Introduction
Lung cancer currently ranks among the foremost
causes of cancer death, with its incidence and fatality rates
escalating annually (1). In 2012,
~1.8 million new cases of lung cancer were reported globally,
resulting in 1.6 million fatalities (1). Epidemiological data suggest an
estimated 230,000 new cases of lung cancer in the USA in 2022.
Furthermore, the mortality rate attributed to lung cancer is
anticipated to surpass the combined rates of breast, prostate and
colon cancer (2). These conditions
impose substantial economic and psychological burdens on society
and families. While smoking is a well-established risk factor for
lung cancer, accounting for 80-90% of the cases, it is noteworthy
that only 15% of smokers ultimately develop lung cancer. This
suggests that hereditary predisposition might also significantly
contribute to lung cancer (1,3).
Histologically, lung cancer is broadly classified into small cell
lung cancer (SCLC) and non-small cell lung cancer (NSCLC) (4). A total of ~80-85% lung cancers are
NSCLC, encompassing various subtypes such as squamous cell
carcinoma, adenocarcinoma, large cell carcinoma, adeno-squamous
carcinoma and sarcomatoid carcinoma (4). Multiple genetic alterations have
shown the potential to disrupt biological process or functionals,
thereby facilitating the onset of lung cancer. Hence, the
identification of susceptibility genes for lung cancer holds
paramount clinical significance.
Cleft-lip and palate transmembrane protein-1-like
(CLPTM1L) protein is identified as a protein associated with
cisplatin resistance (5,6). The gene is situated on chromosome
5p15.33 and encodes a protein comprising 538 amino acids (6) with a molecular weight of 62 KDa
(3,7). Functionally, the protein encoded by
CLPTM1L is a membrane entity implicated in cisplatin resistance,
where its overexpression in cisplatin-sensitive cells triggers
apoptosis (8). Remarkably, both
human and murine proteins exhibit 99% sequence identity, featuring
538 amino acids (5). Notably,
within the chromosomal region 5p15, housing two genes, telomerase
reverse transcriptase (TERT) and CLPTM1L, a susceptibility locus
for lung cancer has been identified, with a potential role
attributed to NKX2.4(9).
Nonetheless, the interaction mechanism between these genes remains
elusive. Moreover, CLPTM1L has been implicated in modulating
inflammatory responses (10).
Given the profound link between malignancy and heightened
inflammation (11), numerous
studies have scrutinized the potential of CLPTM1L mutation in
precipitating tumorigenesis. Genome-wide association studies (GWAS)
have pinpointed specific single nucleotide polymorphisms (SNPs)
within the CLPTM1L gene (such as rs401681 and rs402710) as
intricately intertwined with the onset and progression of various
malignancies spanning lung cancer, breast cancer, pancreatic
cancer, nasopharyngeal carcinoma and bladder cancer among others
(3,7,12).
In addition, clinical analyses have revealed that the expression
levels of CLPTM1L in lung cancer tissue surpass those in healthy
lung tissue, particularly evident in adenocarcinoma cells (5). Adenocarcinoma cells exhibit an
average 2.15-fold elevation in CLPTM1L expression over normal
immortalized cell lines (P = 3.59x10-7) (13). Since its initial identification of
association with lung cancer susceptibility, numerous subsequent
investigations have delved into the relationship between lung
cancer risk and the rs401681 variant. Nevertheless, subsequent
studies have yielded disparate findings (14-20).
To comprehensively elucidate the association between lung cancer
and rs401681, a meta-analysis encompassing correlational studies
published prior to 2023 was undertaken.
Materials and methods
Literature search
A meticulous literature search was performed across
multiple electronic databases, comprising PubMed (https://pubmed.ncbi.nlm.nih.gov/), Embase
(https://www.embase.com), Cochrane Library
(https://www.cochranelibrary.com/),
Medline (https://www.nlm.nih.gov/medline/medline_overview.html),
Google Scholar (https://scholar.google.com/), Wanfang (http://www.wanfangdata.com/) and Chinese National
Knowledge Infrastructure (https://oversea.cnki.net/index/). Employing requisite
combinations of keywords, such as ‘LC’ or ‘lung carcinoma’ or ‘lung
neoplasm’ or ‘lung cancer’ and ‘rs401681 or CLPTM1L’, facilitated
comprehensive retrieval. Moreover, additional relevant studies were
identified through thorough examination of the reference lists of
the included studies.
Selection criteria
The inclusion criteria for eligible studies were as
follows: i) Studies comprising both control and case groups; ii)
studies investigating the association between CLPTM1L rs401681 and
lung cancer susceptibility; iii) studies providing adequate data on
genotype frequencies, or odds ratio (OR) with 95% confidence
interval (95% CI) for lung cancer risk assessment and iv)
investigations involving human subjects. A study that meets any of
the following criteria was deemed to be non-compliant with the
present meta-analysis: i) Comments, abstracts for conference, case
reports, editorials or reviews; ii) insufficient data for OR
calculation; iii) animal studies; and iv) studies lacking control
groups.
Quality assessment
The quality of all included studies was assessed
utilizing the Newcastle-Ottawa Scale (NOS). The NOS assesses
studies across three key dimensions: The selection of the study
groups; the comparability of these groups; and the ascertainment of
either the exposure or outcome of interest for case-control or
cohort studies respectively. The scale yields a maximum score of
9(21). Finally, the studies with
a total score surpassing 6 are considered high quality, those
scoring between 4 and 6 are considered medium quality, and studies
with a total score below 4 are categorized as low quality.
Data extraction
Data extraction was independently carried out by two
investigators. Any discordant opinion was solved through discussing
to reach a final consensus. The data obtained from each included
study encompassed the first author's name, publication year,
region, ethnicity, disease type, sample size, genotype and/or
allele frequencies of case and control subjects, and testimony of
Hardy-Weinberg equilibrium in the control group.
Statistical analysis
The statistical analyses were conducted using STATA
software (version 17.0, StataCorp LP). The effect size of the
correlation between the CLPTM1L rs401681 polymorphism and lung
cancer risk was evaluated by pooled ORs with corresponding 95% CIs.
χ2-based Q-test and I2 statistic were used to
assess heterogeneity among studies. The random-effects model was
used for the present meta-analysis. The significant heterogeneity
was defined as P<0.05 for the Q test or I2>50%.
Moreover, subgroup analysis was used to investigate potential
sources of substantial heterogeneity. Publication bias was assessed
by Egger's linear regression test. To test the robustness of the
pooled outcomes, sensitivity analysis was employed using the
leave-one-out method to identify the influence of individual
studies on the overall effect estimate. P<0.05 was considered to
indicate a statistically significant.
In silico analysis
Aiming to elucidate the impact of polymorphisms on
CLPTM1L, the authors embarked on an exploration of the association
between the rs401681 and CLPTM1L expression levels via
Genotype-Tissue Expression (GTEx) Analysis Release V6 (dbGaP
Accession phs000424.v7.p2) data (22). To further investigate the potential
influence of CLPTM1L expression on tumorigenesis, the authors
endeavored to mine evidence from The Cancer Genome Atlas (TCGA)
database (https://www.cancer.gov/ccg/research/genome-sequencing/tcga)
by using the Kaplan-Meier. Additionally, the recently developed
interactive platform, Gene Expression Profiling Interactive
Analysis, was utilized to augment the inquiry (23-25).
Results
Characteristics of included
studies
Through the investigation of public databases, a
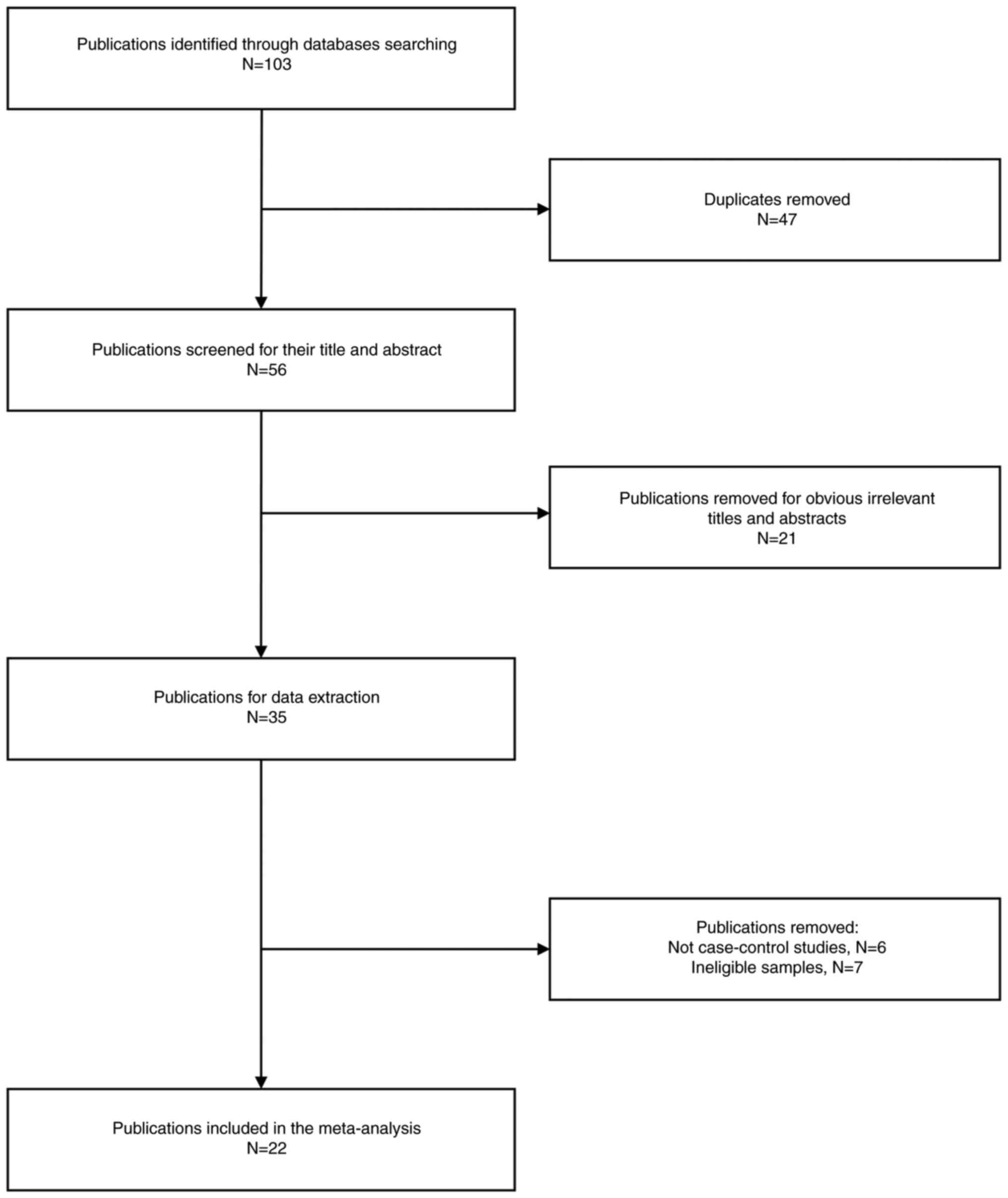
total of 103 potential papers according to the search criteria were
identified, from which 47 were subsequently excluded due to
duplication (Fig. 1). Following
this, a total of 22 studies were excluded due to unrelated topics.
Moreover, 13 additional studies were excluded for lack of alignment
with the case control design (n=6) and for including ineligible
samples (n=7). Finally, 22 eligible articles were included
(14,16,18,20,26-43).
Among these studies, data pertaining to Caucasian populations were
presented in 5 studies (26,29,30,38,42),
whereas 17 studies included Asian populations. A more comprehensive
overview of the studies included in the present meta-analysis is
provided in Tables I and II.
 | Table IMain characteristics of the included
studies in the present meta-analysis. |
Table I
Main characteristics of the included
studies in the present meta-analysis.
| | Sample size | Case genotype | Control
genotype | |
|---|
| First author,
year | Race | Case | Control | CC | CT | TT | CC | CT | TT | NOS score | HWE | (Refs.) |
|---|
| Wang et al,
2008 | Caucasian | 2,396 | 3,051 | 868 | 1,134 | 394 | 994 | 1,506 | 551 | 7 | Y | (38) |
| Amos et al,
2008 | Caucasian | 1,153 | 1,137 | 396 | 570 | 187 | 336 | 596 | 205 | 8 | Y | (26) |
| Hung et al,
2008 | Caucasian | 1,920 | 2,517 | 710 | 949 | 261 | 857 | 1,239 | 421 | 8 | Y | (30) |
| Zienolddiny et
al, 2009 | Caucasian | 341 | 431 | 107 | 177 | 57 | 117 | 224 | 90 | 8 | Y | (42) |
| Yoon et al,
2010 | Asian | 431 | 341 | 117 | 224 | 90 | 107 | 177 | 57 | 8 | Y | (40) |
| Bae et al,
2012 | Asian | 1,086 | 1,079 | 545 | 434 | 107 | 499 | 484 | 96 | 7 | Y | (27) |
| Chen et al,
2012 | Asian | 195 | 228 | 95 | 90 | 10 | 126 | 77 | 25 | 7 | Y | (28) |
| Li et al,
2013 | Asian | 464 | 536 | 218 | 205 | 41 | 244 | 234 | 58 | 8 | Y | (14) |
| Myneni et
al, 2013 | Asian | 350 | 441 | 181 | 135 | 34 | 194 | 200 | 47 | 8 | Y | (35) |
| Wang et al,
2013 | Asian | 492 | 486 | 245 | 201 | 46 | 215 | 203 | 68 | 8 | Y | (37) |
| de Mello et
al, 2013 | Caucasian | 144 | 144 | 40 | 77 | 27 | 44 | 75 | 25 | 8 | Y | (29) |
| Jiang et al,
2013 | Asian | 726 | 860 | 371 | 289 | 66 | 395 | 378 | 87 | 8 | Y | (31) |
| Ke et al,
2013 | Asian | 611 | 1,062 | 324 | 231 | 56 | 495 | 459 | 108 | 8 | Y | (33) |
| Sun et al,
2013 | Asian | 400 | 200 | 186 | 188 | 26 | 95 | 86 | 19 | 8 | Y | (36) |
| Lv et al,
2013 | Asian | 602 | 1,060 | 315 | 231 | 56 | 493 | 459 | 108 | 8 | Y | (43) |
| Zhao et al,
2014 | Asian | 951 | 954 | 436 | 424 | 91 | 437 | 424 | 93 | 8 | Y | (20) |
| Xun et al,
2014 | Asian | 228 | 299 | 112 | 102 | 14 | 132 | 132 | 35 | 8 | Y | (18) |
| Zhang et al,
2014 | Asian | 366 | 364 | 182 | 149 | 35 | 167 | 169 | 28 | 8 | Y | (41) |
| Liang et al,
2014 | Asian | 309 | 308 | 145 | 139 | 25 | 137 | 134 | 37 | 8 | Y | (16) |
| Liu et al,
2015 | Asian | 292 | 319 | 157 | 112 | 23 | 138 | 146 | 35 | 8 | Y | (34) |
| Jin et al,
2016 | Asian | 554 | 695 | 276 | 234 | 44 | 308 | 302 | 85 | 8 | Y | (32) |
| Xiao et al,
2017 | Asian | 199 | 220 | 85 | 99 | 15 | 128 | 79 | 13 | 8 | Y | (39) |
 | Table IIComparative analysis of genotype
frequencies and the risk of lung cancer in the subgroups. |
Table II
Comparative analysis of genotype
frequencies and the risk of lung cancer in the subgroups.
| | Sample size | Case genotype | Control
genotype | |
|---|
| First author,
year | Race | Histology | Case | Control | CC | CT | TT | CC | CT | TT | (Refs.) |
|---|
| Zienolddiny et
al, 2009 | Caucasian | NSCLC | 341 | 431 | 107 | 177 | 57 | 117 | 224 | 90 | (42) |
| Yoon et al,
2010 | Asian | NSCLC | 431 | 341 | 117 | 224 | 90 | 107 | 177 | 57 | (40) |
| Chen et al,
2012 | Asian | SCLC | 16 | 228 | 8 | 8 | 0 | 126 | 77 | 25 | (28) |
| Chen et al,
2012 | Asian | NSCLC | 180 | 228 | 87 | 82 | 10 | 126 | 77 | 25 | (28) |
| Li et al,
2013 | Asian | NSCLC | 464 | 536 | 218 | 205 | 41 | 244 | 234 | 58 | (14) |
| Wang et al,
2013 | Asian | SCLC | 99 | 486 | 45 | 45 | 9 | 215 | 203 | 68 | (37) |
| Wang et al,
2013 | Asian | NSCLC | 393 | 486 | 200 | 156 | 37 | 215 | 203 | 68 | (37) |
| de Mello et
al, 2013 | Caucasian | NSCLC | 144 | 144 | 40 | 77 | 27 | 44 | 75 | 25 | (29) |
| Ke et al,
2013 | Asian | NSCLC | 427 | 1,062 | 229 | 155 | 37 | 493 | 459 | 108 | (33) |
| Sun et al,
2013 | Asian | NSCLC | 400 | 200 | 186 | 188 | 26 | 95 | 86 | 19 | (36) |
| Zhao et al,
2014 | Asian | SCLC | 703 | 954 | 334 | 301 | 68 | 437 | 424 | 93 | (20) |
| Zhao et al,
2014 | Asian | NSCLC | 139 | 954 | 59 | 68 | 12 | 437 | 424 | 93 | (20) |
| Zhang et al,
2014 | Asian | NSCLC | 366 | 364 | 182 | 149 | 35 | 167 | 169 | 28 | (41) |
Susceptibility to lung cancer
The association between the CLPTM1L rs401681 and
lung cancer is illustrated in Table
III. OR<1 denotes reduced lung cancer risk associated with
the exposed genotype compared with the non-exposed counterpart. The
findings underscored a significant link between CLPTM1L rs401681
and altered lung cancer risk across all genetic comparisons. These
comparisons encompass allele T vs. allele C (OR=0.93, 95%
CI=0.88-0.99, P<0.001), TT + CT vs. CC (OR=0.91,
95%CI=0.87-0.96, P<0.001), TT vs. CC + CT (OR=0.88, 95%
CI=0.80-0.96, P<0.001), TT vs. CC (OR=0.84, 95% CI=0.78-0.90,
P<0.001), and CT vs. CC (OR=0.84, 95% CI=0.75-0.94,
P=0.008).
 | Table IIIResult of meta-analysis for CLPTM1L
rs401681 polymorphism and lung cancer risk. |
Table III
Result of meta-analysis for CLPTM1L
rs401681 polymorphism and lung cancer risk.
| | T vs. C | TT + CT vs. CC | TT vs. CC + CT | TT vs. CC | CT vs. CC |
|---|
| Variables | N | Case/control | OR (95% CI) |
Ph/I2(%)/Pz | PE | OR (95% CI) |
Ph/I2(%)/Pz | PE | OR (95% CI) |
Ph/I2(%)/Pz | PE | OR (95% CI) |
Ph/I2(%)/Pz | PE | OR (95% CI) |
Ph/I2(%)/Pz | PE |
|---|
| Total | 22 | 14195/16732 | 0.93
(0.88-0.99) |
0.000/60.9/0.000 | 0.406 | 0.91
(0.87-0.96) |
0.000/63.7/0.000 | 0.229 | 0.88
(0.80-0.96) | 0.054/35/0.000 | 0.617 | 0.84
(0.78-0.90) |
0.008/46.9/0.000 | 0.886 | 0.84
(0.75-0.94) |
0.000/46.9/0.008 | 0.162 |
| Ethnicity |
| Caucasian | 5 | 5954/7280 | 0.89
(0.85-0.93) | 0.756/0/0.000 | / | 0.86
(0.80-0.92) | 0.722/0/0.000 | / | 0.85
(0.78-0.93) | / | / | 0.79
(0.71-0.87) | 0.688/0/0.000 | | 0.79
(0.71-0.91) | 0.688/0/0.000 | / |
| Asian | 17 | 8241/9452 | 0.95
(0.88-1.03) |
0.000/66.2/0.046 | / | 0.96
(0.90-1.02) |
0.000/68.2/0.151 | / | 0.88
(0.76-1.01) | / | / | 0.89
(0.80-0.99) |
0.005/53.8/0.069 | | 0.86
(0.73-1.01) |
0.005/53.8/0.737 | / |
| Histology |
| NSCLC | 10 | 3849/4746 | 0.93
(0.85-1.01) |
0.123/35.7/0.087 | / | 0.92
(0.82-1.04) |
0.114/36.8/0.178 | / | 0.88
(0.74-1.05) |
0.131/34.6/0.154 | / | 0.85
(0.70-1.03) |
0.121/35.9/0.099 | | 0.95
(0.84-1.07) |
0.096/39.3/0.383 | / |
| SCLC | 3 | 254/1668 | 0.93
(0.87-1.00) |
0.241/20.1/0.756 | / | 0.94
(0.85-1.04) |
0.189/25.3/0.601 | / | 0.87
(0.74-1.01) |
0.214/22.7/0.190 | / | 0.84
(0.72-1.00) |
0.227/21.4/0.318 | | 0.97
(0.87-1.09) |
0.124/32.3/0.280 | / |
Heterogeneity and subgroup
analysis
Q test and I2 findings revealed
significant heterogeneity across four comparisons including allele
T vs. allele C, TT+CT vs. CC, TT vs. CC, and CT vs. CC (Table III). Subgroup analysis results
(Table III; Fig. S1, Fig. S2, Fig. S3, Fig. S4, Fig. S5, Fig. S6, Fig. S7, Fig. S8, Fig. S9 and Fig. S10) suggested that racial
disparities may contribute to the considerable heterogeneity
observed in the majority, if not all, of the comparisons. Upon
stratification by ethnicity, a notable reduction in heterogeneity
within the Caucasian subgroup was observed, although heterogeneity
persisted within the Asian subgroup. Moreover, significant
alterations in lung cancer risks were also detected in all genetic
comparisons among Caucasians, whereas such alterations were
statistically significant only in the TT vs. CC comparison within
the Asian subgroup.
Furthermore, a total of 10 studies focusing on NSCLC
and 3 studies focusing on SCLC furnished adequate data regarding
genotype distribution, enabling the conduction of an additional
subgroup analysis. The results showed that the association between
rs401681 and the risks associated with NSCLC or SCLC did not attain
statistical significance across all comparisons (Table III).
Sensitivity analysis
From the results of sensitivity analysis in various
comparisons (Fig. S11, Fig. S12, Fig. S13, Fig. S14 and Fig. S15), no substantial alterations in
the re-obtained ORs were observed when compared with the initial
ORs. This indicated the robustness and consistency of these
findings.
Publication bias
The P-value of Egger's linear regression test
(Table III) indicated the
absence of noteworthy publication bias across all comparisons.
In silico analysis
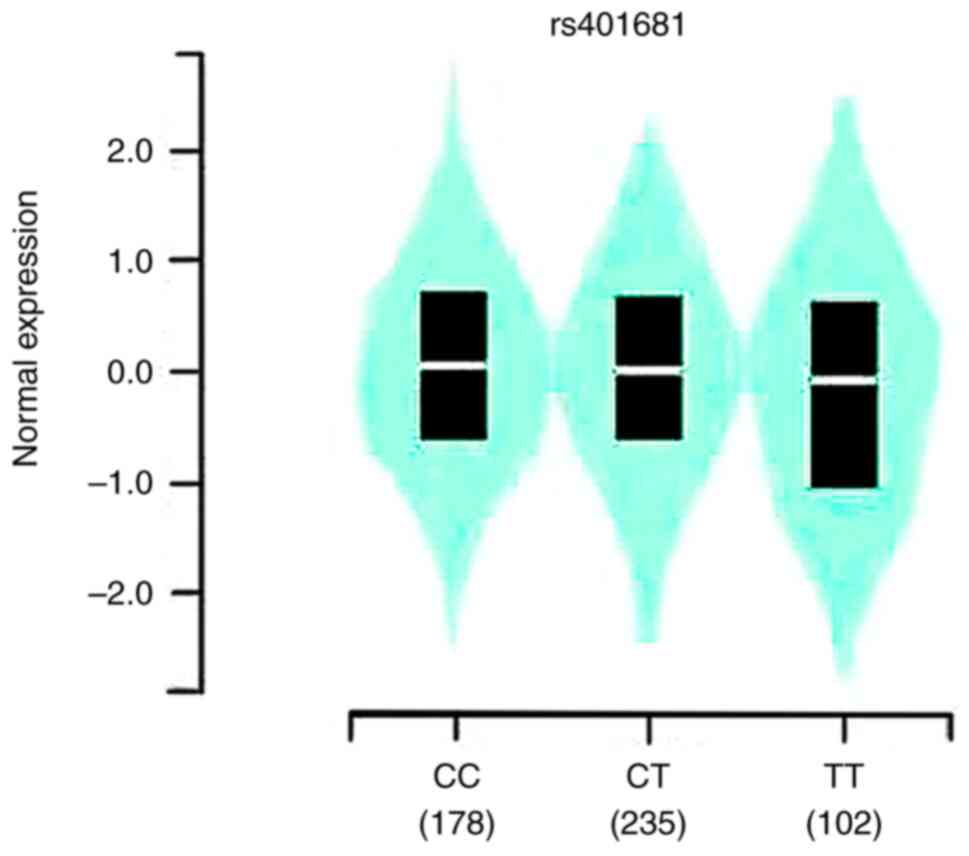
Based on the GTEx portal data, the analysis revealed
that the mutant allele was associated with increased expression of
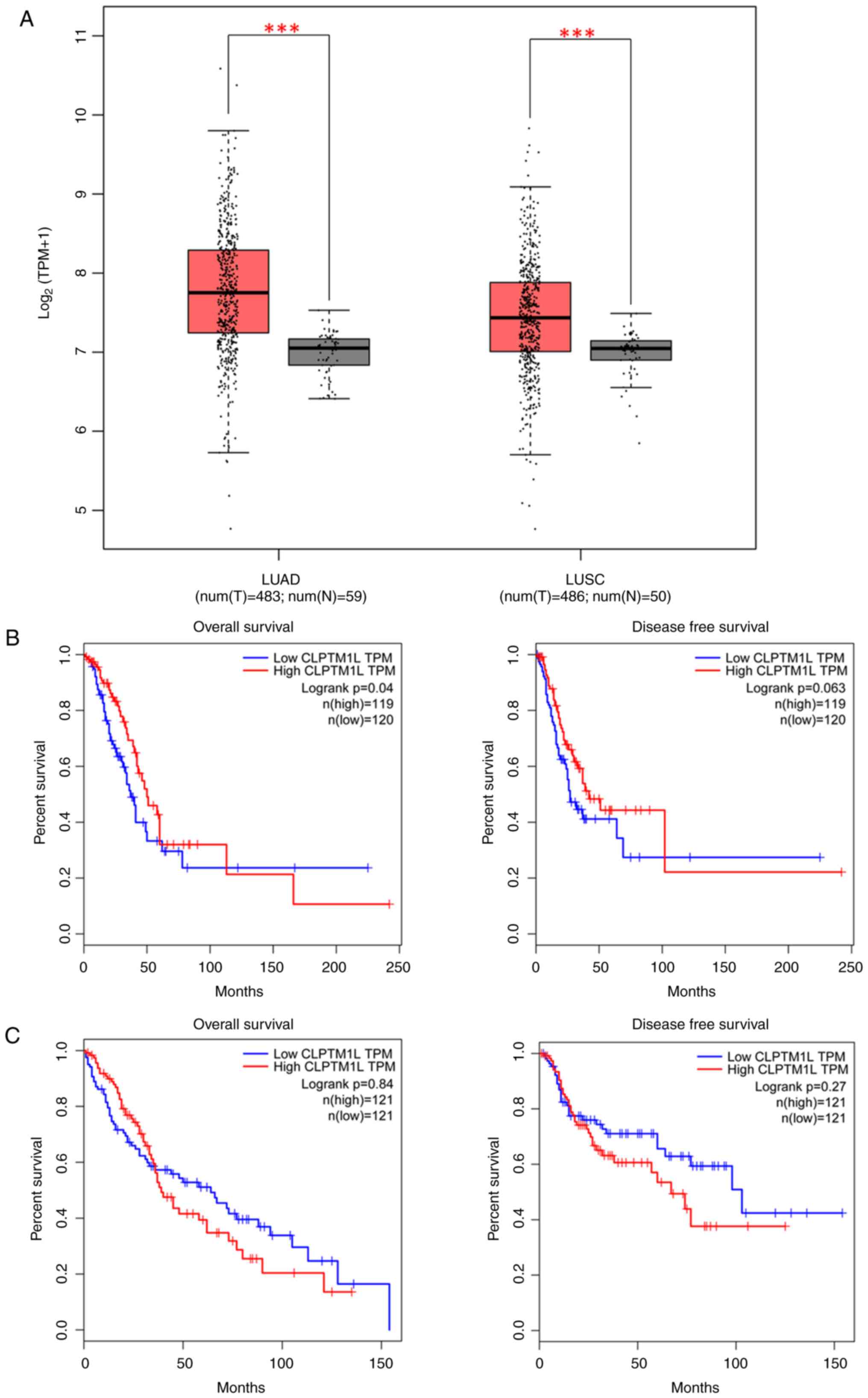
CLPTM1L mRNA at rs401681 (P=0.02) (Fig. 2). Evidence gleaned from TCGA
database further underscored this finding, demonstrating elevated
CLPTM1L expression levels in lung adenocarcinoma (LUAD) tissue
compared with normal tissue [CLPTM1L
expression=(5.5x103) vs. (2.8x103),
P<0.001], as well as in lung squamous cell carcinoma (LUSC)
[CLPTM1L expression=(4.6x103) vs. (2.9x103),
P<0.001] (Fig. 3A).
Moreover, whether the expression level of CLPTM1
influences the disease-free survival (DFS) and overall survival
(OS) of lung cancer was investigated. The Kaplan-Meier estimations
revealed a notable discrepancy in OS between the low and high
CLPTM1 transcripts per kilobase million (TPM) groups in LUAD
(log-rank P=0.04), albeit a difference in DFS close to statistical
significance (log-rank P=0.063). On the other hand, no distinction
was observed between high and low CLPTM1 TPM groups in LUSC
(log-rank P=0.84 for OS and 0.27 for DFS). Details are presented in
Fig. 3B and C.
Discussion
In the present study, the impact of CLPTM1L rs401681
within the locus 5p15.33 on lung cancer incidence was scrutinized,
and it was found that it had a remarkable association with onset of
lung cancer. Prior investigations yield a discordant landscape
regarding the role of CLPTM1L rs401681 in the occurrence of lung
cancer. There was no overall correlation between CLPTM1L rs401681
and the risk of lung cancer in several previous studies (20,28),
whereas others reported that allele T of CLPTM1L rs401681 decreases
the susceptibility of lung cancer (33,39,40,43).
A precedent meta-analysis hinted at an association between CLPTM1L
rs401681 and lung cancer (14),
albeit with a caveat on sample size constraints acknowledged by the
authors. Subsequently, emerging studies exploring the relationship
between CLPTM1L rs401681 and lung cancer risk have been published
(16,18,20,32,34,39,41,44,45).
To further determine the relationship between CLPTM1L rs401681
polymorphism and lung cancer risk, the authors embarked on a
meta-analysis of all the literature published before 2023. In the
present study, the association between CLPTM1L rs401681
polymorphism and lung cancer risk was systematically reviewed and
was based on ten case-control studies, encompassing 14,195 cases
and 16,732 controls. The meta-analysis demonstrated that CLPTM1L
rs401681 polymorphism was associated with the occurrence of lung
cancer in all the genetic comparisons investigated, suggestive of a
protective effect conferred by the T allele of CLPTM1L rs401681
mutation against lung cancer. Nevertheless, the I2
statistic and statistical Q test revealed significant
heterogeneity, with ethnic origin emerging as a plausible primary
contributor to this observed heterogeneity.
Upon stratification by race, a statistically
significant association of CLPTM1L rs401681 with lung cancer risk
was observed in each genetic comparison among Caucasians, whereas
it was only notable for the TT vs. CC comparison among Asians.
Although the exact cause for racial disparities is not entirely
clear, one possible cause may lie within the genetic context. When
considering subgroups based on histology, it was found that the
association between rs401681 and NSCLC or SCLC risks was not
statistically significant for all comparisons. This suggested that
CLPTM1L rs401681 variant genotypes may not exert an effect on the
pathological differentiation of lung cancer. Future investigations
should aim to elucidate the underlying mechanisms, taking into
account factors such as histology or race heterogeneity, to provide
a more nuanced understanding of the impact of CLPTM1L gene
polymorphism on lung cancer risk and to tailor personal
therapy.
The association between lung cancer and the CLPTM1L
rs401681 polymorphism may be confounded by other influential SNPs,
particularly those situated within TERT-CLPTM1L region. This is
especially pertinent for SNPs in close linkage disequilibrium with
rs401681, such as rs31490 and rs414965, or those directly
implicated in lung cancer. In addition, variations in matching
criteria, selection bias, and limited data availability in the
present study may lead to statistical insufficiency to identify
subtle differences, and a fluctuated risk estimate. Several
findings underscored the critical importance of assessing genetic
influences across diverse populations for disease onset and
progression.
The present study possessed several limitations.
Primarily, the intricate interplay between gene-gene and
gene-environment interactions in lung cancer risk poses challenges
for evaluating genotype polymorphism alongside potential
psychological and environmental factors. Consequently, the
assessment primarily focused on delineating the risk coefficient
associated with aberrant genotypes. Secondly, the present study
only included Chinese and English studies, potentially introducing
unacknowledged language bias. Thirdly, the possibility of chance
findings cannot be entirely discounted, given the limited patient
cohorts within the present stratified analyses. These subgroup
associations necessitate validation within larger cohorts for
enhanced precision. Lastly, due to lacking certain subgroup data of
included studies, some important subgroups such as sex are not
analyzed in the present meta-analysis. In future research,
investigators should focus more on this important issue.
In conclusion, the current meta-analysis
demonstrates a significant association between the CLPTM1L rs401681
polymorphism and the risk of lung cancer, wherein the T allele
exhibits a protective effect. These findings offer preliminary
insights into assessing the risk of lung cancer based on the SNP
rs401681 genotype, thus laying the groundwork for personalized
medicine endeavors.
Supplementary Material
Forest plot of the relationship
between the cleft-lip and palate transmembrane protein-1-like
rs401681 polymorphism and lung susceptibility (including racial
subgroup analysis) in the allele T vs. allele C. The squares and
horizontal lines correspond to the study-specific OR and 95% CI.
The area of a square reflects the weight (reciprocal of variance).
The diamond represents the total OR and 95% CI. OR, odds ratio; CI,
confidence interval.
Forest plot of the relationship
between the cleft-lip and palate transmembrane protein-1-like
rs401681 polymorphism and lung susceptibility for racial subgroup
analysis in the allele T vs. allele C. The squares and horizontal
lines correspond to the study-specific OR and 95% CI. The area of a
square reflects the weight (reciprocal of variance). The diamond
represents the summary OR and 95% CI. OR, odds ratio; CI,
confidence interval.
Forest plot of the relationship
between the cleft-lip and palate transmembrane protein-1-like
rs401681 polymorphism and lung susceptibility (including racial
subgroup analysis) in the TT + CT vs. CC. The squares and
horizontal lines correspond to the study-specific OR and 95% CI.
The area of a square reflects the weight (reciprocal of variance).
The diamond represents the summary OR and 95% CI. OR, odds ratio;
CI, confidence interval.
Forest plot of the relationship
between the cleft-lip and palate transmembrane protein-1-like
rs401681 polymorphism and lung susceptibility for racial subgroup
analysis in the TT + CT vs. CC. The squares and horizontal lines
correspond to the study-specific OR and 95% CI. The area of a
square reflects the weight (reciprocal of variance). The diamond
represents the summary OR and 95% CI. OR, odds ratio; CI,
confidence interval.
Forest plot of the relationship
between the cleft-lip and palate transmembrane protein-1-like
rs401681 polymorphism and lung susceptibility (including racial
subgroup analysis) in the TT vs. CC + CT. The squares and
horizontal lines correspond to the study-specific OR and 95% CI.
The area of a square reflects the weight (reciprocal of variance).
The diamond represents the summary OR and 95% CI. OR, odds ratio;
CI, confidence interval.
Forest plot of the relationship
between the cleft-lip and palate transmembrane protein-1-like
rs401681 polymorphism and lung susceptibility for racial subgroup
analysis in the TT vs. CC + CT. The squares and horizontal lines
correspond to the study-specific OR and 95% CI. The area of a
square reflects the weight (reciprocal of variance). The diamond
represents the summary OR and 95% CI. OR, odds ratio; CI,
confidence interval.
Forest plot of the relationship
between the cleft-lip and palate transmembrane protein-1-like
rs401681 polymorphism and lung susceptibility (including racial
subgroup analysis) in the TT vs. CC. The squares and horizontal
lines correspond to the study-specific OR and 95% CI. The area of a
square reflects the weight (reciprocal of variance). The diamond
represents the summary OR and 95% CI. OR, odds ratio; CI,
confidence interval.
Forest plot of the relationship
between the cleft-lip and palate transmembrane protein-1-like
rs401681 polymorphism and lung susceptibility for racial subgroup
analysis in the allele TT vs. CC. The squares and horizontal lines
correspond to the study-specific OR and 95% CI. The area of a
square reflects the weight (reciprocal of variance). The diamond
represents the summary OR and 95% CI. OR, odds ratio; CI,
confidence interval.
Forest plot of the relationship
between the cleft-lip and palate transmembrane protein-1-like
rs401681 polymorphism and lung susceptibility (including racial
subgroup analysis) in the CT vs. CC. The squares and horizontal
lines correspond to the study-specific OR and 95% CI. The area of a
square reflects the weight (reciprocal of variance). The diamond
represents the summary OR and 95% CI. OR, odds ratio; CI,
confidence interval.
Forest plot of the relationship
between the cleft-lip and palate transmembrane protein-1-like
rs401681 polymorphism and lung susceptibility for racial subgroup
analysis in the CT vs. CC. The squares and horizontal lines
correspond to the study-specific OR and 95% CI. The area of a
square reflects the weight (reciprocal of variance). The diamond
represents the summary OR and 95% CI. OR, odds ratio; CI,
confidence interval.
Sensitivity analysis for testing the
stability of overall estimates of allele T vs. allele C.
Sensitivity analysis for testing the
stability of overall estimates of TT + CT vs. CC.
Sensitivity analysis for testing the
stability of overall estimates of TT vs. CC + CT.
Sensitivity analysis for testing the
stability of overall estimates of TT vs. CC.
Sensitivity analysis for testing the
stability of the overall estimate in the CT vs. CC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
GZ and YW conceived and designed the study. YW
provided administrative support. FL provided study materials or
recruited patients. ZD, YW and FL collected and assembled the data.
ZF, YW and FL analyzed and interpreted the data. All authors wrote
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nasim F, Sabath BF and Eapen GA: Lung
cancer. Med Clin North Am. 103:463–473. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Oliver AL: Lung cancer: Epidemiology and
screening. Surg Clin North Am. 102:335–344. 2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Spitz MR, Wei Q, Dong Q, Amos CI and Wu X:
Genetic susceptibility to lung cancer: The role of DNA damage and
repair. Cancer Epidemiol Biomarkers Prev. 12:689–698.
2003.PubMed/NCBI
|
|
4
|
Rodriguez-Canales J, Parra-Cuentas E and
Wistuba II: Diagnosis and molecular classification of lung cancer.
Cancer Treat Res. 170:25–46. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Pande M, Spitz MR, Wu X, Gorlov IP, Chen
WV and Amos CI: Novel genetic variants in the chromosome 5p15.33
region associate with lung cancer risk. Carcinogenesis.
32:1493–1499. 2011.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Yamamoto K, Okamoto A, Isonishi S, Ochiai
K and Ohtake Y: A novel gene, CRR9, which was up-regulated in
CDDP-resistant ovarian tumor cell line, was associated with
apoptosis. Biochem Biophys Res Commun. 280:1148–1154.
2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Petersen GM, Amundadottir L, Fuchs CS,
Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, Arslan AA,
Bueno-de-Mesquita HB, Gallinger S, Gross M, et al: A genome-wide
association study identifies pancreatic cancer susceptibility loci
on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 42:224–228.
2010.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Tian J, Wang Y, Dong Y, Chang J, Wu Y,
Chang S and Che G: Cumulative evidence for relationships between
multiple variants in the TERT and CLPTM1L region and risk of cancer
and non-cancer disease. Front Oncol. 12(946039)2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Derrien AC, Houy A, Ganier O, Dingli F,
Ningarhari M, Mobuchon L, Espejo Díaz MI, Loew D, Cassoux N,
Cussenot O, et al: Functional characterization of 5p15.33 risk
locus in uveal melanoma reveals rs452384 as a functional variant
and NKX2.4 as an allele-specific interactor. Am J Hum Genet.
109:2196–2209. 2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Petkov S and Chiodi F: Distinct
transcriptomic profiles of naïve CD4+ T cells distinguish HIV-1
infected patients initiating antiretroviral therapy at acute or
chronic phase of infection. Genomics. 113:3487–3500. 2021.
|
|
11
|
Aktas G, Sit M, Karagoz I, Erkus E, Ozer
B, Kocak MZ, Yaman S, Keyif F, Altinordu R, Erkol H and Savli H:
Could red cell distribution width be a marker of thyroid cancer? J
Coll Physicians Surg Pak. 27:556–558. 2017.PubMed/NCBI
|
|
12
|
Speedy HE, Di Bernardo MC, Sava GP, Dyer
MJ, Holroyd A, Wang Y, Sunter NJ, Mansouri L, Juliusson G, Smedby
KE, et al: A genome-wide association study identifies multiple
susceptibility loci for chronic lymphocytic leukemia. Nat Genet.
46:56–60. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
13
|
James MA, Wen W, Wang Y, Byers LA, Heymach
JV, Coombes KR, Girard L, Minna J and You M: Functional
characterization of CLPTM1L as a lung cancer risk candidate gene in
the 5p15.33 locus. PLoS One. 7(e36116)2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li C, Yin Z, Wu W, Li X and Zhou B:
Genetic variants in TERT-CLPTM1L genetic region associated with
several types of cancer: A meta-analysis. Gene. 526:390–399.
2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kachuri L, Amos CI, McKay JD, Johansson M,
Vineis P, Bueno-de-Mesquita HB, Boutron-Ruault MC, Johansson M,
Quirós JR, Sieri S, et al: Fine mapping of chromosome 5p15.33 based
on a targeted deep sequencing and high density genotyping
identifies novel lung cancer susceptibility loci. Carcinogenesis.
37:96–105. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Liang Y, Thakur A, Gao L, Wang T, Zhang S,
Ren H, Meng J, Geng T, Jin T and Chen M: Correlation of CLPTM1L
polymorphisms with lung cancer susceptibility and response to
cisplatin-based chemotherapy in a Chinese Han population. Tumour
Biol. 35:12075–12082. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Pintarelli G, Cotroneo CE, Noci S, Dugo M,
Galvan A, Delli Carpini S, Citterio L, Manunta P, Incarbone M, Tosi
D, et al: Genetic susceptibility variants for lung cancer:
Replication study and assessment as expression quantitative trait
loci. Sci Rep. 7(42185)2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Xun X, Wang H, Yang H, Wang H, Wang B,
Kang L, Jin T and Chen C: CLPTM1L genetic polymorphisms and
interaction with smoking and alcohol drinking in lung cancer risk:
A case-control study in the Han population from northwest China.
Medicine (Baltimore). 93(e289)2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yang YC, Fu WP, Zhang J, Zhong L, Cai SX
and Sun C: rs401681 and rs402710 confer lung cancer susceptibility
by regulating TERT expression instead of CLPTM1L in east Asian
populations. Carcinogenesis. 39:1216–1221. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhao MM, Zhang Y, Shen L, Ren YW, Li XL,
Yin ZH and Zhou BS: Genetic variations in TERT-CLPTM1L genes and
risk of lung cancer in a Chinese population. Asian Pac J Cancer
Prev. 15:2809–2813. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Cook DA and Reed DA: Appraising the
quality of medical education research methods: The medical
education research study quality instrument and the
Newcastle-Ottawa scale-education. Acad Med. 90:1067–1076.
2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Fan S, Meng J, Zhang L, Zhang X and Liang
C: CAV1 polymorphisms rs1049334, rs1049337, rs7804372 might be the
potential risk in tumorigenicity of urinary cancer: A systematic
review and meta-analysis. Pathol Res Pract. 215:151–158.
2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Meng J, Fan X, Zhang M, Hao Z and Liang C:
Do polymorphisms in protein kinase catalytic subunit alpha-1 gene
associated with cancer susceptibility? a meta-analysis and
systematic review. BMC Med Genet. 19(189)2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Meng J, Wang S, Shen X, Bai Z, Niu Q, Ma
D, Xu Y and Liang C: Polymorphism of MMP-9 gene is not associated
with the risk of urinary cancers: Evidence from an updated
meta-analysis. Pathol Res Pract. 214:1966–1973. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Amos CI, Wu X, Broderick P, Gorlov IP, Gu
J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, et al:
Genome-wide association scan of tag SNPs identifies a
susceptibility locus for lung cancer at 15q25.1. Nat Genet.
40:616–622. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Bae EY, Lee SY, Kang BK, Lee EJ, Choi YY,
Kang HG, Choi JE, Jeon HS, Lee WK, Kam S, et al: Replication of
results of genome-wide association studies on lung cancer
susceptibility loci in a Korean population. Respirology.
17:699–706. 2012.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chen XF, Cai S, Chen QG, Ni ZH, Tang JH,
Xu DW and Wang XB: Multiple variants of TERT and CLPTM1L constitute
risk factors for lung adenocarcinoma. Genet Mol Res. 11:370–378.
2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
de Mello RA, Ferreira M, Soares-Pires F,
Costa S, Cunha J, Oliveira P, Hespanhol V and Reis RM: The impact
of polymorphic variations in the 5p15, 6p12, 6p21 and 15q25 loci on
the risk and prognosis of portuguese patients with non-small cell
lung cancer. PLoS One. 8(e72373)2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hung RJ, McKay JD, Gaborieau V, Boffetta
P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N,
Lissowska J, Rudnai P, et al: A susceptibility locus for lung
cancer maps to nicotinic acetylcholine receptor subunit genes on
15q25. Nature. 452:633–637. 2008.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Jiang M, Wu H and Qin C: Genetic variant
rs401681 at 5p15.33 modifies susceptibility to lung cancer but not
esophageal squamous cell carcinoma. PLoS One.
8(e84277)2013.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Jin T, Li B, He N, Zhang Y, Xia R, Kang L,
Ding Y and Yuan D: CLPTM1L polymorphism as a protective factor for
lung cancer: A case-control study in southern Chinese population.
Tumour Biol. 37:10533–10538. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ke J, Zhong R, Zhang T, Liu L, Rui R, Shen
N, Sun Y, Liu L, Cheng L and Miao XP: Replication study in Chinese
population and meta-analysis supports association of the 5p15.33
locus with lung cancer. PLoS One. 8(e62485)2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Liu SG, Ma L, Cen QH, Huang JS, Zhang JX
and Zhang JJ: Association of genetic polymorphisms in TERT-CLPTM1L
with lung cancer in a Chinese population. Genet Mol Res.
14:4469–4476. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Myneni AA, Chang SC, Niu R, Liu L,
Ochs-Balcom HM, Li Y, Zhang C, Zhao B, Shi J, Han X, et al: Genetic
polymorphisms of TERT and CLPTM1L and risk of lung cancer-a
case-control study in a Chinese population. Lung Cancer.
80:131–137. 2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Sun Y, Zhang YJ and Kong XM: No
association of XRCC1 and CLPTM1L polymorphisms with non-small cell
lung cancer in a non-smoking Han Chinese population. Asian Pac J
Cancer Prev. 14:5171–5174. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wang H, Zhao Y, Ma J, Zhang G, Mu Y, Qi G,
Fang Z, Wang L, Fan Q and Ma Z: The genetic variant rs401681C/T is
associated with the risk of non-small cell lung cancer in a Chinese
mainland population. Genet Mol Res. 12:67–73. 2013.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Wang Y, Broderick P, Webb E, Wu X,
Vijayakrishnan J, Matakidou A, Qureshi M, Dong Q, Gu X, Chen WV, et
al: Common 5p15.33 and 6p21.33 variants influence lung cancer risk.
Nat Genet. 40:1407–1409. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
39
|
Xiao X and He W: Genetic polymorphisms in
the TERT-CLPTM1L region and lung cancer susceptibility in Chinese
males. Oncol Lett. 14:1588–1594. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Yoon KA, Park JH, Han J, Park S, Lee GK,
Han JY, Zo JI, Kim J, Lee JE, Takahashi A, et al: A genome-wide
association study reveals susceptibility variants for non-small
cell lung cancer in the Korean population. Hum Mol Genet.
19:4948–4954. 2010.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Zhang Y, Zhao M, Shen L, Ren Y, Su L, Li
X, Yin Z and Zhou B: Genetic polymorphisms of TERT and CLPTM1L and
risk of lung cancer: A case-control study in northeast Chinese male
population. Med Oncol. 31(18)2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zienolddiny S, Skaug V, Landvik NE, Ryberg
D, Phillips DH, Houlston R and Haugen A: The TERT-CLPTM1L lung
cancer susceptibility variant associates with higher DNA adduct
formation in the lung. Carcinogenesis. 30:1368–1371.
2009.PubMed/NCBI View Article : Google Scholar
|
|
43
|
XZ L: The CLPTM1L variant rs401681 is
associated with lung cancer risk. Huazhong University of Science
and Technology, 2013. https://xueshu.baidu.com/usercenter/paper/show?paperid=a716ba54558bed957b7463d7e5bf6c75&site=xueshu_se.
|
|
44
|
Liu C, Wang Y, Huang H, Wang C and Zhang
H, Kong Y and Zhang H: Association between CLPTM1L-TERT rs401681
polymorphism and pancreatic cancer risk among Chinese Han
population. Tumor Biol. 35:5453–5457. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Vaiciulis P, Liutkeviciene R, Liutkevicius
V, Vilkeviciute A, Gedvilaite G and Uloza V: Association of
relative leucocyte telomere length and gene single nucleotide
polymorphisms (TERT, TRF1, TNKS2) in laryngeal squamous cell
carcinoma. Cancer Genomics Proteomics. 17:431–439. 2020.PubMed/NCBI View Article : Google Scholar
|