Introduction
Prostate cancer (PCa) is the second most frequently
diagnosed malignancy in men, following lung cancer, with >1.5
million new cases reported globally in 2020. It remains one of the
leading causes of cancer-related mortality in males, accounting for
>375,000 deaths worldwide in the same year (1). Early and accurate detection of PCa is
critical for improving clinical outcomes and reducing mortality.
However, conventional diagnostic tools such as prostate-specific
antigen (PSA) testing have marked limitations in sensitivity and
specificity. PSA levels can be influenced by non-malignant
conditions, including infections or benign prostatic hyperplasia,
leading to false-positive results, overdiagnosis and unnecessary
interventions. These limitations underscore the urgent need for
more reliable biomarkers to enhance diagnostic accuracy and provide
improved risk stratification for patients with PCa (2).
Recent advancements in molecular biology have
identified microRNAs (miRNAs/miRs) as promising diagnostic and
prognostic biomarkers in cancer. miRNAs are small, single-stranded,
non-coding RNA molecules, typically 15-27 nucleotides in length,
that regulate gene expression at the post-transcriptional level.
They serve pivotal roles in several biological processes, including
cell proliferation, differentiation, migration and apoptosis.
Aberrant miRNA expression has been associated with oncogenesis,
tumour progression and metastasis in several cancers, including PCa
(3). Calin et al (4) was the first to propose the
involvement of miRNAs in cancer development and progression in
2004, identifying a genomic region at 13q14 that includes miR-15a
and miR-16-1, which is often deleted in leukaemia. Since then,
abnormal expression of miRNAs has been observed in several cancers,
driven by both genetic alterations and epigenetic mechanisms.
The initial comprehensive profiling study of miRNA
expression in PCa, published in 2007, signified a pivotal
advancement in comprehending their function in oncogenesis
(5). Certain miRNAs act as
oncogenes (oncomiRs) or tumour suppressors, depending on their
target genes. For instance, miR-21, one of the most studied
oncomiRs, is consistently upregulated in multiple cancers,
including PCa. It promotes tumour progression by targeting key
tumour suppressor genes such as PTEN and programmed cell death 4
(PDCD4), thereby enhancing cell proliferation and inhibiting
apoptosis (6). Furthermore, the
expression of miR-21 in the PCa DU145 cell line has been reported
to enhance the expression of hypoxia-inducible factor-1α and
vascular endothelial growth factor, which are pivotal factors in
tumour growth and angiogenesis (7). Similarly, miR-221 has been reported
to be upregulated in PCa PC3 cells, where it serves a notable role
in promoting cell proliferation. One of its mechanisms involves
targeting cyclin dependent kinase inhibitor 1B (p27 Kip1), a key
cyclin-dependent kinase inhibitor that regulates the cell cycle. By
inhibiting p27 Kip1, miR-221 facilitates progression through the
cell cycle, leading to increased cellular proliferation. This
process underscores the potential of miR-221 as an oncogenic factor
in PCa development and progression (8). Overexpression of miR-221 exerts a
pivotal influence on the growth potential of LNCaP cells, inducing
a notable transition from the G1 to the S phase of the cell cycle.
This transition accelerates cell cycle progression, which in turn
enhances cellular proliferation. Furthermore, the colony-forming
potential of LNCaP cells in soft agar is markedly increased by
miR-221 overexpression (9). Other
oncogenic miRNAs, such as miR-125b, contribute to cancer cell
survival and growth by directly repressing the tumour suppressor
gene TP53. TP53, often referred to as the ‘guardian of the genome’,
serves a critical role in regulating cell cycle arrest, DNA repair
and apoptosis in response to cellular stress or DNA damage. By
downregulating TP53, miR-125b enables cancer cells to evade these
protective mechanisms, promoting increased cell survival,
uncontrolled proliferation and resistance to apoptosis (10).
Conversely, several miRNAs function as tumour
suppressors in PCa. For example, miR-34a inhibits cancer cell
proliferation and induces apoptosis by targeting oncogenes such as
c-Myc and BCL-2. By downregulating these oncogenes, miR-34a
effectively disrupts the pathways that promote uncontrolled cell
proliferation and survival, thereby inducing apoptosis in cancer
cells (11,12). Similarly, miR-200c suppresses
epithelial-to-mesenchymal transition (EMT) and metastasis in PCa by
downregulating zinc finger E-box binding homeobox (ZEB)1 and ZEB2,
key transcription factors involved in EMT (13). The stability of miRNAs in body
fluids and tissues, along with their differential expression
patterns between cancerous and non-cancerous tissues, highlights
their potential as minimally invasive diagnostic biomarkers for
PCa.
The association between miR-21 and miR-221 in PCa
and clinicopathological parameters has not been previously assessed
in Moroccan men, to the best of our knowledge. The aim of the
present study was to evaluate the diagnostic potential of miRNAs,
specifically miR-21 and miR-221, as biomarkers for PCa and their
association with clinicopathological parameters in Moroccan men.
From the analysis of tissue samples, the findings of the present
study offer valuable insights that advance diagnostic strategies
and enhance the understanding of PCa progression.
Patients and methods
Study subjects
Between April 2023 and March 2024, patients with a
PCa diagnosis were recruited from the Urology Department of
Mohammed V Military Hospital (Rabat, Morocco). Control cases
without a history or current symptoms related to PCa were also
recruited during this period. The inclusion criterion for the
patients with PCa was histologically-confirmed PCa. Patients with
PCa who were receiving chemotherapy and/or radiotherapy, as well as
those with any other malignant diseases, were excluded from the
present study. Controls were age-matched to the PCa cases and had
no history of malignancy; however, they were diagnosed with benign
inflammatory disorders.
A total of 50 tumor tissues from patients with
diagnosed PCa and 50 control tissues were included in the present
study. After the collection of tissue samples, samples were placed
in RNAlater™ Stabilization Solution (Thermo Fisher
Scientific, Inc.) and were kept at -80˚C until RNA extraction.
Biopsies were obtained by physicians directly performed according
to standard protocols. The clinical, histological and
epidemiological characteristics of the recruited patients were also
obtained. The clinical characteristics of the patients in the PCa
and control groups are provided in Table I. Informed consent was obtained
from all participants, and the study was approved by the Moroccan
Biomedical Research Ethics Committee (approval no. 3/2018; April
30, 2018).
 | Table IThe clinical characteristics of the
patients in the prostate cancer and control groups. |
Table I
The clinical characteristics of the
patients in the prostate cancer and control groups.
| Clinical
characteristics | Cases, n (%) | Controls, n
(%) |
|---|
| Total cases | 50 | 50 |
| Age at
diagnosis/surgery | | |
|
<60
years | 13(26) | 15(30) |
|
≥60
years | 37(74) | 35(70) |
| Prostate-specific
antigen (ng/ml) | | |
|
<2.5 | 2(4) | 35(70) |
|
2.5-10 | 12(24) | 15(30) |
|
≥10 | 36(72) | 0 (0) |
| Pathological
Gleason score | | |
|
≤6 | 14(28) | NA |
|
>6 | 36(72) | NA |
| Pathological
T-stage | | |
|
T1 | 17(34) | NA |
|
T2 X | 29(58) | NA |
|
T3 X | 1(2) | NA |
|
T4 | 3(6) | NA |
| Alcohol
consumption | | |
|
Yes | 28(56) | 20(40) |
|
No | 12(24) | 20(40) |
|
Weaned | 10(20) | 10(20) |
| Smoking | | |
|
Yes | 32(64) | 20(40) |
|
No | 11(22) | 25(50) |
|
Weaned | 7(14) | 5(10) |
RNA extraction
Total RNA, including miRNAs, was extracted using the
mirVana™ miRNA Isolation Kit (Thermo Fisher Scientific,
Inc.), according to the manufacturer's instructions. The purity and
concentration of extracted RNA were assessed using a
NanoDrop™ 2000 Spectrophotometer (NanoDrop Technologies;
Thermo Fisher Scientific, Inc.), with A260/A280 ratios >2.0
indicating high RNA purity. Extracted RNA was stored at -80˚C until
used.
cDNA synthesis
For miRNA detection, cDNA was synthesized using the
TaqMan™ MicroRNA assay (Applied Biosystems; Thermo
Fisher Scientific, Inc.) alongside reagents from the
TaqMan™ MicroRNA Reverse Transcription Kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The primer sequences
used for miR-21 and miR-221 detection are provided in Table SI.
The reverse transcription reactions were performed
using 5 µl total RNA in a 10-µl reaction volume according to the
manufacturer's instructions. The reaction mixture composed of 0.15
µl of 100 mM dNTPs mix, 1 µl of 50 U/µl reverse transcriptase, 1.5
µl of 10X reverse transcriptase buffer, 0.19 µl of 20 U/µl RNase
inhibitor, and 3 µl of 5X RT miR primers. The reaction volume was
made up to a total of 15 µl with nuclease-free water (4.16 µl
ultra-pure H2O). Reactions were incubated at 16˚C for 30
min, 42˚C for 30 min and 85˚C for 5 min using the
GeneAmp® PCR System 2400 (Scientific Support, Inc.).
cDNA was stored at -20˚C until further use.
Reverse transcription-quantitative PCR
(RT-qPCR) of miR-21 and miR-221
RT-qPCR was performed in a volume of 20 µl composed
of 10 µl TaqMan™ Universal PCR Master Mix (2X) (Applied
Biosystems; Thermo Fisher Scientific, Inc.), 1 µl TaqMan miRNA
Assay (20X), 7 µl Nuclease Free Water, and 2 µl cDNA used a
StepOnePlus™ Real-Time PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The internal control U6 small
nuclear (sn) RNA was used. The thermal cycling conditions were as
follows: Initial activation at 95˚C for 10 min, followed by 40
cycles of 95˚C for 15 sec, then 60˚C for 1 min.
The expression levels of the 2 miRNAs candidates,
relative to the internal control, U6 snRNA (reference gene), were
calculated using the 2-ΔΔCq method
(14). Negative controls were used
to detect any possible contamination in the reagents.
Statistical analysis
Statistical analysis was performed using jamovi
software 2022 (www.jamovi.org). The difference in
miRNA expression between the tumour and normal tissues was
evaluated using one-way ANOVA, followed by Tukey's post-hoc test.
Moreover, the association between miRNA expression and several
clinicopathological characteristics was assessed using the
χ2 test or Fisher's exact test. The analysis plotted
sensitivity (true positive rate) against 1-specificity (false
positive rate) across different thresholds of miRNA expression. The
Youden index was used to determine the optimal cut-off values. The
sensitivity analysis measured the ability to correctly identify
cancerous tissues, whilst the specificity analysis measured the
ability to correctly identify normal tissues. The area under the
curve (AUC) was calculated to estimate overall diagnostic accuracy,
and higher AUC values reflected an improved performance of the
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Study population
In the present prospective cohort study, 50 patients
with PCa were included and compared with 50 control subjects. The
median age of the PCa group was 68 years (range, 55-82 years),
whilst the median age of the control group was 63 years (range,
52-78 years).
Expression levels of miR-21 and
miR-221
RT-qPCR analysis revealed that the expression level
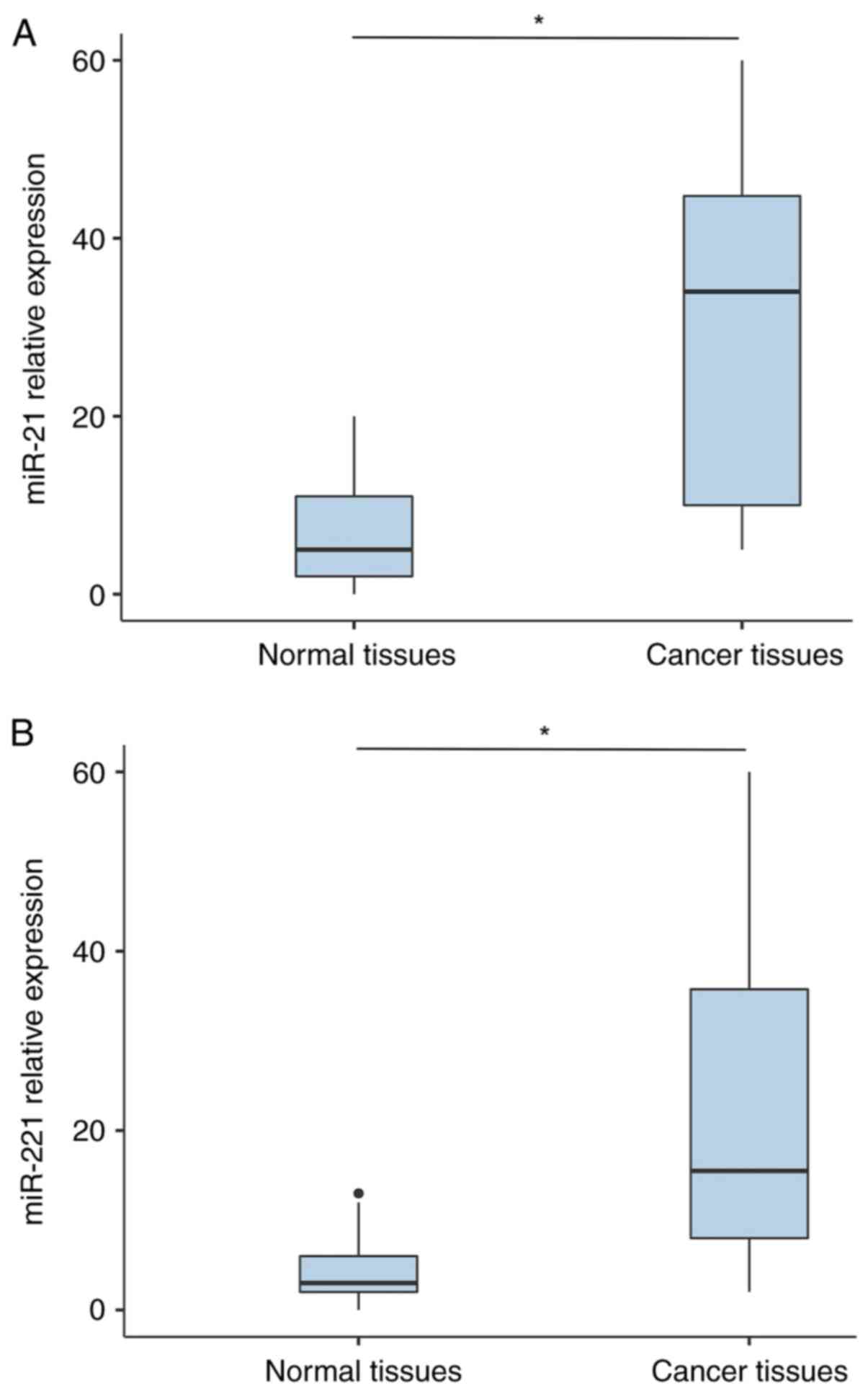
of miR-21 in PCa tissues was significantly higher than that in
normal tissues (P<0.001; Fig.
1A). Similarly, miR-221 expression level was significantly
higher in patients with PCa compared with that of controls
(P<0.001; Fig. 1B).
Diagnostic performance of miR-21 and
miR-221
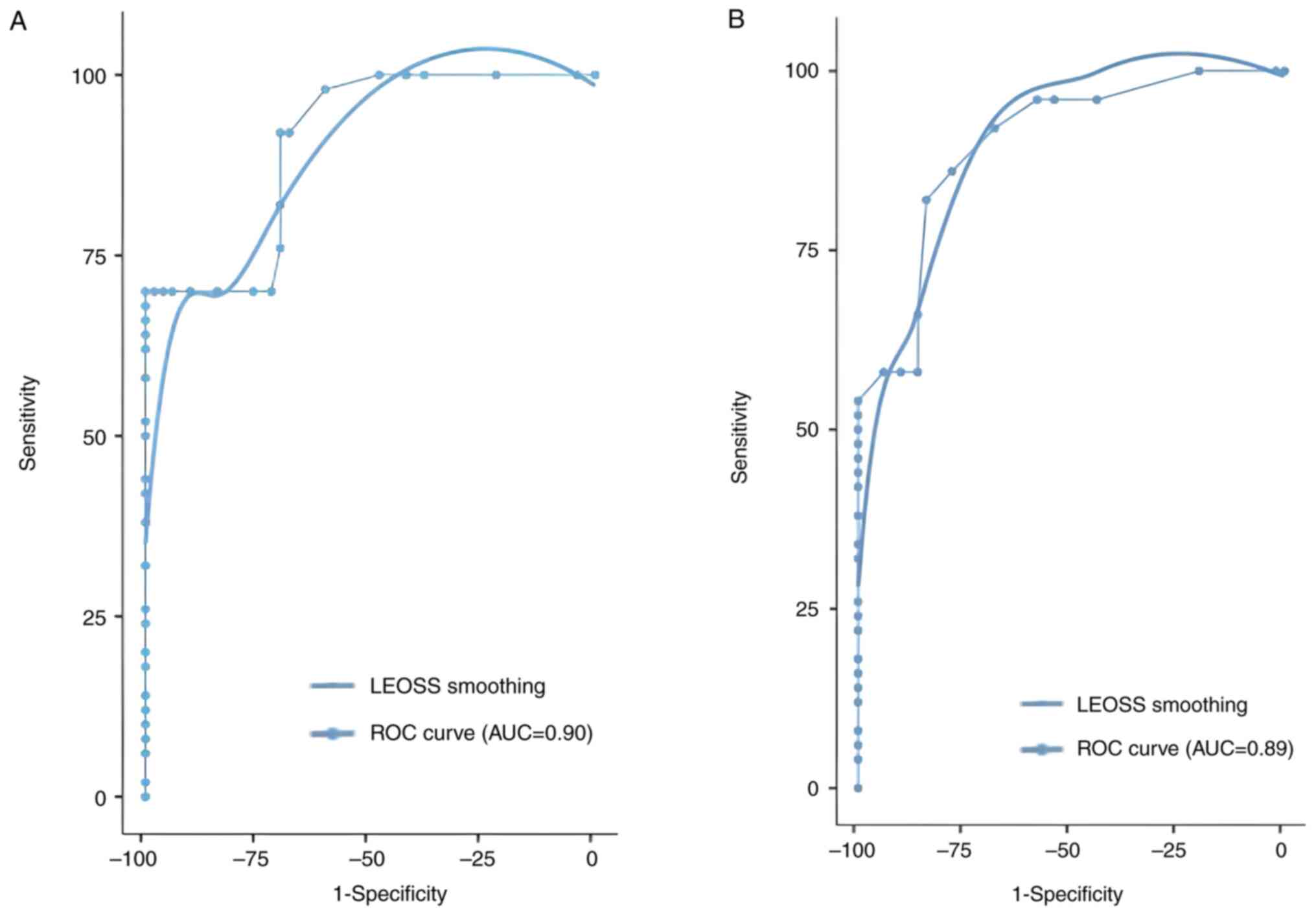
A receiver operating characteristic (ROC) curve was
used to evaluate the diagnostic value of miR-21 and miR-221 as PCa
biomarkers. Both miRNAs were revealed to have significant
diagnostic value for differentiating cancer from non-cancer
tissues. For miR-21, sensitivity and specificity were demonstrated
to be 70 and 96%, respectively, and an AUC of 0.90 (P<0.001).
miR-221 also presented an important diagnostic performance, with
sensitivity and specificity demonstrated to be 86 and 78%,
respectively, and an AUC of 0.89 (P<0.001). The cut-off value
was determined on the basis of Youden's index (maximum=sensitivity
+ specificity-1) to maximize both sensitivity and specificity for
each ROC analysis (Table II;
Fig. 2).
 | Table IIReceiver operating characteristic
curves tested for specificity and sensitivity and the AUC of miR-21
and miR-221. |
Table II
Receiver operating characteristic
curves tested for specificity and sensitivity and the AUC of miR-21
and miR-221.
| miRNA | AUC | Sensitivity | Specificity | P-value | Youden index |
|---|
| miR-21 | 0.90 | 70 | 96 | <0.001 | 0.66 |
| miR-221 | 0.89 | 86 | 78 | <0.001 | 0.64 |
Association with clinicopathological
parameters
Patients were categorized into two groups based on
high and low miRNA expression levels. The high expression levels of
miR-21 and miR-221 were further analysed. An assessment of the
association between these miRNA expression levels and
clinicopathological parameters revealed that although both miR-21
and miR-221 were significantly associated with higher pathological
Gleason scores (>6) and earlier tumour stages (T2X and T1)
(P<0.01), their expression levels did not demonstrate a
statistically significant association with PSA levels (P>0.05).
Notably, for patients with PSA levels of ≥10 ng/ml, the expression
levels of miR-21 and miR-221 were markedly higher compared with
those with lower PSA levels, although this association was not
statistically significant. Additionally, no significant
associations were observed between miRNA expression levels and
other clinical factors, including age at diagnosis, smoking status
or alcohol consumption (P>0.05; Table III).
 | Table IIIAssociation of miRNAs expression with
various clinicopathological parameters of patients with prostate
cancer. |
Table III
Association of miRNAs expression with
various clinicopathological parameters of patients with prostate
cancer.
| Clinicopathological
parameters | Number of
cases | High miR-21
expression (n=35) | Low miR-21
expression (n=15) | P-value | High miR-221
expression (n=30) | Low miR-221
expression (n=20) | P-value |
|---|
| Age at
diagnosis/surgery | | | | | | | |
|
<60
years | 13 | 8 | 5 | 0.44 | 6 | 7 | 0.24 |
|
≥60
years | 37 | 27 | 10 | | 24 | 13 | |
| Prostate-specific
antigen (ng/ml) | | | | | | | |
|
<2.5 | 2 | 1 | 1 | 0.63 | 0 | 2 | 0.13 |
|
2.5-10 | 12 | 8 | 4 | | 6 | 6 | |
|
≥10 | 36 | 26 | 10 | | 24 | 12 | |
| Pathological
Gleason score | | | | | | | |
|
≤6 | 14 | 6 | 8 | 0.009 | 4 | 10 | 0.009 |
|
>6 | 36 | 29 | 7 | | 26 | 10 | |
| Pathological
T-stage | | | | | | | |
|
T1 | 17 | 8 | 9 | 0.007 | 5 | 12 | 0.001 |
|
T2 X | 29 | 24 | 5 | | 22 | 7 | |
|
T3 X | 1 | 1 | 0 | | 1 | 0 | |
|
T4 | 3 | 2 | 1 | | 2 | 1 | |
| Alcohol
consumption | | | | | | | |
|
Yes | 28 | 20 | 8 | 0.54 | 17 | 11 | 0.79 |
|
No | 12 | 7 | 5 | | 8 | 4 | |
|
Weaned | 10 | 8 | 2 | | 5 | 5 | |
| Smoking | | | | | | | |
|
Yes | 32 | 24 | 8 | 0.43 | 10 | 22 | 0.16 |
|
No | 11 | 6 | 5 | | 7 | 4 | |
|
Weaned | 7 | 5 | 2 | | 3 | 4 | |
Discussion
miRNAs are small regulatory molecules involved in
post-transcriptional gene silencing, which serve an essential role
as modulators of key biological processes such as cell
proliferation, apoptosis or differentiation (15). However, this may depend on the
targets they regulate. In cancer, miRNAs can function as either
tumour suppressors or oncogenes (16). Currently, miR-21 and miR-221 have
emerged as two critical upregulated oncomiRs in PCa that promote
tumorigenesis through their high expressions in cancerous tissues
(17,18).
By using RT-qPCR, it was demonstrated that the
expression of miR-21 and miR-221 was upregulated in PCa tissues
compared with that in normal control tissues, which is consistent
with other published studies. Thus, these high levels of expression
suggest that miR-21 and miR-221 may serve as potential biomarkers
for both diagnosis and prognosis prediction of PCa. Previous
studies have highlighted the role of miR-21 as a potential
candidate biomarker for early detection of PCa. miR-21
downregulates a number of important tumour suppressors, such as
PTEN and PDCD4, leading to increased proliferation and
resistance to apoptosis (19).
Wang et al (20) and
Gunawan et al (21)
reported that miR-21 was differentially expressed in patients with
PCa and therefore they concluded that it could be useful as a
potential biomarker for PCa. Similarly, Agaoglu et al
(22) demonstrated that miR-21
levels in patients with PCa were significantly higher than in
controls (P<0.001). Other research on miRNA-221 indicated its
expression was increased in PCa PC3 cells and it may promote cell
growth by targeting p27 Kip1, which blocks the cell cycle (9). Similarly, Kachris et al
(23), Sun et al (24) and Song et al (25) reported that miR-221 was upregulated
in PCa. However, downregulation of miR-221 has also been reported
(26,27).
The findings from the present study demonstrated
that associations between the levels of miR-21 and miR-221 in PCa
tissues vary significantly between key clinicopathological
parameters, particularly Gleason score and tumour stage. It was
revealed that higher expression of both miR-21 and miR-221 was
significantly associated with increased Gleason scores and tumour
stages (P<0.05), suggesting that these miRNAs could be
significant in the progression and aggressiveness of PCa. These
results align with previous research linking miR-21 and miR-221
with PCa progression. For instance, Guan et al (28) reported an association between
increased miR-21 expression and higher Gleason scores. Moreover,
Ibrahim et al (29) and a
meta-analysis by Stafford et al (30) concluded that miR-21 expression was
associated with higher Gleason scores as well as advanced tumour
stages, supporting the notion that miR-21 is involved in promoting
tumour aggressiveness in PCa. Furthermore, miR-221 has been
reported to be upregulated in castration-resistant PCa, a more
advanced and treatment-resistant form of the disease, further
indicating its role in cancer progression (31). Certain studies have also identified
an association between high expression of miR-221 and higher
Gleason scores (29). However,
another study reported that the level of miR-221 was not associated
with a high Gleason score or other clinicopathological parameters
(32).
Although PSA continues to be an important biomarker
for PCa screening, the present study identified no association
between PSA levels and the expression of these miRNAs. This
suggests the potential for enhancing the precision of PCa
evaluation through the incorporation of miR-21 and miR-221 into
diagnostic or prognostic models. These findings are consistent with
the findings of Porzycki et al (33), which highlights the potential for
the use of miRNAs as independent markers for tumour progression.
The absence of associations between the expression of these miRNAs
and other indicators, such as age, smoking status and alcohol
consumption, suggests that miR-21 and miR-221 are likely
independent of these variables. This suggests that they may serve
as specific biomarkers for these indicators, offering insights into
PCa progression.
The diagnostic potential of miR-21 and miR-221
demonstrated in the present study is in agreement with that
reported in previous studies. The meta-analysis by Zhou and Zhu
(34) revealed that miR-21 had an
AUC of 0.95, with a pooled sensitivity and specificity of 91 and
88%, respectively. Likewise, Purnomo et al (35) reported a sensitivity and
specificity of 91 and 89%, respectively, with an AUC of 0.97. On
the other hand, Ibrahim et al (29) reported an AUC of 0.872 for miR-221,
with sensitivity and specificity values of 82 and 72%,
respectively, indicating its potential as a diagnostic biomarker.
In another study, increased expression of miR-221 was associated
with a sensitivity of 57% and a specificity of 100%, with an AUC of
0.74 in ROC analysis (36). These
results support the findings of the present study and highlight the
clinical importance of miR-21 and miR-221 in the diagnosis of
PCa.
Studies on miR-21 as a biomarker for diagnosis and
prognosis are widespread for other cancers such as breast, gastric
and endometrial cancer. In breast cancer, a meta-analysis
evaluating the diagnostic value of miR-21 as a serum biomarker
reported a sensitivity of 79%, specificity of 85% and an AUC of
0.89(37). In gastric cancer,
miR-21 demonstrated diagnostic potential with a sensitivity of
82.9%, a specificity of 85.7% and an AUC of 0.96(38). In endometrial cancer, miR-21
demonstrated a sensitivity of 84.51% and specificity of 86.79%,
with an AUC of 0.925(39).
miR-221 has also been demonstrated to be upregulated
in several cancers, including breast and thyroid cancer, targeting
tumour suppressor genes such as p27 Kip1 and oestrogen receptor-α
in breast cancer (40). In breast
cancer, ROC curve analysis revealed an AUC of 0.769, with a
sensitivity of 61.29% and a specificity of 85.19% (41). For thyroid cancer, a meta-analysis
of 16 studies reported a pooled sensitivity of 82%, specificity of
84% and an AUC of 0.88(42).
However, the reasons for the variable expression of these miRNAs in
different cancer cell types remain unclear.
The results of the present study highlight the
potential of miR-21 and miR-221 as diagnostic and prognostic
biomarkers for PCa. These miRNAs serve not only as predictive
markers for PCa but also as important indicators of cancer
aggressiveness. In particular, the present study demonstrated that
elevated expression of miR-21 and miR-221 was significantly
associated with early-stage PCa and higher Gleason scores, which
are indicative of more aggressive disease. This finding reinforces
their potential not only in the early detection of PCa but also in
assessing prognosis. By focusing on the relationship between miRNA
expression, tumour staging and clinical outcomes, the present study
highlights their value in predicting disease progression and
guiding treatment decisions.
In the present study, the control group was defined
as individuals with benign inflammatory disorders, reflecting the
common occurrence of low-level inflammation in the general
population, particularly with aging, a phenomenon known as
‘inflammaging’ (43). The
procurement of completely healthy, inflammation-free tissue
presents significant ethical challenges and practical difficulties,
and it could introduce potential selection bias (44,45).
By employing controls with benign inflammatory conditions, our
objective was to emulate real-world scenarios, as inflammation is
frequently observed in tumour microenvironments and contributes to
cancer progression (46).
Despite the promising results of the present study,
there are certain limitations. First, the sample size, of 50
patients with PCa and 50 controls is relatively small, which may
affect the degree to which the results apply to a larger
population. Additionally, the cross-sectional design of the study
restricts the ability to track miRNA expression changes over time
and establish a direct cause-and-effect relationship between miR-21
and mirR-221 and cancer progression across different disease
stages. Lastly, RT-qPCR was used but more advanced techniques, such
as western blotting and flow cytometry, could improve the precision
and reliability of biomarker investigations. While these findings
are encouraging, further research is needed to validate the
potential of miR-21 and miR-221 as biomarkers, and larger and more
diverse cohorts are needed to confirm the generalizability of the
results. Future research should also utilise non-invasive samples,
such as blood or urine, to assess the use of less invasive
diagnostic approaches and pave the way for personalized and
effective PCa management. Furthermore, integrating miR-21 and
miR-221 with other molecular markers into a multi-biomarker panel
could enhance diagnostic accuracy and refine risk stratification,
ultimately improving outcomes for patients with PCa.
In conclusion, the significant overexpression of
miR-21 and miR-221 in PCa tissues compared with control tissues,
coupled with their association with critical clinicopathological
parameters such as Gleason score, tumour stage and patient
prognosis, emphasizes their pivotal role in both the initiation and
progression of PCa. These miRNAs are not only integral to key
cellular processes such as cell cycle regulation, apoptosis and
metastasis but also act as potential modulators of the tumour
microenvironment, further highlighting their relevance in cancer
biology. Furthermore, given their stable presence in body fluids
and tissues, miR-21 and miR-221 hold potential as non-invasive
biomarkers for the early detection, prognosis and monitoring of
disease progression. Their ability to serve as molecular signatures
for aggressive forms of PCa provides a unique opportunity to
improve the sensitivity and specificity of current diagnostic
tools, surpassing the limitations of traditional methods such as
PSA testing. Furthermore, the identification of these miRNAs could
enhance risk stratification, offering a more personalized approach
to treatment planning. However, as the field of miRNA research
continues to evolve, it is crucial to delve deeper into the
molecular mechanisms through which miR-21 and miR-221 exert their
oncogenic effects, as well as their potential interactions with
other biomarkers and therapeutic targets. Understanding these
interactions could lead to the development of miRNA-based
therapies, potentially inhibiting tumour progression or sensitizing
tumours to existing treatments. Ultimately, the findings of the
present study could contribute to a paradigm shift in PCa
management, fostering the development of more accurate, effective
and personalized diagnostic and therapeutic strategies that improve
patient outcomes and reduce mortality rates globally.
Supplementary Material
Sequences of miRNAs used in the
present study.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
IM conceptualized the study, developed methodology,
wrote the manuscript and performed formal analysis. KAT performed
formal analysis, reviewed and edited the manuscript. MA conducted
investigation and conceptualized the study. AL, AA, KE and AEG
contributed to sample collection and patient data acquisition. MME
supervised the study, performed project administration, developed
methodology and validated analysis and interpretation of data. All
authors read and approved the final version of the manuscript. MA
and KAT confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Informed consent was obtained from all participants.
The present study was approved by the Moroccan Biomedical Research
Ethics Committee (approval no. 3/2018; approval date: April 30,
2018; Rabat; Morocco).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang L, Lu B, He M, Wang Y, Wang Z and Du
L: Prostate cancer incidence and mortality: Global status and
temporal trends in 89 countries from 2000 to 2019. Front Public
Heal. 10(811044)2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bratt O, Auvinen A, Arnsrud Godtman R,
Hellström M, Hugosson J, Lilja H, Wallström J and Roobol MJ:
Screening for prostate cancer: Evidence, ongoing trials, policies
and knowledge gaps. BMJ Oncol. 2(e000039)2023.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chakrabortty A, Patton DJ, Smith BF and
Agarwal P: miRNAs: Potential as biomarkers and therapeutic targets
for cancer. Genes (Basel). 14(1375)2023.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Calin GA, Liu CG, Sevignani C, Ferracin M,
Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al:
MicroRNA profiling reveals distinct signatures in B cell chronic
lymphocytic leukemias. Proc Natl Acad Sci USA. 101:11755–11760.
2004.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Porkka KP, Pfeiffer MJ, Waltering KK,
Vessella RL, Tammela TL and Visakorpi T: MicroRNA expression
profiling in prostate cancer. Cancer Res. 67:6130–6135.
2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Rhim J, Baek W, Seo Y and Kim JH: From
molecular mechanisms to therapeutics: Understanding MicroRNA-21 in
cancer. Cells. 11(2791)2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Liu LZ, Li C, Chen Q, Jing Y, Carpenter R,
Jiang Y, Kung HF, Lai L and Jiang BH: MiR-21 induced angiogenesis
through AKT and ERK activation and HIF-1α expression. PLoS One.
6(e19139)2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Vanacore D, Boccellino M, Rossetti S,
Cavaliere C, D'Aniello C, Di Franco R, Romano FJ, Montanari M, La
Mantia E, Piscitelli R, et al: Micrornas in prostate cancer: An
overview. Oncotarget. 8:50240–50251. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Galardi S, Mercatelli N, Giorda E,
Massalini S, Frajese GV, Ciafrè SA and Farace MG: miR-221 and
miR-222 expression affects the proliferation potential of human
prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem.
282:23716–23724. 2007.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Amir S, Ma AH, Shi XB, Xue L, Kung HJ and
Devere White RW: Oncomir miR-125b suppresses p14(ARF) to modulate
p53-dependent and p53-independent apoptosis in prostate cancer.
PLoS One. 8(e61064)2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kalfert D, Ludvikova M, Pesta M, Ludvik J,
Dostalova L and Kholová I: Multifunctional roles of miR-34a in
cancer: A review with the emphasis on head and neck squamous cell
carcinoma and thyroid cancer with clinical implications.
Diagnostics (Basel). 10(563)2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yamamura S, Saini S, Majid S, Hirata H,
Ueno K, Deng G and Dahiya R: MicroRNA-34a modulates c-Myc
transcriptional complexes to suppress malignancy in human prostate
cancer cells. PLoS One. 7(e29722)2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cavallari I, Ciccarese F, Sharova E, Urso
L, Raimondi V, Silic-Benussi M, D'Agostino DM and Ciminale V: The
miR-200 family of microRNAs: Fine tuners of epithelial-mesenchymal
transition and circulating cancer biomarkers. Cancers (Basel).
13(5874)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ahmad J, Hasnain SE, Siddiqui MA, Ahamed
M, Musarrat J and Al-Khedhairy AA: MicroRNA in carcinogenesis &
cancer diagnostics: A new paradigm. Indian J Med Res. 137:680–694.
2013.PubMed/NCBI
|
|
16
|
Menon A, Abd-Aziz N, Khalid K, Poh CL and
Naidu R: miRNA: A promising therapeutic target in cancer. Int J Mol
Sci. 23(11502)2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Folini M, Gandellini P, Longoni N, Profumo
V, Callari M, Pennati M, Colecchia M, Supino R, Veneroni S,
Salvioni R, et al: miR-21: An oncomir on strike in prostate cancer.
Mol Cancer. 9(12)2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Kneitz B, Krebs M, Kalogirou C, Schubert
M, Joniau S, van Poppel H, Lerut E, Kneitz S, Scholz CJ, Ströbel P,
et al: Survival in patients with high-risk prostate cancer is
predicted by miR-221, which regulates proliferation, apoptosis, and
invasion of prostate cancer cells by inhibiting IRF2 and SOCS3.
Cancer Res. 74:2591–2603. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Huang W, Kang XL, Cen S, Wang Y and Chen
X: High-level expression of microRNA-21 in peripheral blood
mononuclear cells is a diagnostic and prognostic marker in prostate
cancer. Genet Test Mol Biomarkers. 19:469–475. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang W, Li J, Zhu W, Gao C, Jiang R, Li W,
Hu Q and Zhang B: MicroRNA-21 and the clinical outcomes of various
carcinomas: A systematic review and meta-analysis. BMC Cancer.
14(819)2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gunawan RR, Astuti I and Danarto HR:
miRNA-21 as high potential prostate cancer biomarker in prostate
cancer patients in Indonesia. Asian Pac J Cancer Prev.
24:1095–1099. 2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yaman Agaoglu F, Kovancilar M, Dizdar Y,
Darendeliler E, Holdenrieder S, Dalay N and Gezer U: Investigation
of miR-21, miR-141, and miR-221 in blood circulation of patients
with prostate cancer. Tumor Biol. 32:583–588. 2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kachris S, Papadaki C, Rounis K, Tsitoura
E, Kokkinaki C, Nikolaou C, Sourvinos G and Mavroudis D:
Circulating miRNAs as potential biomarkers in prostate cancer
patients undergoing radiotherapy. Cancer Manag Res. 13:8257–8271.
2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sun T, Yang M, Chen S, Balk S, Pomerantz
M, Hsieh CL, Brown M, Lee GM and Kantoff PW: The altered expression
of MiR-221/-222 and MiR-23b/-27b is associated with the development
of human castration resistant prostate cancer. Prostate.
72:1093–1103. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Song C, Chen H, Wang T, Zhang W, Ru G and
Lang J: Expression profile analysis of microRNAs in prostate cancer
by next-generation sequencing. Prostate. 75:500–516.
2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Coarfa C, Fiskus W, Eedunuri VK,
Rajapakshe K, Foley C, Chew SA, Shah SS, Geng C, Shou J, Mohamed
JS, et al: Comprehensive proteomic profiling identifies the
androgen receptor axis and other signaling pathways as targets of
microRNAs suppressed in metastatic prostate cancer. Oncogene.
35:2345–2356. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Spahn M, Kneitz S, Scholz CJ, Stenger N,
Rüdiger T, Ströbel P, Riedmiller H and Kneitz B: Expression of
microRNA-221 is progressively reduced in aggressive prostate cancer
and metastasis and predicts clinical recurrence. Int J Cancer.
127:394–403. 2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Guan C, Zhang L, Wang S, Long L, Zhou H,
Qian S, Ma M, Bai F, Meng QH and Lyu J: Upregulation of MicroRNA-21
promotes tumorigenesis of prostate cancer cells by targeting KLF5.
Cancer Biol Ther. 20:1149–1161. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ibrahim NH, Abdellateif MS, Kassem SH, Abd
El Salam MA and El Gammal MM: Diagnostic significance of miR-21,
miR-141, miR-18a and miR-221 as novel biomarkers in prostate cancer
among Egyptian patients. Andrologia. 51(e13384)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Stafford MYC, Willoughby CE, Walsh CP and
McKenna DJ: Prognostic value of miR-21 for prostate cancer: A
systematic review and meta-analysis. Biosci Rep.
42(BSR20211972)2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Sun T, Wang Q, Balk S, Brown M, Lee GS and
Kantoff P: The role of microRNA-221 and microRNA-222 in
androgen-independent prostate cancer cell lines. Cancer Res.
69:3356–3363. 2009.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kang SG, Ha YR, Kim SJ, Kang SH, Park HS,
Lee JG, Cheon J and Kim CH: Do microRNA 96, 145 and 221 expressions
really aid in the prognosis of prostate carcinoma? Asian J Androl.
14:752–757. 2012.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Porzycki P, Ciszkowicz E, Semik M and
Tyrka M: Combination of three miRNA (miR-141, miR-21, and miR-375)
as potential diagnostic tool for prostate cancer recognition. Int
Urol Nephrol. 50:1619–1626. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhou H and Zhu X: MicroRNA-21 and
microRNA-30c as diagnostic biomarkers for prostate cancer: A
meta-analysis. Cancer Manag Res. 11:2039–2050. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Seputra KP, Purnomo BB, Susianti H, Kalim
H and Purnomo AF: miRNA-21 as reliable serum diagnostic biomarker
candidate for metastatic progressive prostate cancer: Meta-analysis
approach. Med Arch. 75:347–350. 2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Hekımler Öztürk K, Mutlu İçduygu F, Özgöz
A and Özorak A: miR-221, miR-650 and miR-4534 as diagnostic markers
in prostate cancer and their relationship with lymphatic invasion.
Turk J Biochem. 47:435–443. 2022.
|
|
37
|
Li S, Yang X, Yang J, Zhen J and Zhang D:
Serum microRNA-21 as a potential diagnostic biomarker for breast
cancer: A systematic review and meta-analysis. Clin Exp Med.
16:29–35. 2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Rihane FE, Erguibi D, Abumsimir B,
Charoute H, Chehab F and Ennaji MM: Expression of microRNAs in
gastric cancerous tissues and their association with Helicobacter
pylori and Epstein-Barr virus infections. World Acad Sci J.
4(1)2022.
|
|
39
|
Lamsisi M, Li G, Benhessou M, El Mzibri M,
Bouziyane A, Wakrime L, Laraqui A, Ennaji Y, Abumsimir B, El
Karroumi M, et al: Long noncoding RNA HOTAIR as a biomarker for the
detection of cervical cancer and cervical intraepithelial
neoplasia. Indian J Gynecol Oncol. 19(69)2021.
|
|
40
|
Shah MY and Calin GA: MicroRNAs miR-221
and miR-222: A new level of regulation in aggressive breast cancer.
Genome Med. 3(56)2011.PubMed/NCBI View
Article : Google Scholar
|
|
41
|
Li F: Expression of miR-221 and miR-489 in
breast cancer patients and their relationship with prognosis. Oncol
Lett. 19:1523–1529. 2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Liang L, Zheng X, Hu M, Cui Y, Zhong Q,
Wang S and Huang F: MiRNA-221/222 in thyroid cancer: A
meta-analysis. Clin Chim Acta. 484:284–292. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Franceschi C and Campisi J: Chronic
inflammation (inflammaging) and its potential contribution to
age-associated diseases. J Gerontol A Biol Sci Med Sci. 69 (Suppl
1):S4–S9. 2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Grivennikov SI, Greten FR and Karin M:
Immunity, inflammation, and cancer. Cell. 140:883–899.
2010.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Beskow LM and Dean E: Informed consent for
biorepositories: Assessing prospective participants' understanding
and opinions. Cancer Epidemiol Biomarkers Prev. 17:1440–1451.
2008.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Mantovani A, Allavena P, Sica A and
Balkwill F: Cancer-related inflammation. Nature. 454:436–444.
2008.PubMed/NCBI View Article : Google Scholar
|