Introduction
Colorectal cancer, also known as colon cancer, is a
type of cancer that occurs in the colon or rectum. Cells that
undergo mutation may grow and divide at an unusual rate and fail to
undergo standard apoptosis. Thus, such cells may develop into
unnatural growths known as polyps, which if left alone, can develop
into cancerous tumors (1).
According to the American Society of Clinical Oncology, the current
treatment methods for colorectal cancer can be categorized into
three types: Surgeries, radiation therapies and medication
therapies (2). Surgeries typically
involve the physical removal of the cancer and are also used to
mitigate certain symptoms of colorectal cancer, such as in cases
where the large intestine is blocked/damaged by cancer, preventing
proper exocrine function. However, such operations can also result
in side effects such as constipation and diarrhea. Radiation
therapy, on the other hand, removes cancer using high-frequency
waves. This is often used to complement surgeries, such as by
removing small tumors that are difficult to remove surgically. Side
effects range from fatigue to bleeding in the rectum. Finally,
treatments with medication utilize various drugs to eliminate or
prevent the replication of cancer cells through chemotherapy or
targeted therapy. Some medications also aim to increase the
capabilities of the immune system of the body. Side effects tend to
be drug-specific (2).
Naproxen (NS) is typically known for its use as a
non-steroidal anti-inflammatory drug (NSAID) and has been a subject
of research due to its potential use as a medical treatment in the
form of chemotherapy. In 2014, a study by the Women's Health
Initiative (3) concluded that
NSAIDs were associated with reduced risks of colorectal cancer and
could have chemopreventive effects. Possible chemopreventive
effects of a hydrogen sulfide-releasing NS were also supported by a
study that used a xenograft mouse model. The study also revealed
that NS could inhibit growth via downregulation of NF-κB (4). In 2019, an in vivo study on
rats further demonstrated the potential for aspirin and NS to fight
colorectal cancer, including through enabling apoptosis via
caspase-3(5). A previous study on
the effects of NS in patients with Lynch syndrome, which increases
the probability of developing colorectal cancer, found that NS was
safe to use, boosted immune activity in humans and exerted
chemopreventive effects in mice (6). While studies such as the
aforementioned ones have demonstrated that NS may have
chemopreventive properties, fewer studies have analyzed the
association between NS and the activation of specific pathways in
colon cells, such as apoptosis. In the present study, it was
hypothesized that NS may have a variety of beneficial effects in
fighting and limiting colorectal cancer. The present study aimed to
reveal the impacts of NS on relevant biological cell pathways and
its effects on cancer cell survival and proliferation.
Materials and methods
Cell culture
Colo320 (Colo 320DM; cat. no. CCL-220) and CCD-18
cells (CCD-18Co; cat. no. CRL-1459) were obtained from American
Type Culture Collection and used in the experiments. The cells were
cultured in cell culture flasks with Eagle's Minimum Essential
Medium (MEM) supplemented with 10% FBS, both of which were supplied
by Thermo Fisher Scientific, Inc. MEM was replaced weekly. Cells
were incubated at 37˚C with 5% CO2. For the assays,
96-well plates contained 100 µl MEM and ~10,000 cells per well, and
6-well plates contained 2 ml MEM and ~200,000 cells per well.
Molecular docking
PyRx version 0.8 (https://pyrx.sourceforge.io/) was used for molecular
docking analysis to help identify potential targets for NS in
colorectal cancer cells. Images were generated of the interactions
via BIOVIA Discovery Studio Visualizer v21.1.0.20298 (https://discover.3ds.com/discovery-studio-visualizer-download).
Dilutions
NS caplets (Sigma-Aldrich; Merck KGaA) dissolved in
water were obtained at a concentration of 83.5 mM. Serial dilutions
were used to obtain four more different concentrations of NS. These
five total concentrations were then used for each assay,
occasionally further diluted by differing quantities due to the
differing procedures of each specific assay.
Caspase-3 assay
The caspase-3 assay (cat. no. BAQ009; G-Biosciences)
was performed according to the manufacturer's protocol (7). Cells were cultured in a 6-well plate
and treated with 10 µl per well of the respective chemicals at
various concentrations. After treatment and incubation, the cells
were collected in 1.5-ml tubes with 50 µl caspase lysis buffer and
stored at -20˚C for 24 h. The assay was performed in a 96-well
plate in triplicates per sample. Each well of the 96-well plate
contained 50 µl caspase assay buffer, 45 µl caspase lysis buffer, 5
µl lysed sample and 5 µl caspase-3 substrate. The application
‘microplate manager 6’ version 3.2.1 by Bio-Rad Laboratories, Inc.
was used to obtain absorbance data at a wavelength of 415 nm. Data
points were collected every 15 min for 60 min. Percent changes in
caspase-3 activity were calculated using the following equation:
[(Treatment group absorbance-control absorbance)/control
absorbance] x100%=% change in caspase activity.
MTT assay
Colo320 cells were seeded into a 96-well plate. Each
well received 5 µl of the respective treatments. The plate was
divided into six groups of 16 wells each for the five
concentrations of treatments and the control. The plate was then
placed in an incubator at 37˚C for two different time periods of 24
h and 1 week. For assays lasting 1 week, media were replaced and
treatment was re-added at the same concentration on the fourth day.
After the respective time period, 10 µl MTT was added to each well.
After 90 min, 80 µl DMSO was added to each well. After ~10 min, the
wells were transferred to a spectrophotometer, and ‘microplate
manager 6’ was used to obtain absorbance data at a wavelength of
595 nm. The percentage cell survival was calculated using the
following equation: (Treatment group absorbance/control absorbance)
x100%=% cell survival.
Cell migration assay
The cell migration assay was performed only on the
cancerous cell line because the cancerous cell line was the one
relevant to NS's effects on the migratory ability of colorectal
cancer. Colo320 cells were seeded into two 6-well plates at ~80%
confluency. One plate received no treatment and the other received
10 µl of 41.75 µM NS. A total of three relatively straight,
horizontal scratch lines were made on each well using a 200-µl
pipette tip. Each well was provided with 2 ml Eagle's Minimum
Essential Medium (MEM) supplemented with 5% fetal bovine serum
(FBS), after which the plates were placed in an incubator and
stored for 24 h at 37˚C. Images were captured of each well at 0 and
24 h. Measurements were made regarding the vertical distance of
each scratch mark in each image using a compound light microscope
[AMScope version x64, 4.8.15934.20191112 (https://amscope.com/) (AmScope) and MS Paint
(Microsoft Corporation)]. The percentage of wound closure was
calculated for each group using the following equation: [(0 h
average distance-24 h average distance)/0 h average distance]
x100%=% increase in cell distance.
RNA sequencing
RNA sequencing was selected over DNA microarray due
to a variety of advantages, including avoiding systematic biases,
increased number of detectable splices, and ability to detect a
wider range of expression changes, as demonstrated by studies over
the past decade (8,9). Similar to cell migration, the test
was performed only on the cancerous cell line because the cancerous
cell line was the one relevant to NS's effects on the gene
expression of colorectal cancer. RNA sequencing services were
provided by Azenta, Inc. Sequencing was performed using the methods
listed online (https://fs.hubspotusercontent00.net/hubfs/3478602/13002-SD-2%201221%20RNA-Seq%20Technical%20Specs%20Sell%20Sheet.pdf).
ELISA
ELISA kits [Human MUC5AC (Mucin 5 Subtype AC) ELISA
kit, cat.no. E-EL-H2279; and human MUC5B (Mucin 5 Subtype B) ELISA
kit, cat.no. E-EL-H2280] were obtained from Elabscience
Biotechnology, Inc. All reagents, apart from DI water, were
provided in the kits. The ELISA was performed according to the
manufacturer's protocol (10). The
assay was performed in 34 wells of a 96-well plate. A total of 16
wells contained 100 µl of varying concentrations of standard; six
more contained control, 4.175 or 41.75 µM NS treatments of Colo320
cells. The plate was incubated for 90 min at 37˚C. Subsequently,
100 µl Biotinylated Detection Ab working solution was added to each
well, followed by 1 h of incubation. The wells were then washed
three times before 100 µl HRP Conjugate working solution was added
to each well and incubation was performed for another 30 min. After
five more washes, 90 µl Substrate Reagent was added to each well,
followed by 15 min of incubation. Finally, 50 µl Stop Solution was
added to each well before the optical density was measured using
‘microplate manager 6’. The mucin 5AC, oligomeric mucus/gel-forming
(MUC5AC) concentration was calculated for each treatment and the
control via the equation provided by the standard curve was
generated by the 16 wells of standard.
Statistical analysis
Experiments were repeated 3 times. Data were
presented as the mean ± 5-10% error. For all statistical analyses
including multiple groups, two-tailed one-factor ANOVA tests were
performed in Microsoft Excel (Microsoft Corporation) with P<0.05
considered to indicate a statistically significant difference. For
statistically significant ANOVA results, Dunnett's was used in
post-hoc tests with control and performed in Microsoft Excel. In
cases where ANOVA was not applicable, two-tailed t-tests were
performed in Google Sheets with a 95% confidence interval and
P<0.05.
Results
Binding of NS to critical
molecules
Molecular docking analysis using PyRx demonstrated
the potential for NS to bind to molecules relevant to colorectal
cancer, including Fas receptor with a Vina binding affinity of -6.3
(PDB ID: 1DDF), TRAIL with a Vina binding affinity of -6.2 (PDB ID:
1D2Q), caspase-3 with a Vina binding affinity of -7.2 (PDB ID:
1NMS), and MMP9 with Vina binding affinities of -7 (PDB ID 1L6J),
-6.8 (PDB ID 1GKD) and -6.5 (PDB ID 1GKC), as observed in Fig. S1, Fig. S2, Fig. S3, Fig. S4, Fig. S5 and Fig. S6, respectively.
Effects of NS on cell survival
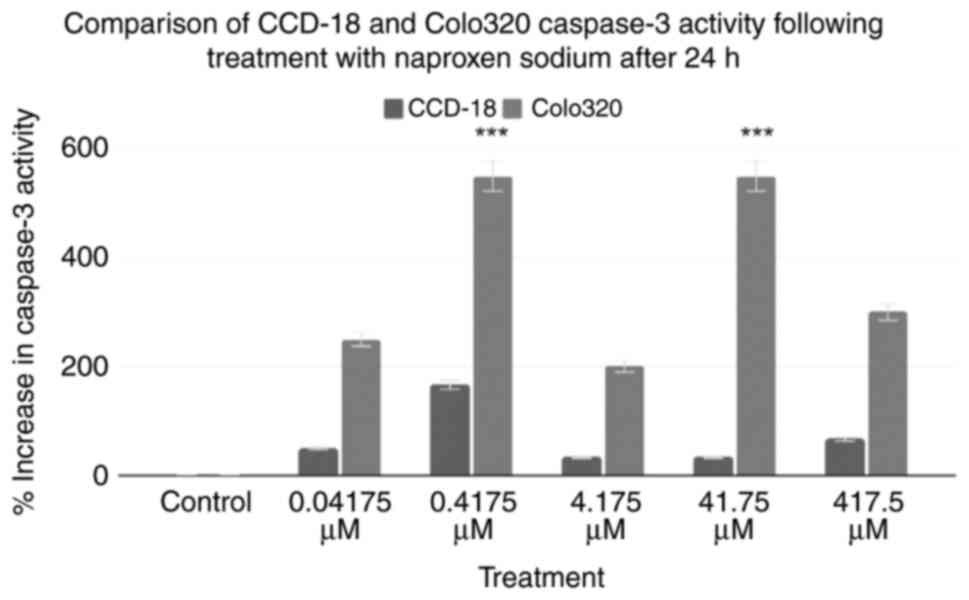
Caspase-3 assays were performed on two cell lines:
CCD-18 and Colo320 cells. The combined results of the caspase-3
assays are shown in Fig. 1. Using
the control as a baseline, all treatment groups exhibited an
increase in caspase-3 activity. The 0.04175, 0.4175, 4.175, 41.75
and 417.5 µM treatments of Colo320 cells increased caspase-3
activity by 250, 550, 200, 550 and 300%, respectively. A two-tailed
ANOVA at P<0.05 demonstrated statistical significance within
groups for Colo320, but not CCD18 (P=0.17, P<0.001,
respectively). Post-hoc Dunnett's tests revealed that only the
0.4175 and 41.75 µM treatments for Colo320 were statistically
significant (P≥0.999, P<0.001, P≥0.999, P<0.001, P=0.052,
respectively).
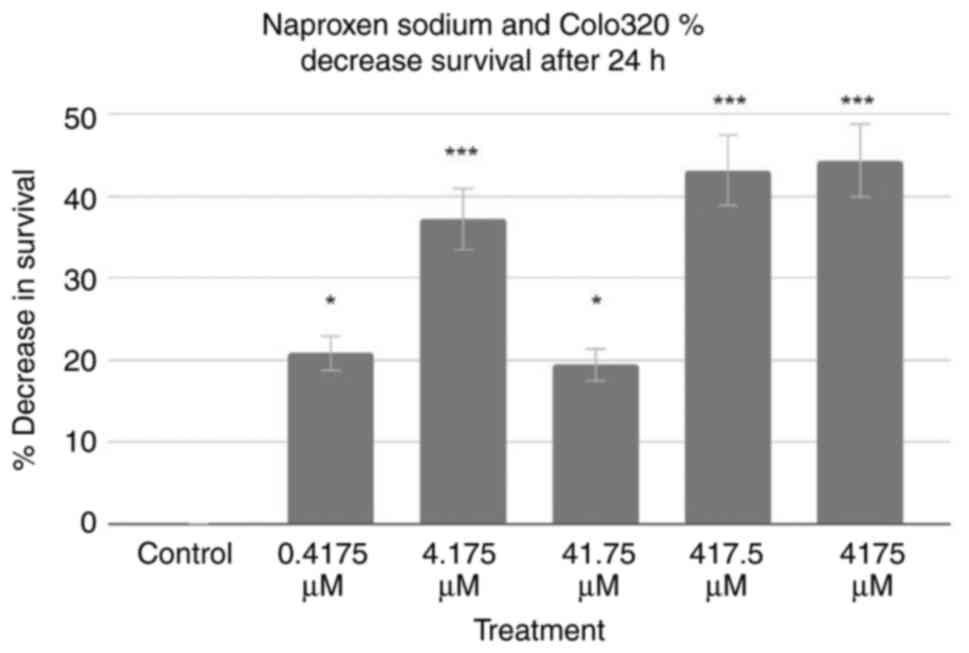
Based on these data, MTT assays for the Colo320 cell
line were performed. The results of 24-h tests are demonstrated in
Fig. 2. Using the control as a
baseline, all treatment groups exhibited a decrease in Colo320 cell
survival. The 0.4175, 4.175, 41.75, 417.5 and 4,175 µM treatments
decreased Colo320 cell survival rates by 20.86, 37.24, 19.45, 43.22
and 44.40%, respectively. A two-tailed ANOVA at P<0.05
demonstrated significance (P<0.001), and post-hoc Dunnett's
tests revealed that every treatment was statistically significant
(P=0.015, P<0.001, P=0.027, P<0.001 and P<0.001,
respectively).
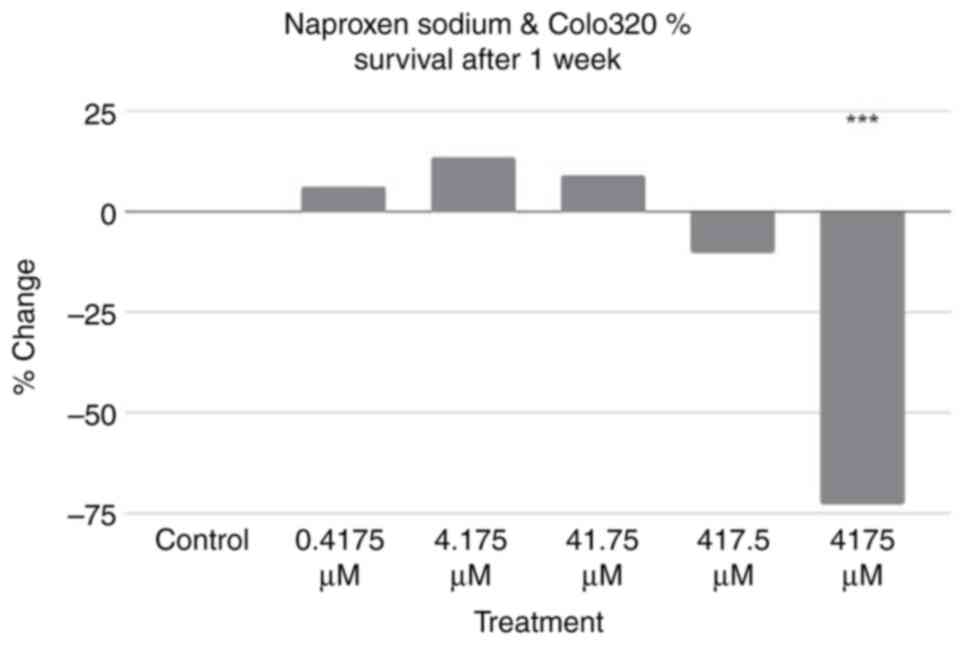
Additional assays using the same treatment
conditions were performed using Colo320 cells incubated for a week
rather than a day. The results are revealed in Fig. 3. Using the control as a baseline,
three treatment groups exhibited an increase in percentage of cell
survival, while two exhibited a decrease in percentage of cell
survival. A two-tailed ANOVA at P<0.05 showed statistical
significance between groups (P<0.001); a post-hoc Dunnett's test
revealed that only the result of the 4,175 µM treatment was
statistically significant (P=≥0.999, P=≥0.999, P=≥0.999, P=≥0.999
and P<0.001, respectively).
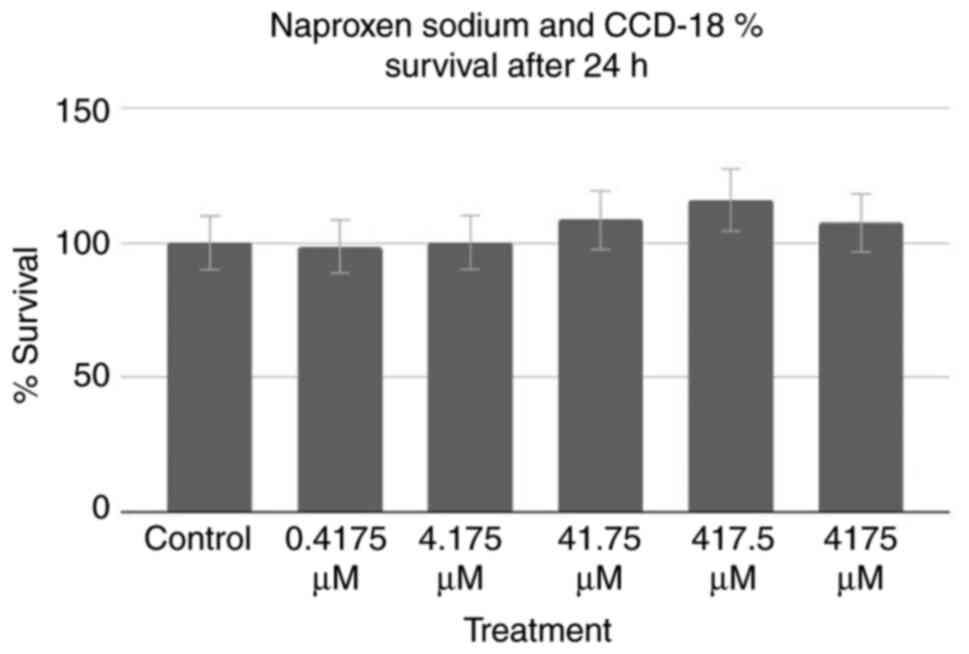
All aforementioned assays were performed on Colo320
cancer cells; however, assays were also performed on healthy cells
to test for potentially harmful effects on surrounding
non-cancerous cells. An MTT assay of healthy CCD-18 cells was
performed, the results of which can be observed in Fig. 4. Using the control as a baseline,
one group (0.4175 µM treatment) exhibited a decrease in percentage
of cell survival, whereas the remaining four groups, and notably
the groups treated with the higher half of concentrations,
exhibited an increase. However, a two-tailed ANOVA at P<0.05
demonstrated no statistical significance (P=0.596).
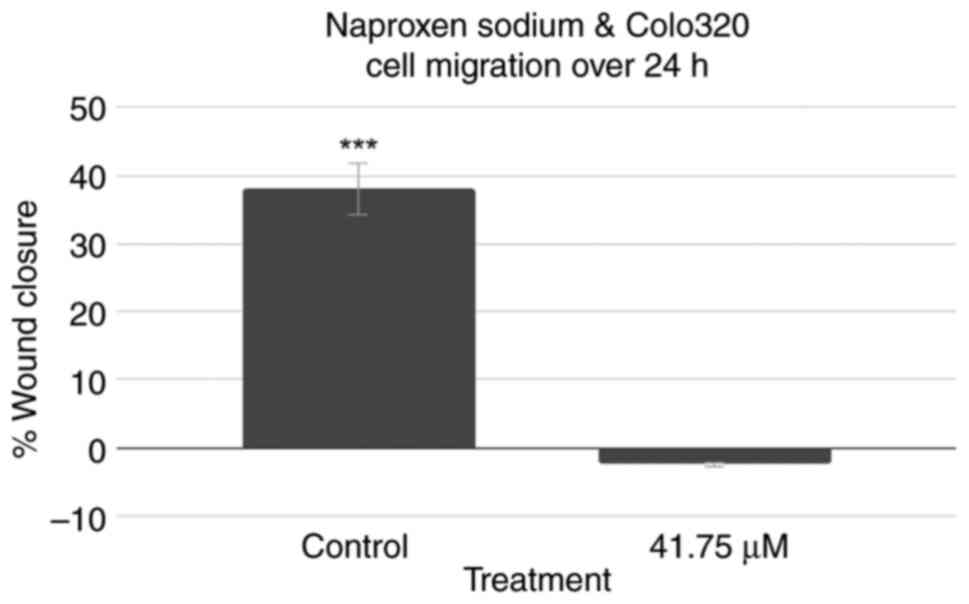
Effects of NS on cell migration
A scratch cell migration assay was also performed
using Colo320 cells to assess the effect of treatment with NS on
the capability of colorectal cancer cells to regenerate and
migrate. The results are demonstrated in Fig. 5. Representative images captured for
the 0-h control, 0-h treatment, 24-h control and 24-h treatment
wells can be observed in Fig. S7,
Fig. S8, Fig. S9 and Fig. S10, respectively. While the
control group exhibited a decrease in cell distance and
demonstrated wound closure, the treatment group instead exhibited a
slight increase in cell distance. A two-tailed t-test at P<0.05
revealed that the control group's results were statistically
significant (P<0.001), while the treatment group's results were
not (P=0.338).
Effects of NS on gene expression
RNA sequencing was performed to analyze the impact
of NS treatment on gene expression of Colo320 cells and its
potential association with the aforementioned outcomes with regards
to cell survival, caspase-3 expression and cell migration. The
results are revealed in Fig. 6.
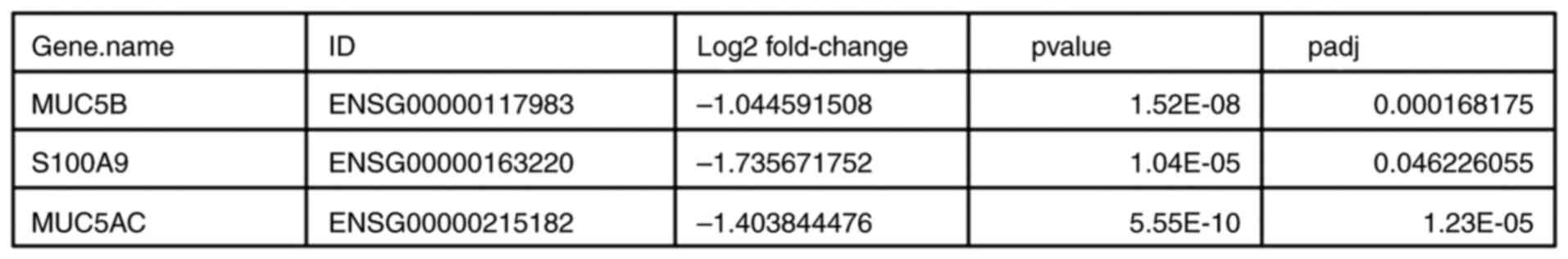
RNA sequencing of cells treated with NS revealed, after adjusting
the P-value, that the difference in expression of exclusively three
genes compared with the control samples was statistically
significant. The three genes were: Mucin 5B, oligomeric
mucus/gel-forming (MUC5B), S100 calcium binding protein A9 (S100A9)
and MUC5AC. The log2 fold change was -1.044591508,
-1.735671752 and -1.403844476, respectively, with P-values of
1.52x10-8, 1.04x10-5 and
5.55x10-10, respectively, and adjusted P-values of
1.68x10-4, 4.62x10-2 and
1.23x10-5, respectively.
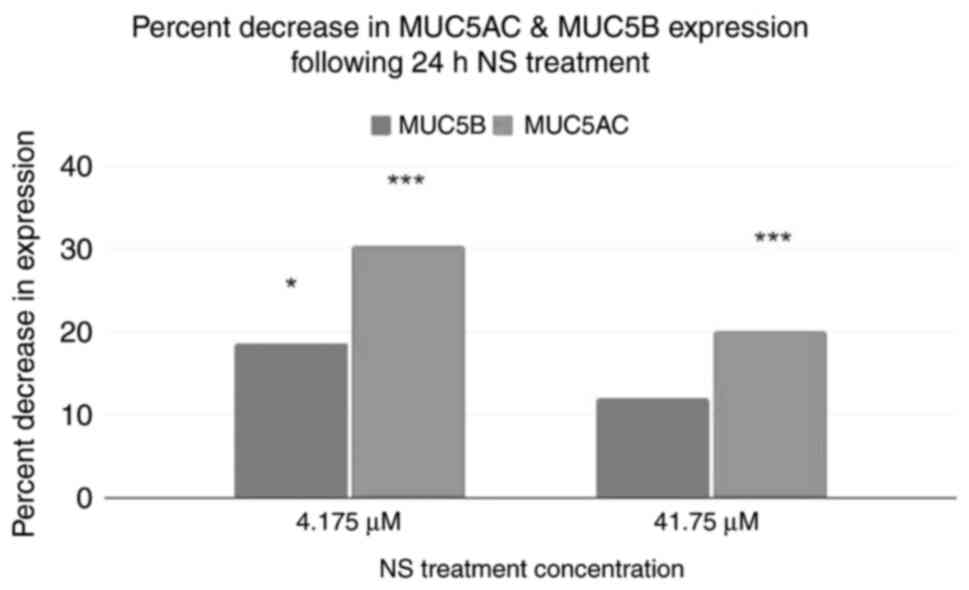
An ELISA was performed to corroborate the results of
RNA sequencing (Fig. 7). Using the
control as a baseline, both treatment groups exhibited a decrease
in MUC5AC and MUC5B protein expression. A two-tailed ANOVA showed
that both MUC5AC and MUC5B levels were significantly different in
response to NS treatment (P<0.001 and P=0.032). Post-hoc
Dunnett's tests demonstrated that the levels of MUC5AC were
significantly different in both treatment groups (P<0.001 and
P<0.001), whereas MUC5B levels were only significantly different
in the 4.175 µM NS group, but not in the 41.75 µM treatment group
(P=0.01733 and P≥0.999).
Discussion
The caspase-3 assay suggested that treatment with NS
increased caspase-3 activity in Colo320 cells, especially since no
treatment resulted in a decrease in caspase-3 activity. However,
this appears to contradict previous findings on the nature of
NSAIDs (11). This apparent
contradiction may have been due to the testing of NS in cancer
cells as opposed to healthy cells, a possibility reinforced by the
lack of a statistically significant increase in caspase-3 activity
in most CCD-18 cell treatment groups. However, the Colo320 cell
results also confirmed the previously mentioned study which found
that aspirin and NS could fight colorectal cancer in rats via
increased caspase-3 activity (5),
as caspase-3 is a known apoptotic marker (12).
Combined with the caspase-3 assay, the results of
the first MTT assay demonstrated that treatment of colorectal
cancer cells with NS induced apoptosis, leading to cell death.
However, the second MTT assay demonstrated that over time, the
effects of the treatment weakened if the treatment was not
maintained at high concentrations. Additionally, the third MTT
assay discovered no evidence that NS caused any change in the
survival of non-cancerous, healthy cells. This finding was also
consistent with the finding of the present study that treatments
with NS had no statistically significant impact on caspase-3
activity in healthy cells. These findings were reflected clearly in
Figs. 1 and 2. The former revealed how all treatments
resulted in a large increase in Caspase-3 activity for Colo320
cells, and how those changes were significantly smaller for CCD-18
cells. The latter revealed how all treatments of Colo320 resulted
in a statistically significant decrease in cell survival.
Cell migration assay results demonstrated that NS
treatment resulted in a lack of change in cell distance, implying
that NS treatment resulted in decreases in the migration of Colo320
cells.
Based on RNA sequencing, the MUC5B, S100A9 and
MUC5AC genes were downregulated significantly, and all three are
known to be upregulated in cancer cells (13-15).
While there is less known about the impact of MUC5B on cancer, it
is known to be a useful biomarker and may be responsible for
increased proliferation and migration of colorectal and lung cancer
cells (13,16). Blocking of S100A9 is known to
suppress colorectal cancer cell stemness as well as general growth
(14). Lastly, overexpression of
MUC5AC results in higher cell invasion and migration and is
additionally known to make cancer cells more resilient by
decreasing the probability of triggering apoptosis and increasing
cell resistance to two important drugs for treating cancer,
fluorouracil and oxaliplatin (15). Together with the caspase-3 and MTT
assays of the present study, NS downregulation of all three of
these genes indicates a novel explanation for NS's chemo preventive
and anticancer effects that was not discovered in previous studies,
which had instead suggested inhibition of COX-2, inhibition of
NF-kB signaling, reduction of PGE2 and multi-caspase inhibition
(3,4,6,11).
This further explains how NS impacts cancerous Colo320 cells but
not healthy CCD-18 colon cells.
The ELISAs further confirmed and quantified the
results of RNA sequencing. Both MUC5B and MUC5AC exhibited
noticeable and statistically significant decreases in expression,
ranging between 11.97 and 30.33%.
These results can be explained by a CD44 and
MMP9-dependent signaling pathway. Changes in MMP9 expression have
been found to lead to corresponding changes in MUC5AC and MUC5B
expression (17,18). Additionally, S100A9 and CD44 are
known to be involved in the regulation of MMP9 (19,20).
Notably, CD44 has been discovered to associate with MMP9 on the
cell surface, and CD44 activity has been revealed to trigger MMP9
expression and lead to invasion (20). Hyaluronic acid-coated NS has
already been tested and revealed to be able to target cells with
high CD44 expression, demonstrating that CD44 may act as the
initial receptor from which NS may selectively affect colorectal
cancer cells (21). Furthermore,
the present molecular docking analysis indicated the potential for
NS to bind directly to MMP9, which may be another way NS could
interact with this pathway to influence MUC5AC and MUC5B
expression.
The present study has notable limitations. Firstly,
there was large variance in the results of the caspase assay, such
that the treatments of 4.175 and 417.5 µM for Colo320 cells were
not statistically significant. Although all treatments resulted in
a large increase in Caspase-3 activity for Colo320 cells, further
study may be warranted to examine why the results were not linear.
A similar question could be posited for the 41.75-µM treatment of
Colo320 cells during the 24 h MTT assay; a linear relationship
between NS concentration and Colo320 cell death would predict the
aforementioned treatment to cause a larger, not a smaller, decrease
in Colo320 cell survival.
Additionally, the only points at which data was
collected for MTT and Caspase assays were at 24 h and 7 days. More
time points between these, or tests for longer periods, could
provide further insight into the present study's findings.
Furthermore, other tests such as a TUNEL assay or
immunohistochemistry assays for apoptotic markers could be used in
the future to confirm these results.
The cell migration assay also was performed on only
one treatment group. Tests utilizing multiple treatment groups of
different concentrations could provide insight on the necessary
concentration of NS to reach the identified effect, as well as the
precise nature of the relationship between NS concentration and
wound healing.
With regards to mechanism, the present study
performed RNA sequencing on only Colo320 cells. RNA sequencing of
CCD-18 could provide further insight into changes in gene
expression between CCD-18 and Colo320, as well as provide further
information on the effects of NS on healthy colorectal cells.
Confirmation for the change in expression of MUC5AC and MUC5A could
also be checked via a DNA microarray. Other studies, such as
loss-of-function or other assays, could further improve on this via
confirmation of results or analysis into other apoptosis-related
genes not examined in the present study, such as Bax, Bcl-2, or
Caspase 7. Such tests could also help confirm the role of the
PI3K/Akt pathway, which the present study was unable to confirm
(22).
Furthermore, while the results of the ELISAs were
able to confirm a decrease in MUC5AC and MUC5B concentration, the
decrease was less marked when the treatment concentration was
increased, resulting in a lack of statistical significance for the
41.75 µM treatment group regarding MUC5B. Future tests analyzing
more treatment concentrations and using larger sample sizes could
be useful for better identifying the precise relationship between
NS concentration and MUC5AC and MUC5B levels.
In conclusion, based on the increased caspase-3
activity and decreased survival of the tested Colo320 colon cancer
cells, treatment with NS could induce apoptosis in colorectal
cancer cells. Furthermore, the present study revealed no evidence
that NS exhibited any such effect on healthy cells. Additionally,
the results of the cell migration assay revealed that NS was
capable of reducing cancer cell migration and regeneration. Based
on RNA sequencing, it was hypothesized that these capabilities are
due to the ability of NS to downregulate the MUC5B, S100A9 and
MUC5AC genes, potentially through a CD44 and MMP9-dependent
pathway, which is responsible for causing decreased cancer cell
proliferation, migration, invasion, stemness and resilience to
treatment with other drugs.
Supplementary Material
BIOVIA image of binding of naproxen
sodium to FAS receptor (PBD ID: 1DDF).
BIOVIA image of binding of naproxen
sodium to TRAIL (PBD ID: 1D2Q).
BIOVIA image of binding of naproxen
sodium to caspase-3 (PBD ID: 1NMS).
BIOVIA image of binding of naproxen
sodium to MMP9 (PBD ID: 1L6J).
BIOVIA image of binding of naproxen
sodium to MMP9 (PDB ID: 1GKD).
BIOVIA image of binding of naproxen
sodium to MMP9 (PDB ID: 1GKC).
Representative image of 0-h control
for cell migration assay. Magnification, x40.
Representative image of 0-h treatment
for cell migration assay. Magnification, x40.
Representative image of 24-h control
for cell migration assay. Magnification, x40.
Representative image of 24-h treatment
for cell migration assay. Magnification, x40.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author. The RNA sequencing data
generated in the present study may be found in the NCBI Sequence
Read Archive under accession no. PRJNA1102838 or at the following
URL: http://www.ncbi.nlm.nih.gov/bioproject/1102838.
Authors' contributions
AC collected all the data, performed statistical
analyses, and wrote the manuscript. WZ was responsible for
experimental design, provided laboratory materials for all
experiments and edited the manuscript. RG was responsible for tech
support, assisted in data collection and analyses, and proofread
the manuscript. All authors read and approved the final manuscript.
AC and WZ confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Centers for Disease Control and
Prevention: What is Colorectal Cancer? Centers for Disease Control
and Prevention. Available from: https://www.cdc.gov/cancer/colorectal/basic_info/what-is-colorectal-cancer.htm.
Accepted at March 19, 2022.
|
|
2
|
American Society of Clinical Oncology.
Colorectal Cancer-Types of Treatment. Cancer.Net. Available
from: https://www.cancer.net/cancer-types/colorectal-cancer/types-treatment.
Accepted at March 19, 2022.
|
|
3
|
Brasky TM, Liu J, White E, Peters U,
Potter JD, Walter RB, Baik CS, Lane DS, Manson JAE, Vitolins MZ, et
al: Non-steroidal anti-inflammatory drugs and cancer risk in women:
Results from the women's health initiative. Int J Cancer.
135:1869–1883. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kodela R, Nath N, Chattopadhyay M, Nesbitt
DE, Velázquez-Martínez CA and Kashfi K: Hydrogen sulfide-releasing
naproxen suppresses colon cancer cell growth and inhibits NF-κB
signaling. Drug Des Devel Ther. 9:4873–4882. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mohammed A, Janakiram NB, Madka V, Zhang
Y, Singh A, Biddick L, Li Q, Lightfoot S, Steele VE, Lubet RA, et
al: Intermittent dosing regimens of aspirin and naproxen inhibit
azoxymethane-induced colon adenoma progression to adenocarcinoma
and invasive carcinoma. Cancer Prev Res (Phila). 12:751–762.
2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Reyes-Uribe L, Wu W, Gelincik O, Bommi PV,
Francisco-Cruz A, Solis LM, Lynch PM, Lim R, Stoffel EM, Kanth P,
et al: Naproxen chemoprevention promotes immune activation in Lynch
syndrome colorectal mucosa. Gut. 70:555–566. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
CasPASE™ Apoptosis Assay-G-Biosciences.
G-Biosciences. Available from: https://cdn.gbiosciences.com/pdfs/protocol/Caspase_Apoptosis_Colorimentric_Assay_BAQ.pdf.
Accepted March 12, 2022.
|
|
8
|
Bumgarner R: Overview of DNA microarrays:
Types, applications, and their future. Curr Protoc Mol Biol Chapter
22:Unit 22.1, 2013.
|
|
9
|
Rao MS, Van Vleet TR, Ciurlionis R, Buck
WR, Mittelstadt SW, Blomme EAG and Liguori MJ: Comparison of
RNA-seq and microarray gene expression platforms for the
toxicogenomic evaluation of liver from short-term RAT toxicity
studies. Front Genet. 9(636)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Elabscience. Human muc5ac(mucin 5 subtype
AC) Elisa Kit. Elabscience. Available from: https://www.elabscience.com/p-human_muc5ac_mucin_5_subtype_ac_elisa_kit-19687.html.
Accepted May 6, 2023.
|
|
11
|
Smith CE, Soti S, Jones TA, Nakagawa A,
Xue D and Yin H: Non-steroidal anti-inflammatory drugs are caspase
inhibitors. Cell Chem Biol. 24:281–292. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Crowley LC and Waterhouse NJ: Detecting
cleaved caspase-3 in apoptotic cells by flow cytometry. Cold Spring
Harb Protoc 2016: 2016.
|
|
13
|
Lahdaoui F, Messager M, Vincent A, Hec F,
Gandon A, Warlaumont M, Renaud F, Leteurtre E, Piessen G,
Jonckheere N, et al: Depletion of MUC5B mucin in gastrointestinal
cancer cells alters their tumorigenic properties: Implication of
the Wnt/β-catenin pathway. Biochem J. 474:3733–3746.
2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Wang Y, Yin K, Tian J, Xia X, Ma J, Tang
X, Xu H and Wang S: Granulocytic myeloid-derived suppressor cells
promote the stemness of colorectal cancer cells through exosomal
S100A9. Adv Sci (Weinh). 6(1901278)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Pothuraju R, Rachagani S, Krishn SR,
Chaudhary S, Nimmakayala RK, Siddiqui JA, Ganguly K, Lakshmanan I,
Cox JL, Mallya K, et al: Molecular implications of MUC5AC-CD44 axis
in colorectal cancer progression and chemoresistance. Mol Cancer.
19(37)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yuan S, Liu Q, Hu Z, Zhou Z, Wang G, Li C,
Xie W, Meng G, Xiang Y, Wu N, et al: Long non-coding RNA MUC5B-AS1
promotes metastasis through mutually regulating MUC5B expression in
lung adenocarcinoma. Cell Death Dis. 9(450)2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Choi YS, Na HG, Bae CH, Song S and Kim Y:
Pepsin exposure in a non-acidic environment upregulates mucin 5AC
(MUC5AC) expression via matrix metalloproteinase 9 (mmp9)/nuclear
factor ΚB (NF-ΚB) in human airway epithelial cells. Int Forum
Allergy Rhinol. 11:894–901. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Song EJ, Bae CH, Kim JY, Kim YW, Park SY,
Song SY and Kim YD: Effect of epigallocatechin-3-gallate on
PMA-induced MUC5B expression in human airway epithelial cells. Clin
Exp Otorhinolaryngol. 6:237–242. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Shi S and Yi JL: S100A8/A9 promotes MMP-9
expression in the fibroblasts from cardiac rupture after myocardial
infarction by inducing macrophages secreting TNFα. Eur Rev Med
Pharmacol Sci. 22:3925–3935. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Murray D, Morrin M and McDonnell S:
Increased invasion and expression of MMP-9 in human colorectal cell
lines by a CD44-dependent mechanism. Anticancer Res. 24:489–494.
2004.PubMed/NCBI
|
|
21
|
Espinosa-Cano E, Huerta-Madroñal M,
Cámara-Sánchez P, Seras-Franzoso J, Schwartz S Jr, Abasolo I, San
Román J and Aguilar MR: Hyaluronic acid (ha)-coated
naproxen-nanoparticles selectively target breast cancer stem cells
through COX-independent pathways. Mater Sci Eng C Mater Biol Appl.
124(112024)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kim MS, Kim JE, Lim DY, Huang Z, Chen H,
Langfald A, Lubet RA, Grubbs CJ, Dong Z and Bode AM: Naproxen
induces cell-cycle arrest and apoptosis in human urinary bladder
cancer cell lines and chemically induced cancers by targeting PI3K.
Cancer Prev Res (Phila). 7:236–245. 2014.PubMed/NCBI View Article : Google Scholar
|