Introduction
The molecular characterization of bladder cancer
(BC) has transformed understanding of BC pathogenesis and paved the
way for new biomarker discoveries. However, translating these
insights into clinical practice remains a challenge due to the
complexity of the disease and the multitude of altered molecular
and pathological pathways (1,2).
Signal transducers and activators of transcription
(STAT) proteins are key players in influencing tumor behavior and
modulating the tumor microenvironment. The STAT family is composed
of seven transcription factors (STAT1, STAT2, STAT3, STAT4, STAT5A,
STAT5B and STAT6), which play a critical and multifaceted role in
regulating vital physiological processes such as cell
proliferation, differentiation, apoptosis, angiogenesis and the
epigenetic organization of immune cells. Beyond these functions,
STATs also contribute to pathological processes by selectively
inducing and maintaining a pro-carcinogenic inflammatory
microenvironment at the initiation of malignant transformation and
during cancer progression (3-6).
Dysregulation in this pathway is associated with poor prognosis in
various cancers and has been recognized as a common driver in BC,
promoting tumor cell proliferation and motility (7).
STAT1 is considered a tumor suppressor, although
there is increasing evidence for its tumor-promoting functions
(8). The depletion of STAT1 in
cancer cells, observed in a broad spectrum of cancers, is often
correlated with a poor prognosis. However, the mechanism behind the
oncogenic or tumor-suppressing role of STAT1 remains unclear
(9). STAT4 is involved in
carcinogenesis and tumor progression (6). Although STAT4 inhibition appears to
promote tumorigenesis and predict poor outcomes in hepatocellular
carcinoma and gastric cancer, its expression was reported to be
significantly increased in colorectal cancer specimens, and
associated with tumor spread, suggesting a pro-tumorigenic role of
STAT4 in this type of cancer (10-12).
In ovarian cancer, activation of STAT4 is abundant in epithelial
cells and its overexpression was associated with poor outcome and
promoted metastasis both in vivo and in vitro,
suggesting a pro-metastatic function of STAT4(13).
Previous studies reported that STAT1 and STAT4
exhibited both complementary and opposite roles. These functions
appear to be regulated in a manner dependent on both cell type and
cancer type (14,15). The molecular events in bladder
tumorigenesis have sparked considerable interest, as they hold
promise for advancing patient diagnosis, prognosis and the
development of targeted therapies. While STAT1 has been extensively
studied in BC due to its well-established antitumor role, the
involvement of STAT4 in tumor development and progression remains
largely unexplored, with only limited insights available.
Considering the significant roles of STAT1 and STAT4
in cancer, as well as the increasing need for improved tools to
stratify patients and develop reliable prognostic and diagnostic
biomarkers, the expression level of STAT1 and STAT4 was evaluated
in primary and recurrent BC patient specimens, spanning the full
range of the disease's grades and stages. Their potential
associations with clinicopathological features and patient clinical
outcomes were also evaluated. Additionally, STAT1/4 expression was
quantified in human bladder cell lines mimicking the four stages of
tumorigenesis. Overall, the present study provides insight into the
expression pattern of STAT1 and STAT4 at the different stages of
tumorigenesis and contributes to the identification of potential
prognostic biomarkers in BC.
Materials and methods
Characteristics of the patients and
tissue samples
A total of 70 fresh frozen tumor samples were
obtained by transurethral resection of the bladder or cystoscopy at
the Urology Department of the University Military Hospital in
Rabat-Morocco between August 2019 and July 2022. The study protocol
was approved (approval no. Ref 82/19) by the Ethics Committee for
Biomedical Research from the Faculty of Medicine and Pharmacy of
Rabat (Rabat, Morocco). Written informed consent was obtained from
each recruited patient before sample collection. Staging and
grading were conducted according to the World Health Organization
Consensus Classification at the Anatomopathology Department of the
Military Hospital Mohammed V. The data collected from patients'
records is summarized in Table I.
Among the 70 recruited patients, 68 men and 2 women were included.
The mean age of patients was 67 years, ranging from 47-85 years. A
total 28 patients (40%) have reported to be active or former
smokers. Most cases were staged ≤ PT1 (74.29%) and had high tumor
grade (61.43%). Among patients with non-muscle invasive BC (NMIBC)
stage collected during the present study, 12 experienced tumor
recurrence (23.08%) and 5 were diagnosed with progression upon the
4-year follow up.
 | Table IClinicopathological characteristics of
patients. |
Table I
Clinicopathological characteristics of
patients.
| Parameter | Total cases | Percentage (%) |
|---|
| Sex | | |
|
Male | 68 | 97.14 |
|
Female | 2 | 2.86 |
| Age, years | | |
|
<50 | 1 | 1.43 |
|
50-70 | 44 | 62.86 |
|
>70 | 25 | 35.71 |
| Smoking
history | | |
|
Yes | 28 | 40 |
|
No | 42 | 60 |
| Tumor stage | | |
|
≤PT1 | 52 | 74.29 |
|
>PT1 | 18 | 25.71 |
| Tumor grade | | |
|
Low
grade | 27 | 38.57 |
|
High
grade | 43 | 61.43 |
| Tumor
recurrence | | |
|
Yes | 12 | 23.08 |
|
No | 40 | 76.92 |
| Tumor
progression | | |
|
Yes | 5 | 9.62 |
|
No | 47 | 90.38 |
Cell lines and cell culture
A total of four human bladder cell lines were used
in the present study: BU68.08, mimicking a NMIBC stage [generated
and kindly provided by Dr Laurent Derré, University Hospital
Lausanne, Switzerland (16)] and
RT4:pT2 (cat. no. HTB-2™), J82:pT3 (cat. no. HTB-1™) and
TCC-SUP:pT4 (cat. no. HTB-5™) corresponding to the three stages of
disease invasiveness, were purchased from the American Type Culture
Collection. Cells were cultured in RPMI-1640 10% heat-inactivated
FCS (Gibco; Thermo Scientific, Inc.) at 37˚C in a 5% CO2
humidified atmosphere. The medium was changed as recommended by the
manufacturer and cells were used for the experiment when confluency
reached 70-80%.
RNA isolation
Total RNA was extracted from 70 fresh frozen
biopsies conserved in RNA Later (Invitrogen; Thermo Fisher
Scientific, Inc.) and from four BC cell lines using TRI Reagent
(Sigma-Aldrich; Merck KGaA), according to the manufacturer's
protocol. The amount and quality of DNA and RNA were measured by
NanoDrop 2000 (Thermo Fisher Scientific, Inc.). The ratio of
absorbance at 260 and 280 nm was used to assess purity. A ratio of
~1.8 was accepted as ‘pure’ for DNA. RNA was considered DNA and
protein free if the ratio of readings at 260/280 nm was ~2.
Gene expression study
A total of 1 µg of each of the 70 RNA samples was
subjected to reverse transcription independently using the
High-capacity cDNA reverse transcription kit according to
manufacturer's protocol (Applied Biosystems; Thermo Fisher
Scientific, Inc.). cDNAs were subsequently used to perform a SYBR
green-based quantitative PCR using the KAPA SYBR FAST Kit (Roche
Diagnostics). Enzyme was first activated at 95˚C for 3 min,
followed by 40 cycles of denaturation at 95˚C for 3 sec, primer
annealing and extension for 20 sec at 60˚C. Samples were amplified
in triplicate, and a non-template control was used for each primer
pair to control for contamination or primer dimerization. STAT1 and
STAT4 levels were normalized to the expression of β2
microglobulin (β2M) used as an internal control gene,
using the 2-ΔΔCq formula (17). Primers' sequences are shown in
Table II.
 | Table IISequences of primers used in the
present study. |
Table II
Sequences of primers used in the
present study.
| Gene name | Primer sequence
(5'-3') |
|---|
| β2M | F:
GAGGCTATCCAGCGTACTCCA |
| | R:
CGGCAGGCATACTCATCTTTT |
| STAT1 | F:
ATCAGGCTCAGTCGGGGAATA |
| | R:
TGGTCTCGTGTTCTCTGTTCT |
| STAT4 | F:
TGTTGGCCCAATGGATTGAAA |
| | R:
GGAAACACGACCTAACTGTTCAT |
Statistical analysis
Statistical analysis of the potential association
between clinicopathological features and patients' clinical
outcomes was performed using Graph Pad Prism software version 10
(Dotmatics). Unpaired student's t-test was used for the comparison
between two groups; for the comparison between multiple groups,
two-way ANOVA or the non-parametric (Mann-Whitney or Kruskal-Wallis
tests) tests were used. Survival analyses were performed using the
Kaplan-Meier method and compared by a log-rank test on Graph Pad
Prism software version 10, and the follow-up period was defined
from the date of first transurethral resection of bladder tumor
treatment to the moment of death for deceased cases, or the date of
the last follow up for survivors. P<0.05 (two-tailed) was
considered to indicate a statistically significant difference.
Results
Differential expression of STAT1 and
STAT4 in early vs. aggressive tumors
In the present prospective study, the mRNA level of
STAT1 and STAT4 genes were evaluated in both BC cases as well as in
BC cell lines mimicking the four state and the four stages of
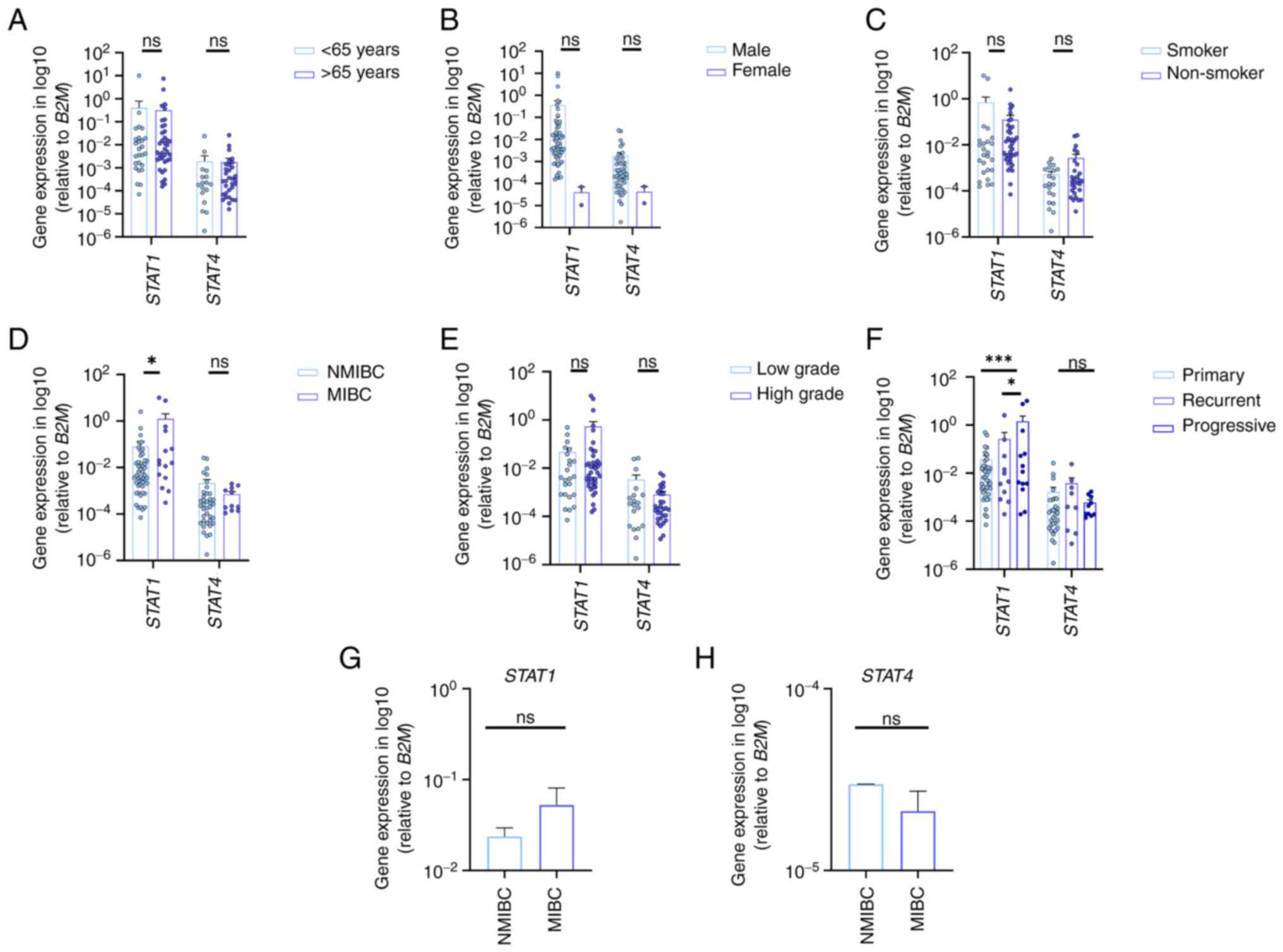
cancer progression. The results are presented in Fig. 1, where the expression of STAT1 and
STAT4 showed no significant association with patient age, sex, or
smoking history (Fig. 1A-C).
STAT1 expression was significantly upregulated in
patients with muscle-invasive BC (MIBC) compared with those with
NMIBC, and in progressive tumors relative to primary and recurrent
cases. By contrast, STAT4 expression was slightly higher in NMIBC
compared with MIBC cases (Fig.
1D), in low-grade tumors compared with higher-grade stages
(Fig. 1E), and in recurrent cases
compared with primary and progressive tumors (Fig. 1F), although these differences were
not statistically significant.
In BC cell lines, STAT1 and STAT4 patterns are
similar to those observed in human primary tissue samples. Indeed,
STAT1 expression scored the highest rate in advanced tumor stages,
while STAT4 was higher in the NMIBC cell lines and its expression
decreased according to tumor invasion (Fig. 1G and H).
STAT1/4 expression as a survival
prognostic biomarker in patients with BC
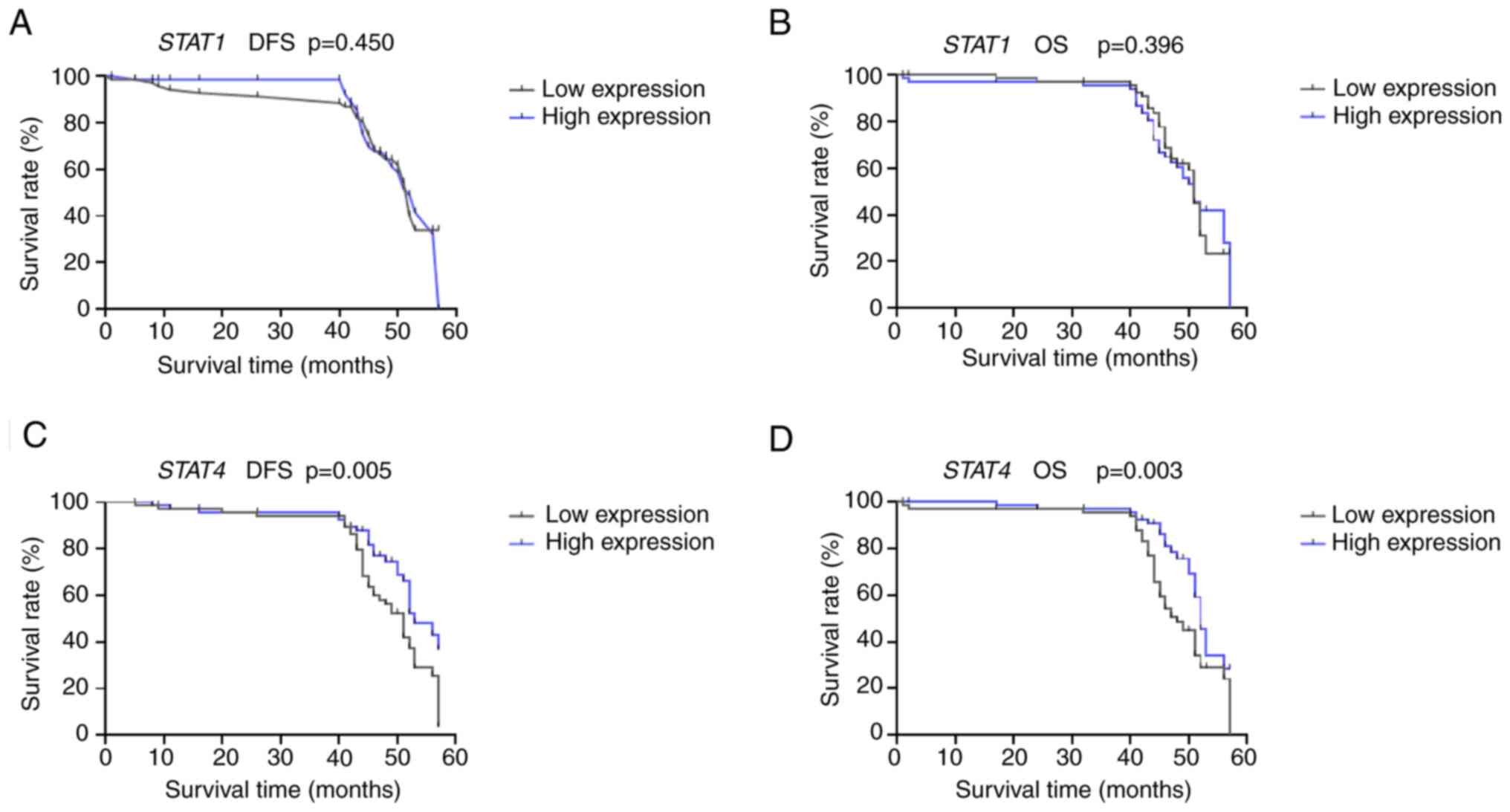
In the present study, the mean and the median of
clinical follow-up periods were 44 and 45 months, respectively.
Kaplan-Meier analyses were performed to examine the possible
correlations between the expression of the studied and patients'
clinical outcomes: Disease-free survival (DFS) and overall survival
(OS). Based on the median value, patients were stratified into two
groups: The low expression group, with mRNA levels below the
cohort's median, and the high expression group, with mRNA levels
equal to or above the median.
For STAT1, the DFS and OS rates were nearly
identical (Fig. 2A and B). The survival curves showed no
significant differences between the low expression and the high
expression groups (DFS, P=0.450; OS, P=0.396). Interestingly,
patients with high STAT4 expression had significantly longer DFS
(P=0.005) and OS (P=0.003) than those with low STAT4 expression
(Fig. 2C and D).
Correlation between STAT1 and STAT4
expression levels
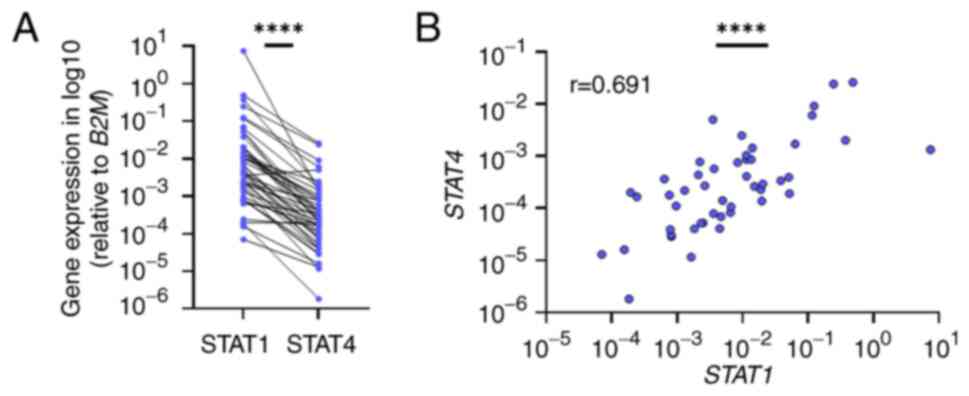
To explore the potential correlation between STAT1
and STAT4 expression in BC, the expression levels of both genes
were compared across patients (Fig.
3). This analysis uncovered a significant positive correlation
between the mRNA levels of STAT1 and STAT4 (r=0.691; P<0.0001;
95% confidence interval, 0,5010-0,8185) (Fig. 3A). The positive correlation was
sustained in both NMIBC and MIBC subtypes, although STAT1 was
upregulated in MIBC and STAT4 in NMIBC. The examination revealed
that most patients with high STAT1 expression had also high STAT4
expression (Fig. 3B).
Discussion
Worldwide, significant efforts have been made to
uncover the molecular mechanisms and pathogenesis of cancer
development and progression. However, the detailed process remains
one of the most unsolved aspects of cancer biology (18). Understanding the molecular events
occurring at different stages of tumorigenesis can pave the way for
the discovery of new biomarkers and improve predictions of disease
progression and patient prognosis factors, as demonstrated by
recent studies on BC (19-22).
In the present study, STAT1 and STAT4 were
investigated as potential biomarkers in BC, building on the concept
that transcription factors can serve as critical prognostic
indicators in various malignancies. For example, transcription
factors such as EGFR (23) and
STAT3 in non-small cell lung cancer (9,24)
and TWIST1/Vimentin have proven also their utility in detecting
urothelial carcinoma of the bladder due to their involvement in key
processes such as epithelial-to-mesenchymal transition and
metastasis. Similarly, E2F family members (E2F3/5/8) have shown
promise as prognostic markers in BC, emphasizing the relevance of
transcription factors in tumor biology and their therapeutic
potential (25).
In the present study, a cohort of 70 samples of BC
obtained from patients receiving the same standard of care were
used for gene expression investigation and survival analyses.
Through the extensive follow-up data that was collected over four
years, it was possible to highlight the opposite roles played by
STAT1 and STAT4 in bladder tumorigenesis. It was identified that
STAT1 is significantly upregulated in advanced stages of BC, in
recurrent and in progressing tumors, emphasizing its role as a
biomarker of aggressiveness and progression. On the other hand,
STAT4 appeared to act as a protective factor in our cohort, with
slightly higher expression observed in early-stage cancer biopsies,
though not reaching statistical significance.
Furthermore, STAT1 was almost 10-fold overexpressed
in the high-grade group. These observations align with a previous
study that has shown significantly higher STAT1 expression across a
panel of 12 cancer types, with elevated STAT1 levels linked to
higher tumor grades in bladder carcinoma (26).
The protective role of STAT4 is experimentally
demonstrated by the high level of mRNA detected in non-invasive
tumor stages and non-relapsing patients over the 4-year follow-up
period. STAT4 has been suggested to induce inflammation and
autoimmune diseases, inhibit tumor growth, or promote tumors via
regulating numerous facets of the innate and adaptive immune
responses (27). According to a
previous study, STAT1 and STAT4 tend to increase simultaneously in
several immune disorders, including inflammatory bowel disease
(15). The present study points to
contrasting roles for these two genes and, if validated in larger
patient cohorts, they could be utilized as prognostic markers.
STAT1 overexpression might indicate aggressive tumors, while high
STAT4 expression could be associated with favorable clinical
outcomes. Our survival analysis revealed that STAT1 expression was
not significantly associated with OS or DFS, as shown in Fig. 2. This unexpected result may reflect
several factors: The heterogeneous nature of BC, the influence of
other molecular pathways compensating for STAT1 activity, or the
sample size limitations of our cohort. It is also possible that
STAT1's role in tumor progression depends on context-specific
factors, such as interactions with the tumor microenvironment or
post-transcriptional modifications affecting its activity. These
findings warrant further validation in larger cohorts and
functional studies to elucidate the interplay between STAT1, STAT4
and the broader immune landscape in BC.
On the other side, the survival analysis revealed a
significantly superior DFS and OS in patients displaying higher
STAT4 expression. This improved outcome may be attributed to the
crucial role of STAT4 in IFN-γ induction, which is essential for
boosting antitumor immunity. Supporting this idea, STAT4 inhibition
has been demonstrated to promote tumorigenesis and predict poor
outcomes in cutaneous T-cell lymphoma, hepatocellular carcinoma and
gastric cancer (10,11,28-30).
The potential link between the expression profiles
of the two genes studied was further investigated. Interestingly, a
positive correlation was found between STAT1 and STAT4 expression,
indicating that patients with higher STAT1 expression also tend to
exhibit higher STAT4 levels. Although STAT1 and STAT4 may have
opposing roles in BC, this association suggests a functional
relationship likely due to co-regulation from a shared genetic
locus. This co-regulation may enable STAT1 and STAT4 to complement
each other, working synergistically to modulate antitumor immune
responses and revealing an interaction that has not been previously
well-defined (15). A similar
expression trend was observed in bladder cell lines, where high
STAT1 expression was detected in cells from advanced tumor stages,
while low STAT1 expression was associated with BU68.08
(representing the NMIBC stage). By contrast, STAT4 expression
exhibited an opposite pattern, with lower levels in advanced-stage
cells and higher levels in non-invasive BC cells.
To the best of our knowledge, the present study is
the first to report differential expression of STAT1 and STAT4 in
human BC, using both primary samples and cell lines. These findings
indicate that STAT gene expression could be a valuable target for
developing new therapies to improve BC management. However, due to
the limited sample size and the complex role of STAT signaling in
cancer progression, further research is essential to confirm STAT1
and STAT4 as reliable prognostic biomarkers. Exploring the distinct
roles of STAT1 and STAT4 in tumor progression and the tumor
microenvironment presents a promising avenue for future research. A
priority will be to perform targeted knockout and overexpression
experiments for STAT1 and STAT4 in BC cell lines. Complementing
these in vitro studies with in vivo experiments using
animal models will provide critical insights into how these genes
influence tumor growth, metastasis and interaction with the tumor
microenvironment in a physiological context. These combined
approaches aim to elucidate the detailed mechanisms underlying
their roles in cell proliferation, migration and invasion, thereby
advancing our understanding of their functional significance and
therapeutic potential in BC.
In conclusion, the present study demonstrated that
STAT1 expression is significantly upregulated in patients with
advanced MIBC and progressing bladder tumors. By contrast, STAT4
expression was higher in NMIBC, although the difference was not
statistically significant. In BC cell lines, the expression trends
of STAT1 and STAT4 mirrored those observed in human primary
tissues. Notably, patients with high STAT4 expression exhibited
significantly improved DFS and OS compared with those with low
expression, likely due to the enhanced immune response,
particularly through the activation of Th1 cells that aid in
controlling tumor growth and recurrence. Additionally, STAT4
influences pro-inflammatory pathways, creating an antitumor
environment that limits tumor progression and ultimately improves
patient prognosis. As a consequence, the current findings suggested
STAT1 and STAT4 as promising biomarkers for prognostic prediction
in BC.
Furthermore, it is anticipated that the present
study would be a catalyst for collaborative research that could
integrate the present findings into clinical trials and potentially
develop STAT1 and STAT4-based diagnostic and prognostic tools.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by a Consolidation
grant (grant no. COG-2023-37) from the HES-SO Leading House for the
Middle East and North Africa (to EAH).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
EAH designed the study, performed the experiments,
analyzed the data, wrote the manuscript, and provided final
approval for publication. EAM performed the experiments and
analyzed the data. AM analyzed the data and contributed to
manuscript writing. HAC, HI and CI performed biostatistical
analysis. TM, AA, ABA and OM provided patient samples and
follow-up. AA, AM and BL provided intellectual contributions and
revised the manuscript. JC and AM provided intellectual
contributions, critically revised the manuscript, and provided
final approval for publication. EAH and AM confirm the authenticity
of the raw data.
Ethics approval and consent to
participate
The study protocol was approved (approval no. Ref
82/19) by the Ethics Committee of Biomedical Research from the
Faculty of Medicine and Pharmacy of Rabat (Rabat, Morocco). Written
informed consent was obtained from each recruited patient prior to
sample collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu Q: Current advances in
N6-methyladenosine methylation modification during bladder cancer.
Front Genet. 12(825109)2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Inamura K: Bladder cancer: New insights
into its molecular pathology. Cancers (Basel).
10(100)2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Becerra-Díaz M, Valderrama-Carvajal H and
Terrazas LI: Signal transducers and activators of transcription
(STAT) family members in helminth infections. Int J Biol Sci.
7:1371–1381. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Verhoeven Y, Tilborghs S, Jacobs J, De
Waele J, Quatannens D, Deben C, Prenen H, Pauwels P, Trinh XB,
Wouters A, et al: The potential and controversy of targeting STAT
family members in cancer. Semin Cancer Biol. 60:41–56.
2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Alunno A, Padjen I, Fanouriakis A and
Boumpas DT: Pathogenic and therapeutic relevance of JAK/STAT
signaling in systemic lupus erythematosus: Integration of distinct
inflammatory pathways and the prospect of their inhibition with an
oral agent. Cells. 8(898)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kitamura H, Ohno Y, Toyoshima Y, Ohtake J,
Homma S, Kawamura H, Takahashi N and Taketomi A: Interleukin-6/STAT
3 signaling as a promising target to improve the efficacy of cancer
immunotherapy. Cancer Sci. 108:1947–1952. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chen CL, Cen L, Kohout J, Hutzen B, Chan
C, Hsieh FC, Loy A, Huang V, Cheng G and Lin J: Signal transducer
and activator of transcription 3 activation is associated with
bladder cancer cell growth and survival. Mol Cancer.
7(78)2008.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Meissl K, Macho-Maschler S, Müller M and
Strobl B: The good and the bad faces of STAT1 in solid tumours.
Cytokine. 89:12–20. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Pensa sara, Regis G, Boselli D, Novelli F
and Poli V: STAT1 and STAT3 in tumorigenesis: Two sides of the same
coin? In: Madame Curie Bioscience Database. Landes Bioscience,
Austin, TX, 2013.
|
|
10
|
Wubetu GY, Utsunomiya T, Ishikawa D,
Yamada S, Ikemoto T, Morine Y, Iwahashi S, Saito Y, Arakawa Y and
Imura S: , et al: High STAT4 expression is a better
prognostic indicator in patients with hepatocellular carcinoma
After Hepatectomy. Ann Surg Oncol. 21 (Suppl 4):S721–S728.
2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhou X, Xia Y, Su J and Zhang G:
Down-regulation of miR-141 induced by helicobacter pylori promotes
the invasion of gastric cancer by targeting STAT4. Cell Physiol
Biochem. 33:1003–1012. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Cheng JM, Yao MR, Zhu Q, Wu XY, Zhou J,
Tan WL and Zhan SH: Silencing of stat4 gene inhibits cell
proliferation and invasion of colorectal cancer cells. J Biol Regul
Homeost Agents. 29:85–92. 2015.PubMed/NCBI
|
|
13
|
Zhao L, Ji G, Le X, Luo Z, Wang C, Feng M,
Xu L, Zhang Y, Lau WB, Lau B, et al: An integrated analysis
identifies STAT4 as a key regulator of ovarian cancer metastasis.
Oncogene. 36:3384–3396. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Anderson K, Ryan N, Volpedo G, Varikuti S,
Satoskar AR and Oghumu S: Immune suppression mediated by STAT4
deficiency promotes lymphatic metastasis in HNSCC. Front Immunol.
10(3095)2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Hedl M, Sun R and Abraham C: Disease
risk-associated genetic variants in STAT1 and STAT4 function in a
complementary manner to increase pattern-recognition
receptor-induced outcomes in human macrophages. J Immunol.
205:1406–1418. 2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Vanoni G, Ercolano G, Candiani S,
Rutigliani M, Lanata M, Derré L, Marcenaro E, Schneider P, Romero
P, Jandus C and Trabanelli S: Human primed ILCPs support
endothelial activation through NF-κB signaling. ELife.
10(e58838)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000.PubMed/NCBI View Article : Google Scholar
|
|
19
|
El Ahanidi H, El Azzouzi M, Hafidi Alaoui
C, Tetou M, Bensaid M, Chaoui I, Benbacer L, Hassan I, Oukabli M,
Michaud K, et al: Immune checkpoint and telomerase crosstalk is
mediated by miRNA-138 in bladder cancer. Front Oncol.
11(795242)2022.PubMed/NCBI View Article : Google Scholar
|
|
20
|
El Azzouzi M, El Ahanidi H, Hassan I,
Tetou M, Ameur A, Bensaid M, Al Bouzidi A, Oukabli M, Alaoui CH,
Addoum B, et al: Comprehensive behavioural assessment of TERT in
bladder cancer. Urol Oncol. 42:451.e19–451.e29. 2024.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang Q: STAT1 as a potential therapeutic
target to treat bladder cancer. Int J Clin Exp Pathol. 17:298–307.
2024.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Gao B, Li X, Li S, Wang S, Wu J and Li J:
Pan-cancer analysis identifies RNA helicase DDX1 as a prognostic
marker. Phenomics. 2:33–49. 2022.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Hashmi AA, Hussain ZF, Irfan M, Khan EY,
Faridi N, Naqvi H, Khan A and Edhi MM: Prognostic significance of
epidermal growth factor receptor (EGFR) over expression in
urothelial carcinoma of urinary bladder. BMC Urol.
18(59)2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hu Y, Dong Z and Liu K: Unraveling the
complexity of STAT3 in cancer: molecular understanding and drug
discovery. J Exp Clin Cancer Res. 43(23)2024.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zhang C, Xu X, Wang T, Lu Y, Lu Z, Wang T
and Pan Z: Clinical performance and utility of a noninvasive
urine-based methylation biomarker: TWIST1/Vimentin to detect
urothelial carcinoma of the bladder. Sci Rep.
14(7941)2024.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang J, Wang F, Liu F and Xu G:
Predicting STAT1 as a prognostic marker in patients with solid
cancer. Ther Adv Med Oncol. 12(1758835920917558)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Yang C, Mai H, Peng J, Zhou B, Hou J and
Jiang D: STAT4: an immunoregulator contributing to diverse human
diseases. Int J Biol Sci. 16:1575–1585. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Litvinov IV, Cordeiro B, Fredholm S, Ødum
N, Zargham H, Huang Y, Zhou Y, Pehr K, Kupper TS, Woetmann A and
Sasseville D: Analysis of STAT4 expression in cutaneous T-cell
lymphoma (CTCL) patients and patient-derived cell lines. Cell
Cycle. 13:2975–2982. 2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhou M, Zhang P, Da M, Yang R, Ma Y, Zhao
J, Ma T, Xia J, Shen G, Chen Y and Chen D: A pan-cancer analysis of
the expression of STAT family genes in tumors and their
relationship to the tumor microenvironment. Front Oncol.
12(925537)2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li C, Zhao J, Kang B, Li S, Tang J, Dong D
and Chen Y: Identification and validation of STAT4 as a prognostic
biomarker in acute myeloid leukemia. Biosci Rep.
44(BSR20231720)2024.PubMed/NCBI View Article : Google Scholar
|