Introduction
Acute myeloid leukemia (AML) is one of the most
prevalent hematological malignancies globally, constituting
one-third of all adult leukemias (1-4).
Despite advances in treatments leading to complete remission for
most patients with AML, drug resistance remains a major factor in
therapy failure and contributes to the short-term survival of these
patients (4). Recently,
accumulating evidence has implicated leukemic stem cells (LSCs), a
dormant subset of leukemic cells, as pivotal in drug resistance and
cancer relapse (5). These cells
express the same markers as normal adult hematopoietic stem cells
(HSCs) (CD34+ and CD38-) and exhibit stemness
characteristics, such as self-renewal, proliferation and
differentiation (5-8).
In the past few years, several microRNAs (miRNAs/miRs), which
represent small non-coding RNAs that act as post-transcriptional
regulators, have been identified as participants in drug resistance
mechanisms and LSC regulation, such as miR-22 and miR-126 (8,9).
Therefore, investigations into miRNA-associated cancer pathogenesis
are crucial for overcoming drug resistance events and regulating
LSCs in AML.
Dysregulation of miR-223 has been documented in
connection with various cancer types, including hepatocellular
carcinoma, breast cancer and leukemia. Furthermore, numerous
targets of miR-223, including insulin-like growth factor-1
receptor, monocytic enhancer factor 2C, microtubule destabilizer
stathmin 1 and forkhead box protein O1A, have been associated with
cancer-related traits, such as cell proliferation, carcinogenesis
and metastasis (10). However, few
studies have explored the role of miR-223 in AML. Previous studies
demonstrated low miR-223 expression in patients with AML,
especially in those with a poor prognosis (11,12).
Moreover, a marked increase in miR-223 expression was reported in
patients with AML after treatment, regardless of whether complete
remission was achieved or not (12). Another study demonstrated that
miR-223 suppressed cell proliferation and enhanced cell apoptosis
in HL-60 and K-562 cell lines by specifically targeting the
expression of F-box/WD repeat-containing protein 7 (FBXW7)
(13).
Protein kinase C ε (PKCε), an isoform of PKC, is
encoded by the PRKCE gene. It phosphorylates a variety of
protein targets and is known to participate in diverse cellular
signaling pathways, including MAPK, ERK and PI3K/AKT pathways,
which regulate various biological functions, such as proliferation,
differentiation and apoptosis (14,15).
PKCε has been implicated in drug resistance in several cancer
types, including gallbladder cancer, lung cancer, renal cell
carcinoma and prostate cancer (14,16-19).
Additionally, a recent study identified high levels of PRKCE
mRNA in gallbladder cancer stem cells (19). Another recent study demonstrated
that PKCε overexpression has been shown to selectively confer
resistance to daunorubicin (DNR) in the AML U937 and HEL cell
lines. Furthermore, patients with high levels of PKCε protein
exhibit a lower rate of complete remission compared with those with
lower PKCε protein levels. Additionally, elevated PKCε expression
has been associated with decreased disease-free survival,
indicating a consistent pattern of treatment resistance (20).
The present study aimed to elucidate the roles of
miR-223 and PKCε in the regulation of drug resistance mechanisms in
LSCs to bridge current gaps in knowledge and offer valuable
insights for the advancement of targeted therapies in AML.
Materials and methods
Cell culture
The human acute myeloblastic leukemia KG-1 cell line
(cat. no. CCL-246) and its quiescent variant KG-1a (cat. no.
CCL-246.1), obtained from the American Type Culture Collection,
served as LSC models in the present study. KG-1a cells display less
mature morphological, cytochemical and functional characteristics
compared with KG-1 cells (21).
Furthermore, the unresponsiveness of KG-1a cells to
colony-stimulating factor and the lack of expression of human
leukocyte antigen contributes to their increased resistance to
differentiation-inducing drugs (22). Both cell lines were cultured in
Iscove's modified Dulbecco's medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 20% FBS (Capricorn Scientific),
1 mM L-glutamine (Cytiva), 100 U/ml penicillin and 100 µg/ml
streptomycin (Gibco; Thermo Fisher Scientific, Inc.). 293T cells,
used for the luciferase activity assay, were kindly provided by Dr
Pinyaphat Khamphikham (an author of the present study). The 293T
cells were cultured in DMEM (Nacalai Tesque, Inc.) containing 10%
FBS, 1 mM L-glutamine, 100 U/ml penicillin and 100 µg/ml
streptomycin. All cell lines were cultured at 37˚C in a humidified
atmosphere with 5% CO2 and were passaged every 2-3 days
or upon reaching 70-80% confluence.
Drug sensitivity assay
KG-1 and KG-1a cells were seeded in 96-well plates
at a density of 1.5x104 cells per well and were exposed
to doxorubicin (DOX) (Fresenius Kabi Oncology, Ltd.) at
concentrations of 0.03125, 0.0625, 0.125, 0.25, 0.5 and 1.0 µg/ml.
For the DNR sensitivity test, small interfering (si)RNA-transfected
KG-1a cells were seeded at the same density and treated with DNR
(APeXBIO Technology LLC) at concentrations of 0.03125, 0.0625,
0.125 and 0.25 µg/ml. Blank wells containing only growth medium
were utilized to subtract background signals. Following 48 h of
incubation at 37˚C with 5% CO2, the Cell Counting Kit
(CCK)-8 assay (Abbkine Scientific, Co., Ltd.) was conducted
following the manufacturer's protocol. After incubation with the
CCK-8 reagent for 2 h at 37˚C, the absorbance at 450 nm was
determined using a microplate reader (Metertech, Inc.). The
experiment was repeated three times, and cell survival was
determined using the following formula: % Cell viability=[mean
optical density (OD) of the treated group-blank/mean OD of the
untreated control group-blank] x100. The half-maximal inhibitory
concentration (IC50) value was established by plotting
the cell viability rate (y-axis) against DOX concentration (x-axis)
to generate a linear equation: y=ax + b. The IC50 value
was calculated as follows: IC50=(50-b)/a.
RNA isolation and reverse
transcription-quantitative (RT-q)PCR
Total RNA was extracted from KG-1 and KG-1a cells
using TRI Reagent® (Molecular Research Center, Inc.).
The concentration of isolated total RNA was measured using a Qubit
4 fluorometer (Invitrogen; Thermo Fisher Scientific, Inc.) with the
Qubit RNA High Sensitivity Assay kit (Invitrogen; Thermo Fisher
Scientific, Inc.). To reverse transcribe mature miR-223, the RNA
samples underwent conversion into U6 and miR-223-specific
cDNA using SuperScript III Reverse Transcriptase (Invitrogen;
Thermo Fisher Scientific, Inc.) with RT primers, according to the
protocol of Varkonyi-Gasic and Hellens (23). For the conversion of mRNA into
cDNA, total RNA (1 µg) was reverse transcribed into cDNA using
ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo Life
Science) following the manufacturer's protocol. The RT reactions
were performed on an MJ Mini thermocycler (Bio-rad Laboratories,
Inc.). qPCR was performed using SensiFAST SYBR No-ROX reagent kit
(Meridian Bioscience, Inc.). The primers used in the present study
are listed in Table I. The qPCR
was performed on a CFX Opus 96 Real-time PCR system (Bio-Rad
Laboratories, Inc.) with the following cycling conditions: 95˚C for
2 min, followed by 40 cycles at 95˚C for 5 sec and 60˚C for 15 sec.
The samples were independently analyzed three times, and relative
expression was assessed using the 2-∆∆Cq method
(24), with U6 and
GAPDH as reference genes for miR-223 and PRKCE,
respectively. As each replicated experiment was conducted
individually at different time points, the expression levels of
miR-223 or PRKCE of the control groups were standardized to
1, and the relative expression of the other groups were normalized
to the expression of the control groups within each replicate.
 | Table IPrimer sequences used for
RT-quantitative PCR. |
Table I
Primer sequences used for
RT-quantitative PCR.
| Gene | Direction | Sequence
(5'-3') |
|---|
| miR-223
RTpa | N/A |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGGGGTA |
| U6
RTpa | N/A |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAAATATG |
|
miR-223a | Forward |
AACACGCTGTCAGTTTGTCAA |
| | Reverse |
GTCGTATCCAGTGCAGGGT |
|
U6a | Forward |
CTCGCTTCGGCAGCACA |
| | Reverse |
AACGCTTCACGAATTTGCGT |
|
GAPDHb | Forward |
AACGGGAAGCTTGTCATCAATGGAAA |
| | Reverse |
GCATCAGCAGAGGGGGCAGAG |
|
PRKCEa | Forward |
AGCCTCGTTCACGGTTCT |
| | Reverse |
TGTCCAGCCATCATCTCG |
Western blot analysis
Cells were lysed, and proteins were extracted using
RIPA buffer composed of 50 mM Tris-HCl, 150 mM NaCl, 1% Triton
X-100, 0.5 mM EDTA, 0.1% SDS and a protease inhibitor cocktail
(HiMEdia Laboratories, LLC). The quantification of the extracted
proteins was determined using Qubit 4 fluorometer and a Qubit
protein assay kit (Invitrogen; Thermo Fisher Scientific, Inc.).
Subsequently, protein samples (50 µg per lane) were combined with
loading buffer, boiled at 95˚C for 5 min, separated on 7.5%
SDS-PAGE gels, and then transferred to PVDF membranes. The
membranes were blocked with 5% skimmed milk in PBS at room
temperature for 3 h, followed by overnight incubation at 4˚C with
primary rabbit antibody targeting PKCε (1:1,000 dilution; cat. no.
2683; Cell Signaling Technology, Inc.) or for 1 h at room
temperature with primary rabbit antibody targeting GAPDH (1:16,000
dilution; cat. no. ABS16; MilliporeSigma). The membranes were
rinsed six times (5 min each time) with 0.1% Tween-PBS solution and
subsequently incubated with HRP-conjugated anti-rabbit IgG
secondary antibody (1:20,000 dilution; cat. no. W401B; Promega
Corporation) for 1 h at room temperature. Immobilon Forte Western
HRP substrate (MilliporeSigma) was used for signal development, and
the density of protein bands was analyzed using Quantity One
software version 4.6.8 (Bio-Rad Laboratories, Inc.). Each
replicated experiment was performed separately at a different time
point. The levels of the control groups were standardized to 100%,
and the relative expression of the other groups was normalized
against the expression of the control group within each
replicate.
Cell transfection
Mimic-negative control (NC) was obtained from
Shanghai GenePharma Co., Ltd., while miR-223 mimic was purchased
from Ambion (Thermo Fisher Scientific, Inc.). For the knockdown of
PKCε, a TriFECTa RNAi kit, comprising siRNAs si-PRKCE#1,
si-PRKCE#2 and si-PRKCE#3, was purchased from
Integrated DNA Technologies, Inc. si-NC was purchased from Ambion;
Thermo Fisher Scientific, Inc. The sequences of miRNA mimics and
siRNAs utilized in the present study are listed in Table II. The transfection of miRNA
mimics or siRNAs into the cells was performed using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) in accordance with the manufacturer's
protocol.
 | Table IImiRNA mimics and siRNAs. |
Table II
miRNA mimics and siRNAs.
| Molecule | Strand | Sequence
(5'-3') |
|---|
| mimic-NC | Sense |
UUCUCCGAACGUGUCACGUTT |
| | Antisense |
ACGUGACACGUUCGGAGAATT |
| miR-223 mimic | Sense |
UGUCAGUUUGUCAAAUACCCCA |
| | Antisense |
GGGUAUUUGACAAACUGACAUU |
| si-NC | Sense |
GGCAAGACCAGCUGCGACAUU |
| | Antisense |
AAUGUCGCAGCUGGUCUUGCC |
|
si-PRKCE#1 | Sense |
CCAGUCUGAAUACAGGUAGAUAUTA |
| | Antisense |
UAAUAUCUACCUGUAUUCAGACUGGAA |
|
si-PRKCE#2 | Sense |
GUCAAUAAUUUUGAGCAAGACUUTA |
| | Antisense |
UAAAGUCUUGGUCAAAAUUAUUGACGU |
|
si-PRKCE#3 | Sense |
UGAAAGCUUUCAUGACGAAGAAUCC |
| | Antisense |
GGAUUCUUCGUCAUGAAAGCUUUCAAG |
For miRNA mimic transfection, KG-1a cells were
transfected with either 25 nM miR-223 mimic or mimic-NC for 24 h at
37˚C. Regarding siRNA transfection, KG-1a cells were transfected
with 25 nM of either si-NC, si-PRKCE#1, si-PRKCE#2 or
si-PRKCE#3 for 48 h at 37˚C. Subsequent experiments were
conducted instantly following transfection.
Dual-luciferase reporter assay
To investigate whether PKCε serves as a direct
target of miR-223, TargetScan software (https://www.targetscan.org/vert_80/) was used to
predict the binding targets of miR-223, one of which was identified
to be PRKCE mRNA. The pmiRGLO plasmid (Promega Corporation)
was used to generate constructs with the 3' untranslated region
(UTR) sequence of PRKCE, encompassing the miR-223 binding
site. These constructs included
pmiRGLO-PRKCE-3'UTR-wild-type (WT) and
pmiRGLO-PRKCE-3'UTR-mutant (Mut). Co-transfection of these
constructed plasmids (200 ng each) with miR-223 mimic or mimic-NC
(10 pmol each) was performed in 293T cells using Lipofectamine 2000
reagent in accordance with the manufacturer's protocol. After 48 h
of transfection at 37˚C, firefly and Renilla luciferase
activities were promptly quantified using the Dual-Glo Luciferase
Assay System kit (Promega Corporation) and a CLARIOstar Plus
microplate reader (BMG Labtech GmbH). The firefly luciferase
activity was normalized to the Renilla luciferase activity
of each sample. This experiment was independently repeated three
times.
Statistical analysis
Data are presented as the mean ± standard deviation.
Data analysis was conducted using SPSS version 20 (IBM Corp.). An
unpaired Student's t-test was used to make comparisons between two
groups. Differences among multiple groups were analyzed using
one-way ANOVA followed by Dunnett's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-223 is downregulated and PKCε is
upregulated in the drug-resistant KG-1a cell line
The assessment of drug resistance in LSC cell lines
involved treating both KG-1 and KG-1a cells with varying
concentrations of DOX. The findings indicated a significantly
higher IC50 in KG-1a cells (0.650±0.004 µg/ml) compared
with that in KG-1 cells (0.435±0.02 µg/ml) (P<0.001), revealing
that KG-1a cells exhibited higher resistance to DOX (Fig. 1A).
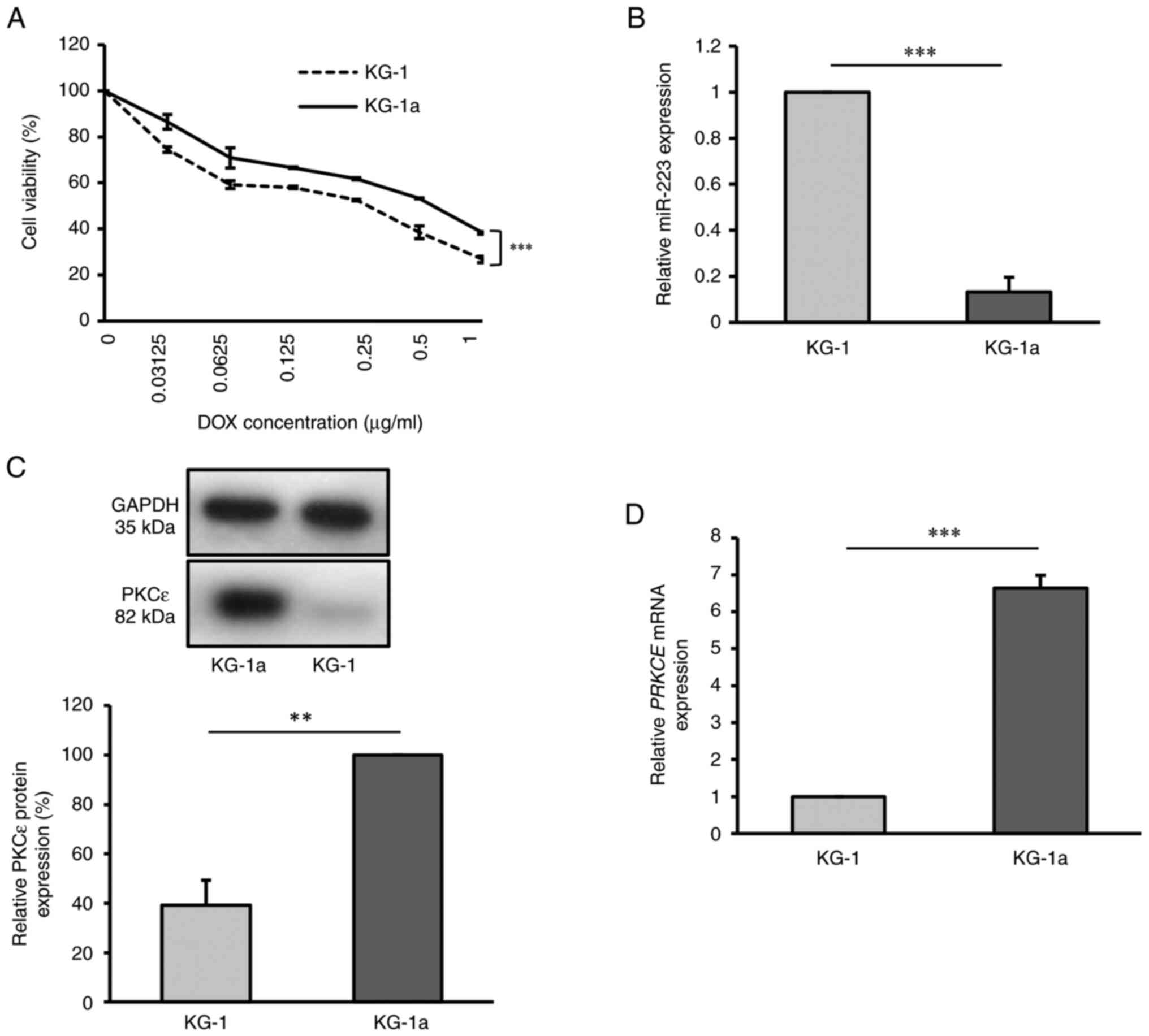 | Figure 1Drug resistant KG-1a cell line
exhibits downregulation of miR-223 and upregulation of PKCε. (A)
Cell viability of KG-1 and KG-1a cell lines after DOX treatment for
48 h, as detected by Cell Counting Kit-8 assay. (B) Relative
miR-223 expression in KG-1 and KG-1a cells, as detected by RT-qPCR.
(C) Relative PKCε protein level in KG-1 and KG-1a cells, as
detected by western blot analysis. (D) Relative PRKCE mRNA
expression in KG-1 and KG-1a cells, as detected by RT-qPCR. The
values of KG-1 group in (B), (C) and (D) were normalized to 1 or
100% without accounting for standard error, due to the use of
different batches for each replicate. **P<0.01;
***P<0.001 (n=3 replicates/group). The data were
analyzed using unpaired Student's t-test. DOX, doxorubicin; miR,
microRNA; PKCε/PRKCE, protein kinase C ε; RT-qPCR, reverse
transcription-quantitative PCR. |
Using the RT-qPCR method, miR-223 levels were
examined in KG-1 and KG-1a cells. There was a significantly higher
expression level of miR-223 in KG-1 cells compared with that in
KG-1a cells (P<0.001), indicating that miR-223 was expressed at
a lower level in the KG-1a cell line (Fig. 1B). To further examine PKCε
expression in KG-1 and KG-1a cells, both RT-qPCR and western blot
analysis were employed. The results demonstrated that PKCε
expression was significantly higher in KG-1a cells at both the mRNA
(P<0.001) and protein levels (P<0.01), compared with that in
the KG-1 cell line (Fig. 1C and
D).
miR-223 targets and inhibits the
expression of PKCε
To investigate the association between miR-223 and
PKCε, KG-1a cells were transfected with miR-223 mimic. After 24 h
of transfection, RT-qPCR analysis revealed a significant increase
in miR-223 expression in the miR-223 mimic group compared with that
in the NC group (P<0.01; Fig.
2A), indicating successful transfection.
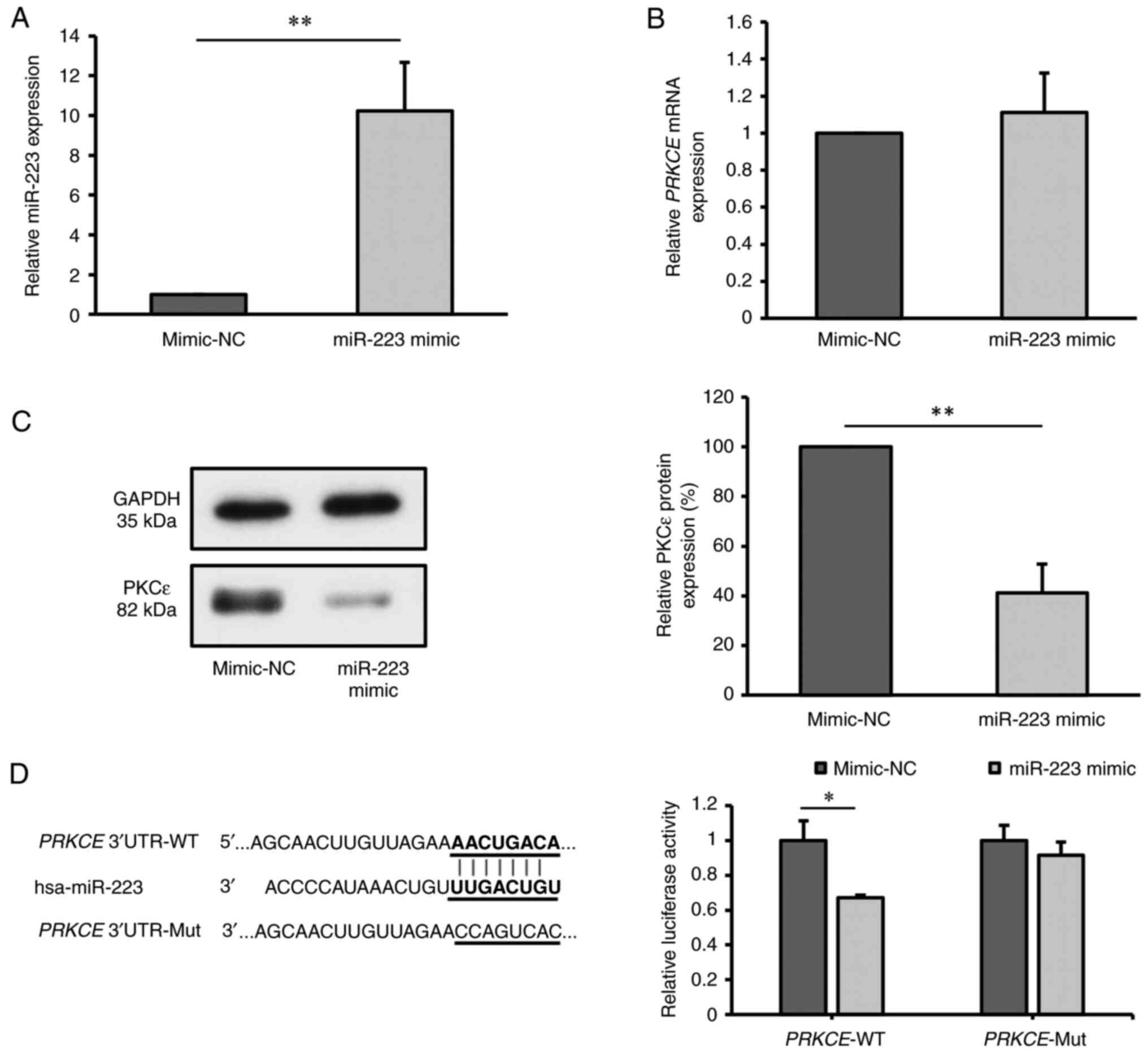 | Figure 2miR-223 targets and inhibits the
expression of PKCε. (A) Relative miR-223 expression in KG-1a cells
after transfection with miR-223 mimic, detected by RT-qPCR;
mimic-NC values were normalized to 1. (B) Relative PRKCE
mRNA expression in KG-1a cells after transfection with miR-223
mimic, as detected by RT-qPCR. (C) Relative PKCε protein level in
KG-1a cells after transfection with miR-223 mimic, as detected by
western blot analysis. (D) Binding sites between miR-223 and
PRKCE mRNA predicted by TargetScan, and relative PKCε
luciferase activity measured by dual-luciferase assay. The values
of mimic-NC group in (B) and (C) were normalized to 1 or 100%
without considering standard error, as each replicate was conducted
using different batches. *P<0.05;
**P<0.01 (n=3 replicates/group). The data were
analyzed using unpaired Student's t-test. miR, microRNA; NC,
negative control; PKCε/PRKCE, protein kinase C ε; WT,
wild-type; Mut, mutant; UTR, untranslated region; RT-qPCR, reverse
transcription-quantitative PCR. |
Subsequent evaluation of PKCε expression at both the
mRNA and protein levels post-transfection showed no significant
difference in PRKCE mRNA levels between the miR-223 mimic
group and mimic-NC group (P=0.455; Fig. 2B). However, there was a significant
decrease in PKCε protein level in the miR-223 mimic group compared
with that in the mimic-NC group (P<0.01; Fig. 2C), suggesting that the miR-223
mimic inhibited the protein expression of PKCε.
Considering the predicted binding site of miR-223 on
PRKCE mRNA, as obtained through the bioinformatics software
TargetScan, a dual-luciferase reporter assay was conducted to
validate this interaction. There was a significant reduction in
luciferase activity in cells co-transfected with
PRKCE-3'UTR-WT and miR-223 mimic (P<0.05), while no
changes were observed in the luciferase activity of cells
co-transfected with PRKCE-3'UTR-Mut and miR-223 mimic,
compared with that in the corresponding mimic-NC group (P=0.132)
(Fig. 2D). These findings indicate
that miR-223 can target and suppress PKCε expression.
Overexpression of miR-223 and
inhibition of PKCε are not associated with the drug sensitivity of
KG-1a cells
To explore the impact of miR-223 overexpression on
drug sensitivity in KG-1a cells, miR-223 mimic-transfected cells
were subjected to varying concentrations of DOX. The results showed
no significant difference in DOX sensitivity between the miR-223
mimic group and the mimic-NC group, with IC50 values of
0.614±0.15 and 0.558±0.08 µg/ml, respectively (P=0.598; Fig. 3A). This suggests that the
overexpression of miR-223 did not affect the DOX sensitivity of the
KG-1a cell line.
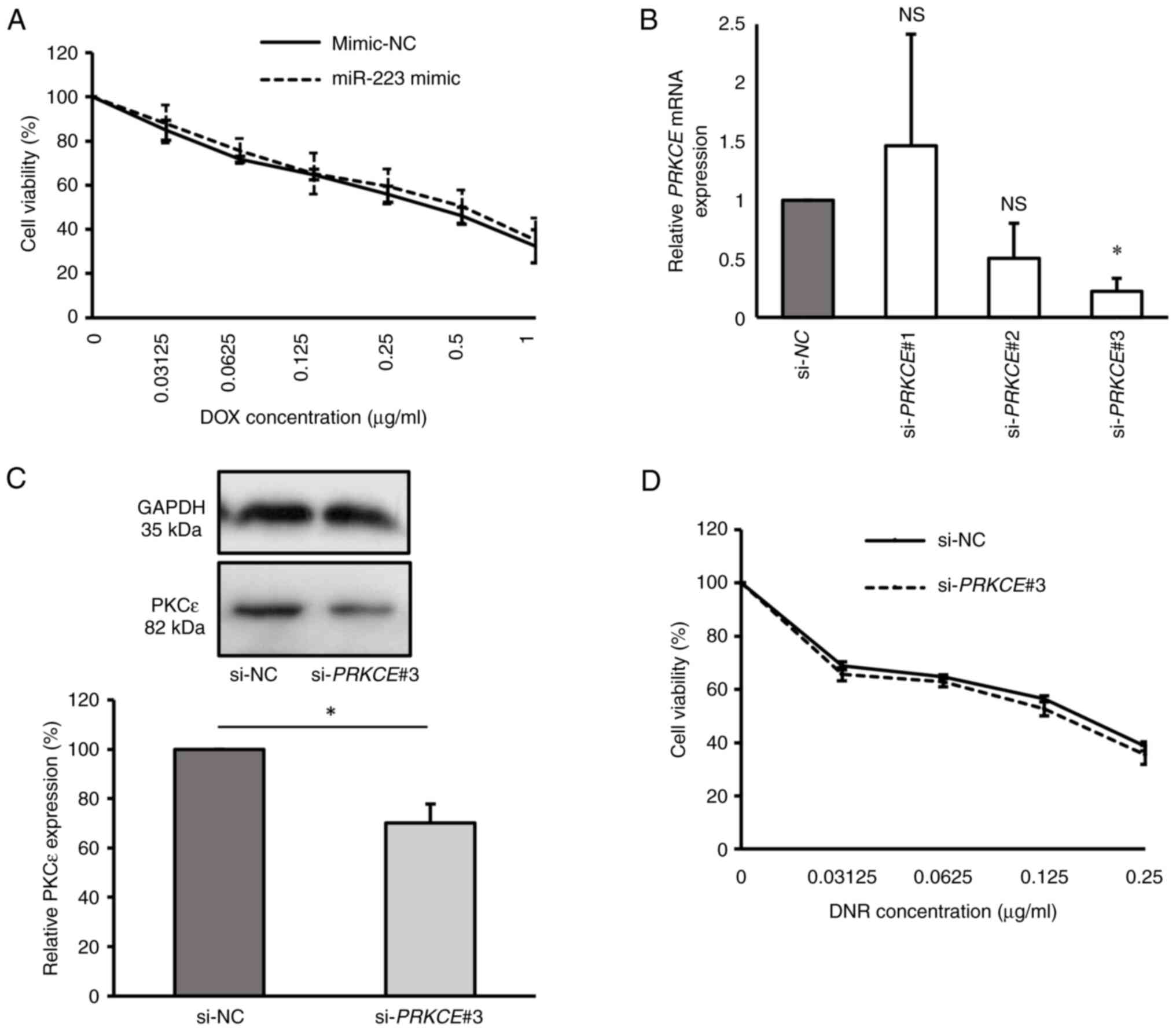 | Figure 3Overexpression of miR-223 and
inhibition of PKCε are not associated with the drug sensitivity of
KG-1a cells. (A) Cell viability of transfected KG-1a cells after
DOX treatment for 48 h, as detected by CCK-8 assay. (B) Relative
PRKCE mRNA expression in KG-1a cells after transfection with
PRKCE siRNAs, as detected by reverse
transcription-quantitative PCR. (C) Relative PKCε protein level in
KG-1a cells after transfection with si-PRKCE#3, as detected
by western blot analysis. (D) Cell viability of PKCε-knocked down
KG-1a cells after DNR treatment for 48 h, as detected by CCK-8
assay. The values of si-NC group in (B) and (C) were normalized to
1 or 100% without considering standard error, as each replicate was
conducted using different batches. *P<0.05 vs. si-NC
(n=3 replicates/group). The data were analyzed using unpaired
Student's t-test or one-way ANOVA followed by Dunnett's post hoc
test. DOX, doxorubicin; miR, microRNA; NC, negative control; si,
siRNA; PKCε/PRKCE; protein kinase C epsilon; DNR,
daunorubicin; CCK-8, Cell Counting Kit-8; NS, non-significant. |
To investigate the role of PKCε in the drug
sensitivity of KG-1a cells, the cells were transfected with si-NC,
si-PRKCE#1, si-PRKCE#2 and si-PRKCE#3 to
assess the effects of PKCε knockdown. The results indicated that
si-PRKCE#3 was the most effective siRNA for suppressing PKCε
expression, resulting in significantly lower PRKCE mRNA
expression compared with the si-NC group (P<0.05). Conversely,
the si-PRKCE#1 (P=0.917) and si-PRKCE#2 (P=0.291)
groups exhibited no significant differences in PRKCE mRNA
expression compared with the si-NC group (Fig. 3B). PKCε protein levels were
evaluated in the si-PRKCE#3 group, confirming a reduction in
PKCε levels in si-PRKCE#3-transfected cells compared with
those in the si-NC group (P<0.05; Fig. 3C). Consequently, KG-1a cells
transfected with si-PRKCE#3 were treated with various
concentrations of DNR, as previous findings have suggested that
PKCε overexpression confers selective resistance to DNR in AML
(20). However, the results
demonstrated that DNR sensitivity was not changed after PKCε
knockdown compared with the si-NC group, with IC50
values of 0.157±0.01 and 0.172±0.01 µg/ml, respectively (P=0.076;
Fig. 3D). These findings suggest
that neither miR-223 overexpression nor PKCε knockdown appears to
be directly associated with drug sensitivity in KG-1a cells.
Discussion
AML is a prevalent global hematological malignancy
(1-4),
with significant advancements in molecular targeted therapy leading
to complete remission in a number of patients. However,
affordability remains a considerable challenge in developing
countries, driven by high treatment expenses, limited
accessibility, inadequate insurance coverage, significant
out-of-pocket costs and income disparities. Furthermore, the lack
of treatment options in certain countries poses a challenge,
considering that targeted therapies may not be provided according
to the standard of care or may not be available for specific cancer
types (25-29).
Additionally, drug resistance remains the primary cause of
chemotherapy failure, impacting patient survival rates (4). Therefore, studies into traditional
chemotherapy remain crucial to provide effective treatment options.
Given the association of miR-223 with various cancer types
(10) and bioinformatics
predictions identifying the PRKCE gene as one of the targets
of miR-223, the present study aimed to elucidate the roles of
miR-223 and PKCε in regulating drug resistance mechanisms in
LSCs.
The current study showed that KG-1a cells exhibit a
higher IC50 for DOX compared with KG-1 cells, which is
consistent with the findings of a previous study that reported that
KG-1 cells were more responsive to DOX than KG-1a cells (30). The difference in the percentage of
CD34+ CD38- LSCs between the two cell lines
(75.95% in KG-1 and 92.82% in KG-1a), as revealed in our previous
study (31), indicates that the
number of CD34+ CD38- LSCs may impact the
chemosensitivity of these cell lines. It is well-established that
LSCs, characterized by dynamic origins, derive from various cell
types within leukemia, demonstrating their complex adaptability in
disease progression and treatment. These cells harbor various
elusive resistance mechanisms, such as inherent dormancy,
overexpression of ATP-binding cassette transporters, defects in
apoptotic signals, resistance to apoptosis and senescent signals,
metabolic reprogramming and epigenetic alternations, which
contribute to differences in chemosensitivity (32).
In the present study, lower miR-223 expression was
observed in KG-1a cells compared with KG-1 cells. Similarly, a
previous study reported lower expression of miR-223 in blast cells
from AML patients within fractions containing
CD34+CD38- LSCs compared with fractions of
more committed leukemic cells lacking the CD34 marker (11). Downregulation of miR-223 has been
observed in patients with AML, particularly those with intermediate
and unfavorable prognoses (11-13).
It is therefore likely that miR-223 functions as a
tumor-suppressing miRNA in AML.
In contrast to miR-223, PKCε was observed to be
upregulated in KG-1a cells at both the mRNA and protein levels,
compared with the KG-1 cell line. The bioinformatics prediction
software TargetScan, along with a luciferase assay, confirmed that
miR-223 binds to the 3'UTR of PRKCE mRNA, resulting in the
inhibition of the luciferase activity. Additionally, overexpression
of miR-223 suppressed PKCε protein expression in KG-1a cells,
although there was no notable impact on PRKCE mRNA levels. A
previous study similarly reported that, in non-small cell lung
cancer cell lines, miR-143 mimic transfection resulted in the
reduction of PKCε protein levels, while mRNA levels remained
unchanged (33). Typically, miRNAs
function by inhibiting translation when they only partially match
the 3'UTR of target genes. However, if there is a perfect match,
miRNAs induce mRNA cleavage of the target genes (34,35).
In animal cells, most interactions between miRNAs and their binding
sequences are not completely complementary, and mismatches usually
occur, especially in the central region of the target sequence
(35). Based on the findings of
the present study, miR-223 likely inhibits PRKCE mRNA
translation. However, a previous study found that when miRNAs are
temporarily overexpressed using mimics, the mRNA levels of target
genes decrease 30 min after transfection, but start to recover
after 12 h, while protein levels begin to recover after 24 h
(36). In the present study,
changes in PRKCE mRNA and protein levels were examined 24 h
after transfection. It cannot therefore be definitively determined
whether miR-223 functions as a translation suppressor for
PRKCE mRNA or if the observed effect is due to the recovery
of PRKCE mRNA following transfection. Previous studies have
also demonstrated the contrasting expression of miR-223 and PKCε.
Reduced miR-223 expression and the accompanying elevation of PKCε
levels were associated with the formation of Gottron's papules in
dermatomyositis (37). Conversely,
in ovarian cancer, increased miR-223 expression was observed
alongside decreased levels of PKCε (38). Together with the results of the
present study, this indicates that miR-223 can target and suppress
PKCε protein expression.
Despite the aforementioned findings, the
overexpression of miR-223 did not affect DOX sensitivity in the
KG-1a cell line. As miRNAs have the capacity to regulate numerous
genes by binding to the 3'UTR of target mRNAs, it is plausible that
miR-223 may target multiple genes apart from PKCε. For example, a
previous study in colorectal cancer cells demonstrated that miR-223
promoted DOX resistance by regulating epithelial-mesenchymal
transition via targeting of FBXW7 (39). Therefore, it is possible that
miR-223 may regulate other genes that have a more significant
impact on drug resistance than PKCε in the KG-1a LSC line.
It was also observed that downregulation of PKCε
using siRNA did not improve the sensitivity to DNR in KG-1a cells.
Despite a recent study by Nicholson et al (20), which demonstrated that inducing
PKCε overexpression in AML cell lines resulted in specific
resistance to DNR through an increase in P-glycoprotein levels, it
was also observed that PKCε knockdown in the AML U937 and MV4-11
cell lines did not impact chemosensitivity. This indicates that
PKCε inhibition alone may be insufficient to restore drug
resistance in AML cells, possibly due to the redundancy among PKC
isoforms. Drug resistance in LSCs can arise from various mechanisms
beyond drug efflux transporters. LSCs employ crucial signaling
pathways, such as Wnt/β-catenin, Hedgehog, NOTCH and PI3K/AKT, to
manage their stemness traits. This regulation induces a state of
dormancy (G0 state), which protects them from cell
cycle-specific factors that target actively proliferating cells.
Dysfunctions of apoptotic signals and senescence mechanisms in LSCs
also contribute to chemotherapy failure. Metabolic reprogramming
enables LSCs to adapt to energy level fluctuations, while
epigenetic modifications and reprogramming further enhance their
stemness properties (32). Some
studies have discussed the role of PKCε in the regulation of
stemness features, such as differentiation and self-renewal. For
example, downregulation of PKCε was indicated to preferentially
promote the differentiation of colorectal cancer cells (40). The phosphorylation of ERK-1/2 and
AKT, which typically promotes differentiation, was reduced through
short hairpin RNA-mediated knockdown of PKCε in human pluripotent
stem cells, resulting in a metastable undifferentiated state
(15). In another study, PKCε
exhibited no notable influence on the efficiency of colony
formation, but eliminated the formation of cobblestone area-forming
cells, indicating that PKCε selectively promotes the quiescence of
HSCs (41). Based on this
evidence, PKCε may preferably function in the regulation of
differentiation or self-renewal signaling pathways rather than by
directly influencing drug sensitivity in AML.
The present study is constrained by certain
limitations. Firstly, the experiments were conducted in
vitro, specifically focusing on a single cell line, KG-1a
cells, which may not fully represent the diversity of LSCs. The
second limitation is the absence of clinical studies to confirm the
results of the in vitro experiments, particularly the
evaluation of miRNA-223 and PKCε levels in newly diagnosed patients
with AML before and after treatment, as well as the therapeutic
response of these patients. Therefore, addressing these limitations
should be a priority for future research endeavors.
Overall, the present study unveiled the
downregulation of miR-223 in human LSC-like KG-1a cells. The
biological function of miRNAs is to regulate target genes. In the
present study, a luciferase reporter assay system and
bioinformatics analysis were utilized to validate PKCε as one of
the target genes of miR-223 in KG-1a LSCs. However, both the
overexpression of miR-223 and PKCε knockdown failed to improve the
chemosensitivity of KG-1a cells, suggesting that the miR-223/PKCε
axis may not be associated with the sensitivity of KG-1a cells to
anthracycline drugs, such as DOX and DNR. Consequently, further
investigations are warranted to elucidate the roles of miR-223 and
PKCε in the drug resistance of LSCs. This understanding is crucial
for unraveling drug resistance mechanisms and achieving objectives
related to using miRNAs as biomarkers for diagnosis and prognosis,
as well as developing targeted therapies in AML.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Master's degree
program in Medical Technology, Faculty of Associated Medical
Science, Chiang Mai University (through the Chiang Mai University
Presidential Scholarship; grant no. 2564-070) and the Fundamental
Fund 2020 from Chiang Mai University (grant no. R000030108).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
MO performed the experiments, and contributed to
data acquisition, statistical analysis and the preparation of the
manuscript. SD contributed to the study conception and design and
the manuscript review. CI performed the experiments regarding the
PKCε knockdown. SA advised on the design of the experiments
regarding cell culture. PK provided the 293T cell line and advised
on the use of molecular techniques during the experiments. SD and
MO confirmed the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Deschler B and Lübbert M: Acute myeloid
leukemia: Epidemiology and etiology. Cancer. 107:2099–2107.
2006.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Pelcovits A and Niroula R: Acute myeloid
leukemia: A review. R I Med J (2013). 103:38–40. 2020.PubMed/NCBI
|
|
3
|
Vakiti A and Mewawalla P: Acute myeloid
leukemia. In: StatPearls [Internet], StatPearls Publishing,
Treasure Island, FL, 2022.
|
|
4
|
Zhang J, Gu Y and Chen B: Mechanisms of
drug resistance in acute myeloid leukemia. Onco Targets Ther.
12:1937–1945. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Marchand T and Pinho S: Leukemic stem
cells: From leukemic niche biology to treatment opportunities.
Front Immunol. 12(775128)2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hanekamp D, Cloos J and Schuurhuis GJ:
Leukemic stem cells: Identification and clinical application. Int J
Hematol. 105:549–557. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Jordan CT: The leukemic stem cell. Best
Pract Res Clin Haematol. 20:13–18. 2007.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang X, Huang S and Chen JL: Understanding
of leukemic stem cells and their clinical implications. Mol Cancer.
16(2)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Phi LTH, Sari IN, Yang YG, Lee SH, Jun N,
Kim KS, Lee YK and Kwon HY: Cancer stem cells (CSCs) in drug
resistance and their therapeutic implications in cancer treatment.
Stem Cells Int. 2018(5416923)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Aziz F, Chakraborty A, Khan I and Monts J:
Relevance of miR-223 as potential diagnostic and prognostic markers
in cancer. Biology (Basel). 11(249)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Gentner B, Pochert N, Rouhi A, Boccalatte
F, Plati T, Berg T, Sun SM, Mah SM, Mirkovic-Hösle M, Ruschmann J,
et al: MicroRNA-223 dose levels fine tune proliferation and
differentiation in human cord blood progenitors and acute myeloid
leukemia. Exp Hematol. 43:858–868.e7. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yu G, Yin Z, He H, Zheng Z, Chai Y, Xuan
L, Lin R, Wang Q, Li J and Xu D: Low serum miR-223 expression
predicts poor outcome in patients with acute myeloid leukemia. J
Clin Lab Anal. 34(e23096)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Xiao Y, Su C and Deng T: miR-223 decreases
cell proliferation and enhances cell apoptosis in acute myeloid
leukemia via targeting FBXW7. Oncol Lett. 12:3531–3536.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ding L, Wang H, Lang W and Xiao L: Protein
kinase C-epsilon promotes survival of lung cancer cells by
suppressing apoptosis through dysregulation of the mitochondrial
caspase pathway. J Biol Chem. 277:35305–35313. 2002.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kinehara M, Kawamura S, Tateyama D, Suga
M, Matsumura H, Mimura S, Hirayama N, Hirata M, Uchio-Yamada K,
Kohara A, et al: Protein kinase C regulates human pluripotent stem
cell self-renewal. PLoS One. 8(e54122)2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Flescher E and Rotem R: Protein kinase C
epsilon mediates the induction of P-glycoprotein in LNCaP prostate
carcinoma cells. Cell Signal. 14:37–43. 2002.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Huang B, Cao K, Li X, Guo S, Mao X, Wang
Z, Zhuang J, Pan J, Mo C, Chen J and Qiu S: The expression and role
of protein kinase C (PKC) epsilon in clear cell renal cell
carcinoma. J Exp Clin Cancer Res. 30(88)2011.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang H, Zhan M, Xu SW, Chen W, Long MM,
Shi YH, Liu Q, Mohan M and Wang J: miR-218-5p restores sensitivity
to gemcitabine through PRKCE/MDR1 axis in gallbladder cancer. Cell
Death Dis. 8(e2770)2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhang GF, Wu JC, Wang HY, Jiang WD and Qiu
L: Overexpression of microRNA-205-5p exerts suppressive effects on
stem cell drug resistance in gallbladder cancer by down-regulating
PRKCE. Biosci Rep. 40(BSR20194509)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Nicholson R, Menezes AC, Azevedo A,
Leckenby A, Davies S, Seedhouse C, Gilkes A, Knapper S, Tonks A and
Darley RL: Protein kinase C epsilon overexpression is associated
with poor patient outcomes in aml and promotes daunorubicin
resistance through p-glycoprotein-mediated drug efflux. Front
Oncol. 12(840046)2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Skopek R, Palusińska M, Kaczor-Keller K,
Pingwara R, Papierniak-Wyglądała A, Schenk T, Lewicki S, Zelent A
and Szymański Ł: Choosing the right cell line for acute myeloid
leukemia (AML) research. Int J Mol Sci. 24(5377)2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Koeffler HP, Billing R, Lusis AJ, Sparkes
R and Golde DW: An undifferentiated variant derived from the human
acute myelogenous leukemia cell line (KG-1). Blood. 56:265–273.
1980.PubMed/NCBI
|
|
23
|
Varkonyi-Gasic E and Hellens RP:
Quantitative stem-loop RT-PCR for detection of microRNAs. In: RNAi
and plant gene function analysis: Methods and protocols. Kodama H
and Komamine A (eds). Humana Press, Totowa, NJ, pp145-157,
2011.
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Lopes Gde L Jr, de Souza JA and Barrios C:
Access to cancer medications in low- and middle-income countries.
Nat Rev Clin Oncol. 10:314–322. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Cherny N, Sullivan R, Torode J, Saar M and
Eniu A: ESMO European consortium study on the availability,
out-of-pocket costs and accessibility of antineoplastic medicines
in Europe. Ann Oncol. 27:1423–1443. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Jain M and Mukherjee K: Economic burden of
breast cancer to the households in Punjab, India. Int J Med Public
Health. 6(13)2016.
|
|
28
|
Ruff P, Al-Sukhun S, Blanchard C and
Shulman LN: Access to cancer therapeutics in low- and middle-income
countries. Am Soc Clin Oncol Educ Book. 35:58–65. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kaiser AH, Rotigliano N, Flessa S, Ekman B
and Sundewall J: Extending universal health coverage to informal
workers: A systematic review of health financing schemes in low-
and middle-income countries in Southeast Asia. PLoS One.
18(e0288269)2023.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chueahongthong F, Tima S,
Chiampanichayakul S, Berkland C and Anuchapreeda S: Co-treatments
of edible curcumin from turmeric rhizomes and chemotherapeutic
drugs on cytotoxicity and FLT3 protein expression in leukemic stem
cells. Molecules. 26(5785)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Panyajai P, Amnajphook N,
Keawsangthongcharoen S, Chiampanichayakul S, Tima S and
Anuchapreeda S: Study of leukemic stem cell population
(CD34+/CD38-) and WT1 protein expression in human leukemic cell
lines. J Assoc Med Sci. 51:38–44. 2018.
|
|
32
|
Niu J, Peng D and Liu L: Drug resistance
mechanisms of acute myeloid leukemia stem cells. Front Oncol.
12(896426)2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhang N, Su Y and Xu L: Targeting PKCε by
miR-143 regulates cell apoptosis in lung cancer. FEBS Lett.
587:3661–3667. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Oliveto S, Mancino M, Manfrini N and Biffo
S: Role of microRNAs in translation regulation and cancer. World J
Biol Chem. 8(45)2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9(402)2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Jin HY, Gonzalez-Martin A, Miletic AV, Lai
M, Knight S, Sabouri-Ghomi M, Head SR, Macauley MS, Rickert RC and
Xiao C: Transfection of microRNA mimics should be used with
caution. Front Genet. 6(340)2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Inoue K, Jinnin M, Yamane K, Makino T,
Kajihara I, Makino K, Honda N, Nakayama W, Fukushima S and Ihn H:
Down-regulation of miR-223 contributes to the formation of
Gottron's papules in dermatomyositis via the induction of PKCε. Eur
J Dermatol. 23:160–167. 2013.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Khan K, Zafar S, Badshah Y, Ashraf NM,
Rafiq M, Danish L, Shabbir M, Trembley JH, Afsar T, Almajwal A and
Razak S: Cross talk of tumor protein D52 (TPD52) with KLF9, PKCε,
and MicroRNA 223 in ovarian cancer. J Ovarian Res.
16(202)2023.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ding J, Zhao Z, Song J, Luo B and Huang L:
MiR-223 promotes the doxorubicin resistance of colorectal cancer
cells via regulating epithelial-mesenchymal transition by targeting
FBXW7. Acta Biochim Biophys Sin (Shanghai). 50:597–604.
2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Gobbi G, Di Marcantonio D, Micheloni C,
Carubbi C, Galli D, Vaccarezza M, Bucci G, Vitale M and Mirandola
P: TRAIL up-regulation must be accompanied by a reciprocal PKCε
down-regulation during differentiation of colonic epithelial cell:
Implications for colorectal cancer cell differentiation. J Cell
Physiol. 227:630–638. 2012.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Nicholson RL, Knapper S, Tonks A and
Darley RL: PKC-Epsilon overexpression is associated with poor
outcomes in AML and promotes chemoresistance and hematopoietic stem
cell quiescence. Blood. 134 (Suppl 1)(S2704)2019.
|

















