH,
Shin Y and Lim SB: Rapid and accurate detection of KRAS mutations
in colorectal cancers using the isothermal-based optical sensor for
companion diagnostics. Oncotarget. 8:83860–83871. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Guo TA, Wu YC, Tan C, Jin YT, Sheng WQ,
Cai SJ, Liu FQ and Xu Y: Clinicopathologic features and prognostic
value of KRAS, NRAS and BRAF mutations and DNA mismatch repair
status: A single-center retrospective study of 1,834 Chinese
patients with Stage I-IV colorectal cancer. Int J Cancer.
145:1625–1634. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Cienfuegos JA, Baixauli J, Arredondo J,
Pastor C, Martínez Ortega P, Zozaya G, Martí-Cruchaga P and
Hernández Lizoáin JL: Clinico-pathological and oncological
differences between right and left-sided colon cancer (stages
I-III): Analysis of 950 cases. Rev Esp Enferm Dig. 110:138–144.
2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Xie MZ, Li JL, Cai ZM, Li KZ and Hu BL:
Impact of primary colorectal Cancer location on the KRAS status and
its prognostic value. BMC Gastroenterol. 19(46)2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Mejri N, Dridi M, El Benna H, Labidi S,
Daoud N and Boussen H: Tumor location impact in stage II and III
colon cancer: Epidemiological and outcome evaluation. J
Gastrointest Oncol. 9:263–268. 2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Salem ME, Weinberg BA, Xiu J, El-Deiry WS,
Hwang JJ, Gatalica Z, Philip PA, Shields AF, Lenz HJ and Marshall
JL: Comparative molecular analyses of left-sided colon, right-sided
colon, and rectal cancers. Oncotarget. 8:86356–86368.
2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Charlton ME, Kahl AR, Greenbaum AA,
Karlitz JJ, Lin C, Lynch CF and Chen VW: KRAS testing, tumor
location, and survival in patients with stage IV colorectal cancer:
SEER 2010-2013. J Natl Compr Canc Netw. 15:1484–1493.
2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Natsume S, Yamaguchi T, Takao M, Iijima T,
Wakaume R, Takahashi K, Matsumoto H, Nakano D, Horiguchi SI,
Koizumi K and Miyaki M: Clinicopathological and molecular
differences between right-sided and left-sided colorectal cancer in
Japanese patients. Jpn J Clin Oncol. 48:609–618. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Dinu D, Dobre M, Panaitescu E, Bîrlă R,
Iosif C, Hoara P, Caragui A, Boeriu M, Constantinoiu S and
Ardeleanu C: Prognostic significance of KRAS gene mutations in
colorectal cancer-preliminary study. J Med Life. 7:581–587.
2017.PubMed/NCBI
|