Introduction
Among the female malignancies, cervical cancer
(1) is the fourth type of cancer and
the second leading cause of cancer-related mortality among the
female population, with high incidence and mortality rates
worldwide, with 604,127 (3.1%) new cases and 341,831 (4.1%) related
deaths (1). In addition, CC is the
leading cause of cancer-related death among females in 36 low- and
middle-income countries (2). This is
mainly due to late detection and poor prognoses, which reduces the
chances of curative surgery.
The different types of cervical cancer are
classified according to the tumor site and gene expression. A
number of critical genes are linked to a higher incidence of the
disease. Therefore, numerous genetic and epigenetic alterations
that inactivate tumor suppressor genes and activate oncogenes play
a major role in the pathogenesis of CC. Consequently, there is a
need to explore novel and effective diagnostic biomarkers to better
distinguish patients with CC in order to enable rapid diagnosis and
thus the effective treatment of CC for improved and earlier cancer
management.
Among the most potential biomarkers is proline-rich
protein 11 (PRR11), a recently discovered gene in the amplification
region of chromosome 17q22. Bioinformatics analysis has revealed
that PRR11 comprises a bivalent nuclear localization signal, a pair
of proline-rich regions and zinc finger domains, which are involved
in the transduction of cell signals and mediate a cascade of
cancer-related processes (3).
Pertinent data have shown that PRR11 is a candidate oncogene in
mammals, often playing a vital role in the initiation and
progression, as well as other carcinogenic processes of various
solid tumors, such as hilar cholangiocarcinoma (4), lung cance (5,6),
pancreatic cancer (7), osteosarcoma
(8), gastric cancer (9), breast cancer (10,11),
colorectal cancer (12), ovarian
cancer (13) and cervical cancer
(14). In addition, PRR11 has been
reported to commonly display extremely high expression levels in
solid tumors and it is strongly related to local tumor recurrence
and metastasis. However, although there is growing evidence that
PRR11 is an influential tumor-related gene, the link between PRR11
and Moroccan women with CC remains questionable.
Patients and methods
Patients and specimens
A total of 100 fresh biopsies (80 tumor tissue
samples and 20 corresponding adjacent normal tissues) were
collected from patients with CC undergoing surgery following a
histopathological examination at the Onco-Gynecology Department of
the Mohammed IV Oncology Center in Casablanca, Morocco, between
January, 2020 and December, 2021. Biopsies were sampled by
physicians following standard protocols and immediately stored at
-80˚C until analysis. Clinicopathologic data from enrolled cases
were also collected according to the STROCSS guidelines (15). Patients who received chemotherapy
and/or radiotherapy were excluded from the study.
The present study was ethically approved by the
Biomedical Research Committee of the Faculty of Medicine and
Pharmacy of Casablanca, Casablanca, Morocco (3/2018 on April 30,
2018). Free oral consent was obtained from all recruited patients
and the confidentiality of their personal information was kept
according to ethical rules.
Total RNA extraction and cDNA
synthesis
Total RNA was extracted from the tissue samples
using TRIzol reagent® (Invitrogen; Thermo Fisher Scientific, Inc.)
as instructed by the manufacturer. The NanoDrop 2000
spectrophotometer (Nanodrop Technologies, Inc.) was utilized to
detect the concentration and purity of mRNA (considering the
concentration of 1-2 µg). The High-capacity cDNA Synthesis Kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for
reverse transcription according to the manual provided by the
manufacturer.
Quantitative qPCR (qPCR)
To assess the relative expression of the PRR11 gene,
TaqMan® Universal PCR Master Mix (2X) (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used, as well as
the TaqMan® Gene Expression Assay (Applied Biosystems;
Thermo Fisher Scientific, Inc.) which consists of a pair of
unlabeled PCR primers and a specific TaqMan probe.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an
internal endogenous control for normalizing gene expression. The
primers used are listed in Table I.
The thermocycling conditions involved an initial denaturation at
95˚C for 2 min, followed by 40 cycles of denaturation at 95˚C for
30 sec, annealing at 57˚C for 30 sec, and extension at 72˚C for 30
sec. The relative expression level of PRR11 was calculated using
the 2-ΔΔCq value, based on the threshold cycle (1) method (16).
 | Table IPrimer sequences of PRR11 and GAPDH
targets. |
Table I
Primer sequences of PRR11 and GAPDH
targets.
| Gene name | Gene symbol | Primer (5-3) |
|---|
| Proline-rich protein
11 | PRR11 | F:
GACTTCCAAAGCTGTGCTTCC |
| | | R:
CTGCATGGGTCCATCCTTTTT |
| Glyceraldehyde
3-phosphate dehydrogenase | GAPDH | F:
GGAGCGAGATCCCTCCAAAAT |
| | | R:
GGCTGTTGTCATACTTCTCATGG |
Statistical analysis
One-way ANOVA was conducted to assess the relative
differential expression of PRR11 in CC tissues compared with
adjacent normal tissues. The Chi-squared test was used examine the
association between PRR11 expression levels and the
clinicopathological features of the patients. Additionally, a ROC
analysis was performed to evaluate the overall diagnostic
performance of PPR11 as a biomarker in patients with CC. The
Kaplan-Meier survival curve method was used analyze the overall
survival probabilities of patients with CC according to the PRR11
expression level, with the Tarone-Ware test applied to analyze
survival differences. Jamovi software, version 21.3.2 was used to
perform all analyses, and a P-value <0.05 was considered to
indicate a statistically significant difference; the confidence
interval (CI) was 95%.
Results
Clinicopathological features of
patients with CC
The clinicopathologic characteristics of the
patients with CC included in the present study are summarized in
Table II. The age at diagnosis of
the patients with CC ranged from 27 to 85 years, with a mean age of
54 years (±10.99). The most common age group was ≥41 years,
accounting for 81.25% of the cases. Clinical staging was performed
according to the International Federation of Gynecology and
Obstetrics (FIGO) classification and this revealed the predominance
of stages I and II (91.25%). The predominant form of CC was
squamous cell carcinoma, accounting for 82.5% of cases, while only
17.5% of the patients had adenocarcinoma.
 | Table IIClinicopathological features of the
patients with cervical cancer (n=80). |
Table II
Clinicopathological features of the
patients with cervical cancer (n=80).
| Features | No. of patients | Percentage |
|---|
| Age at diagnosis
(years); mean age, 54 years | | |
|
<41 | 15 | 18.75 |
|
≥41 | 65 | 81.25 |
| Histopathological
grade | | |
|
I-II | 73 | 91.25 |
|
III-IV | 7 | 8.75 |
| Histological
type | | |
|
Squamous
cell carcinoma | 66 | 82.5 |
|
Adenocarcinoma | 14 | 17.5 |
PRR11 mRNA expression in patients with
CC
RT-qPCR was performed to assess the mRNA expression
level of PRR11 in CC tumor tissues compared to corresponding
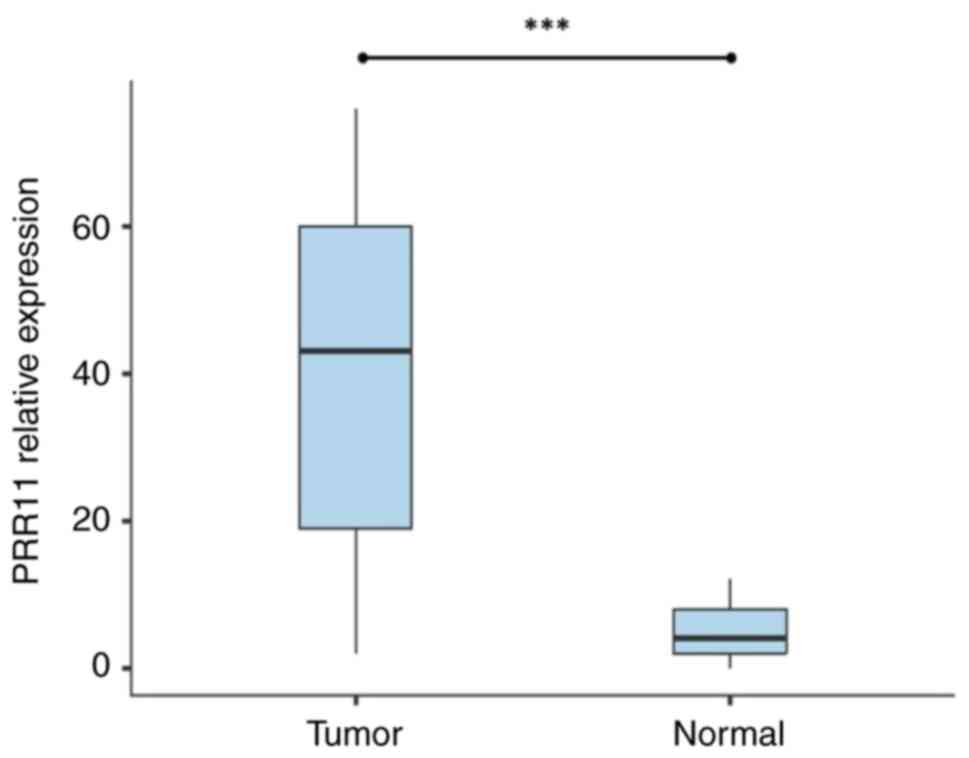
adjacent normal tissues. The results revealed that PRR11 mRNA was
expressed in 77.5% (62/80 cases) of the tumor tissues. It exhibited
a relevant level of expression compared with the adjacent normal
tissues; PRR11 mRNA expression was only found in 20% (4/20) of
normal tissues. The relative expression of PRR11 was significantly
higher in CC tissues than in adjacent normal tissues (P<0.001;
Fig. 1).
Association between PRR11 expression
and clinicopathological characteristics of patients with CC
The results of the analysis of selected
clinicopathological characteristics of patients with CC is
presented in Table III and
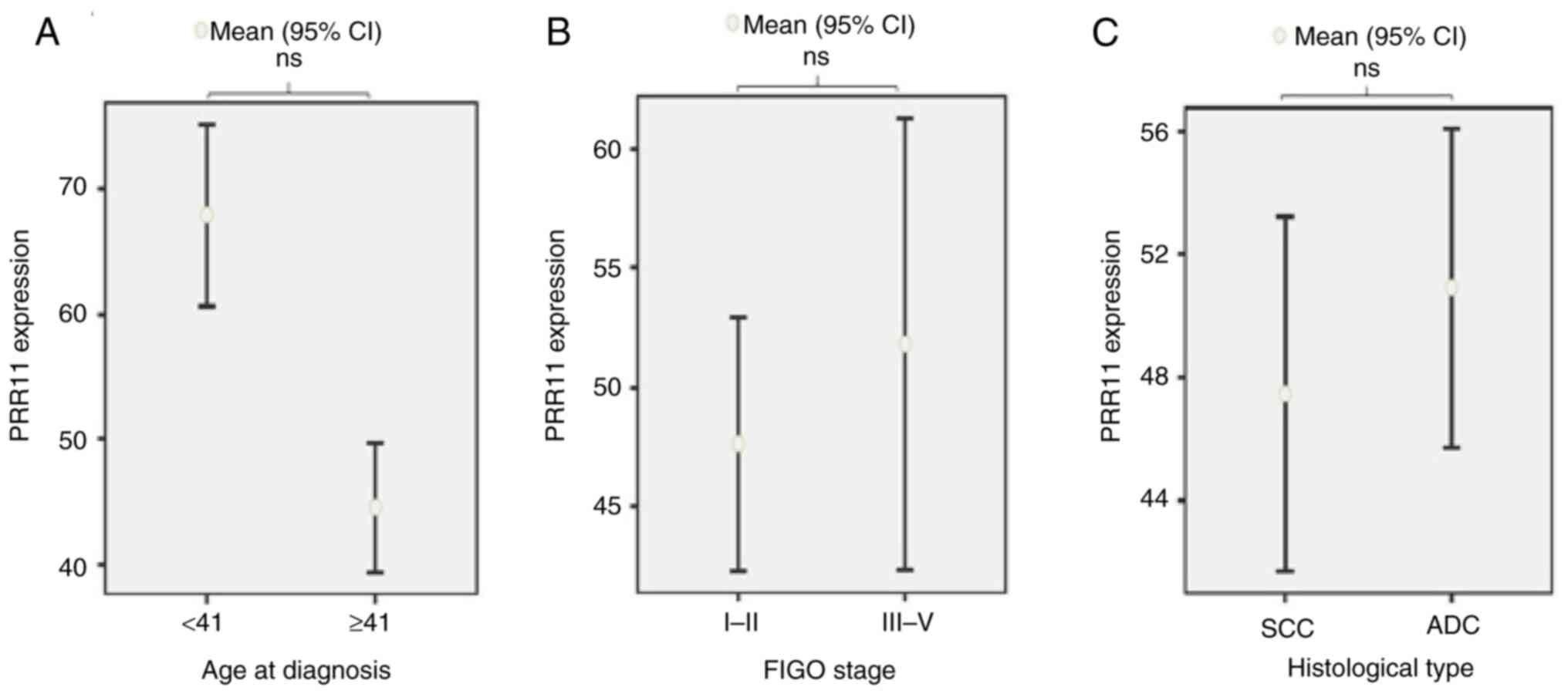
Fig. 2. No significant associations
were observed between PRR11 expression in tumor tissues and
clinicopathological characteristics, including age at diagnosis
(P=0.308), FIGO stage (P=0.999), or histological type
(P=0.506).
 | Table IIIAssociation between PRR11 expression
and some clinicopathological features of patients with cervical
cancer. |
Table III
Association between PRR11 expression
and some clinicopathological features of patients with cervical
cancer.
| | PRR11 positive
expression | PRR11 negative
expression | |
|---|
| Features | Effective | No. of patients | Percentage | No. of patients | Percentage | P-value |
|---|
| Age at diagnosis
(years) | | | | | | |
|
<41 | 15 | 10 | 16.1 | 5 | 27.8 | 0.308 |
|
≥41 | 65 | 52 | 83.9 | 13 | 72.2 | |
| FIGO stage | | | | | | |
|
I-II | 73 | 56 | 90.3 | 17 | 94.4 | 0.999 |
|
III-V | 7 | 6 | 9.7 | 1 | 5.6 | |
| Histological
type | | | | | | |
|
Squamous
cell carcinoma | 66 | 52 | 83.9 | 14 | 77.8 | 0.506 |
|
Adenocarcinoma | 14 | 10 | 16.1 | 4 | 22.2 | |
PRR11 as a potential diagnostic
biomarker for CC
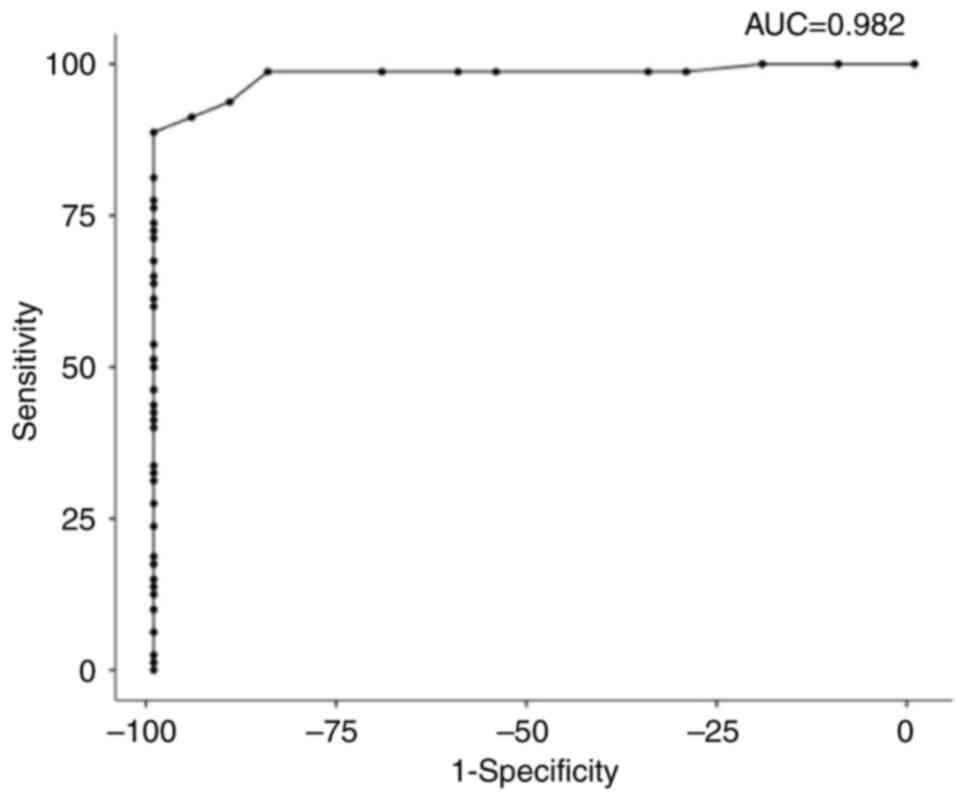
ROC analysis was performed to examine PRR11 as a
potential diagnostic biomarker for CC. The Roc analysis of CC vs.
normal tissues revealed that PRR11 was a good potential diagnostic
biomarker for discrimination between CC and non-tumor tissues. The
sensitivity and specificity were 88.75 and 100%, respectively. The
cut-off value was 13, the area under the ROC curve (AUC) was 0.982,
and the P-value was <0.001. The positive predictive value (PPV)
was 100%, which is the probability that the disease is present when
the test is positive. The negative predictive value (NPV) was
68.97%, the probability that the disease is not present when the
test is negative. The Youden index was 88.7%, which is >50%,
thus indicating that the test satisfies the empirical criteria to
be applied in the diagnosis of CC (Fig.
3 and Table IV).
 | Table IVROC curve analysis for PRR11 in
patients with cervical cancer. |
Table IV
ROC curve analysis for PRR11 in
patients with cervical cancer.
| ROC curve data | Values |
|---|
| Cut-off value | 13 |
| Sensitivity
(%) | 88.75% |
| Specificity
(%) | 100% |
| Positive predictive
value (PPV) (%) | 100% |
| Negative predictive
value (NPV) (%) | 68.97% |
| Youden's index | 0.887 |
| Area under curve
(AUC) | 0.982 |
| P-value | <0.001 |
| No. of tumor tissue
specimens | 80 |
| No. of control
tissue specimens | 20 |
PRR11 overexpression related to the
prognosis of CC
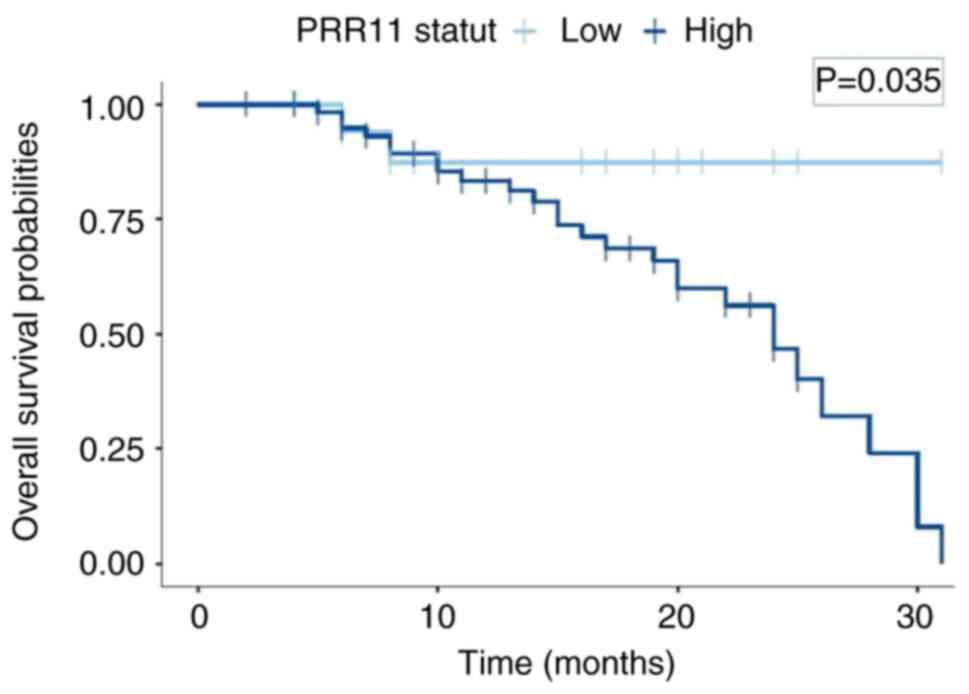
To investigate the prognosis of PRR11 expression in
CC, the Kaplan-Meier method was used to analyze the overall
survival rate of 80 patients with CC followed-up for 36 months. A
total of 30 patients succumbed and 50 patients survived. The median
survival time of the 80 patients with CC was 27 months. Among the
62 patients with CC who were PRR11-positive, 27 succumbed and 35
survived. The median survival time for this event was 24 months. Of
note, 2 patients out of 18 PRR11-negative patients with CC
succumbed and 16 survived. The median survival time for
PRR11-negative patients was 31 months. The survival time of
patients with CC with a positive expression for PRR11 was
significantly lower than that of patients with a negative
expression of PRR11 (P=0.035; Fig.
4), suggesting that PRR11 may be a predictor of a poor
prognosis in patients with CC.
Discussion
CC is the fourth most common type of cancer
affecting the female population. It is a well-known cause of
mortality and is associated with a considerable socioeconomic
burden worldwide (1). Multiple risk
factors, ranging from genetic alterations to hormonal factors,
environmental factors and viral etiology, are linked to the complex
carcinogenesis of CC (1). The
development of the majority of cases of CC is due to genetic
mutations, common to most cancers, which result in either the
overexpression of oncogenes or the inhibition of tumor suppressor
genes (17). Therefore, to improve
early detection and prevention of CC, there is a need to identify
new and more reliable tumor biomarkers.
Over the past decade, researchers have reported that
PRR11, a gene located in the 17q22 region of the chromosome, is a
prominent candidate oncogene in mammals. PRR11 has been shown to be
associated to several types of cancer, including ovarian cancer
(13), gastric cancer (9), breast cancer (10,11),
hilar cholangiocarcinoma (4),
pancreatic cancer (7) and CC
(14).
This has been well documented by the identification
of PRR11 overexpression in these types of cancer and its obvious
involvement in carcinogenesis and several other malignant
biological processes of the cell cycle, such as cell proliferation,
differentiation, migration, invasion, apoptosis, autophagy and cell
resistance to chemotherapy (5,9,13,18-20).
The present study revealed that PRR11 mRNA was
overexpressed in 77.5% of CC tissues vs. 20% of adjacent normal
tissues, exhibiting a significantly higher level of expression in
CC tissues compared with normal tissues (P<0.05). These findings
are in concordance with those in the study by Xu and Chang
(21), who first described that
PRR11 was overexpressed in 76.67% of CC tissues compared with
adjacent non-tumor tissues. Other studies have reported a
significantly high expression of PRR11 in a number of types of
cancer, such as lung cancer (5,6), ovarian
cancer (13), carcinoma of the
tongue (18), gastric cancer
(9), breast cancer (10,11),
hilar cholangiocarcinoma (4),
colorectal cancer (12), pancreatic
cancer (7) and osteosarcoma
(8).
Based on the literature, PRR11 overexpression
affects the cell cycle and promotes lung cancer progression
(15), and the onset and development
of CC (14,21). In addition, previous studies have
indicated that PRR11 overexpression promotes ovarian cancer cell
proliferation, migration and invasion by activating the
PI3K/AKT/β-Catenin pathway (13) and
promoting breast cancer cell progression and invasion by activating
the epithelial-mesenchymal transition (EMT) process (10). Moreover, the functional study by Zhou
et al (10) revealed that
PRR11 decreased the expression of E-cadherin and cytokeratin-18,
and increased the expression of vimentin, N-cadherin and
fibronectin through EMT by targeting transcription factors [Slug,
Snail, zinc finger E-box binding homeobox (ZEB)1 and ZEB2]
(10,20).
However, it has been shown that the inactivation of
PRR11 in CC cell lines increases the occurrence of apoptosis
(21). PRR11 also has the potential
to regulate apoptosis in CC cancer cells by stimulating the
expression of caspase-3 proteins, the executive and irreversible
factors in apoptosis (21),
suggesting that PRR11 protein expression plays a critical oncogenic
role in CC cell development and progression.
In the present study, PRR11 mRNA expression was not
found to be significantly associated with any clinicopathological
features of the patients, whereas opposite results were observed in
the study by Zhao et al (14), which reported that PRR11 was
associated with FIGO stage (P<0.05) in CC. In addition, Zhu
et al (13) found that the
overexpression of PRR11 in ovarian cancer tissues and cells was
significantly associated with an advanced FIGO stage. Moreover, Xu
and Chang (21) found that the
expression level of PRR11 was significantly associated with the
histological type of CC (P<0.05). By contrast, the difference
was not statistically significant between CC and the patient age at
diagnosis (P>0.05) (21), which
is consistent with the results of the present study.
In addition, the results of the ROC analysis
revealed that PRR11 represents a valuable biomarker for the early
diagnosis of CC. This is consistent with previous studies reported
in the literature, including research on lung cancer (5), pancreatic cancer (7), invasive breast cancer (11) and ovarian cancer (13).
On the other hand, the present study demonstrated
that PRR11 has prognostic values and plays a role as a carcinogenic
factor in CC. These findings are consistent with those of previous
studies on PRR11, but in other types of cancer, such as breast
cancer (10,11), hepatocellular carcinoma (22) and gastric cancer (23). Indeed, Wang et al (24) reported in their study that a high
expression of PRR11 was a prognostic risk factor for patients with
tongue squamous cell carcinoma, and this was also reported by Xu
and Chang (21) in their study on
CC.
Furthermore, there is evidence to indicate that
PRR11 is a potential target for anticancer therapies in hilar
cholangiocarcinoma and lung cancer (4,6). In this
regard, it has been suggested that the regulation of PRR11
expression in CC cells can inhibit cell proliferation and promotes
apoptosis through cyclin-D1 and caspase-3 proteins, rendering PRR11
a potential molecular target for CC treatment.
In conclusion, the present study confirmed PRR11
overexpression in CC tissues. Notably, PRR11 is emerging as a
leading diagnostic biomarker candidate, and a factor for a poor
prognosis. However, in order to promote the early identification
and prevention of CC, the detailed underlying mechanisms of PRR11
in the cell cycle and carcinogenesis need to be further
explored.
Acknowledgements
The authors would like to thank the University
Hassan II of Casablanca, Ibn Rochd University Hospital of
Casablanca for their contribution to the study by providing
research facilities, laboratory space, access to technical
equipment and expertise.
Funding
Funding: The authors acknowledge grant support from research
team of Laboratory of virology, oncology, biosciences, environment
and new energies of Mohammedia.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KAT was involved in the conceptualization of the
study, as well as in the study methodology, formal analysis and
investigation, project administration, and in the writing,
reviewing and editing of the original draft of the manuscript. SAS
was involved in the reviewing and editing of the manuscript and in
the formal analysis, including data verification, statistical
testing and interpretation of the results. IT was involved in
obtaining the clinicopathological data of the patients. MA was
involved in the formal analysis, including data verification,
statistical testing and interpretation of the results. AS
participated in the methodology of the study, as well as in the
reviewing and editing of the manuscript. MB provided biological
resources (cervical cancer biopsies and healthy cervical tissue
biopsies), and also participated in the development and validation
of the study methodology. MME was involved in project
administration, in data validation, in the writing, reviewing and
editing of the manuscript, and in project supervision. KAT and MME
confirm the authenticity of the raw data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was ethically approved by the
Biomedical Research Committee of the Faculty of Medicine and
Pharmacy of Casablanca, Casablanca, Morocco (3/2018 on April 30,
2018). Free oral consent was obtained from all recruited patients
and the confidentiality of their personal information was kept
according to ethical rules.
Patient consent for publication
Not applicable.
Competition interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bruni L, Serrano B, Roura E, Alemany L,
Cowan M, Herrero R, Poljak M, Murillo R, Broutet N, Riley LM and de
Sanjose S: Cervical cancer screening programmes and age-specific
coverage estimates for 202 countries and territories worldwide: A
review and synthetic analysis. Lancet Glob Health. 10:e1115–e1127.
2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ai Q, Bu YQ, Liu Z, Lan H, Ji Y, Du G,
Yang ZM, Liu GL and Song FZ: Structural and functional analysis of
human PRR11 promoter. Chin J Biochem Mol Biol. 27:356–363. 2011.(In
Chinese).
|
|
4
|
Chen Y, Cha Z, Fang W, Qian B, Yu W, Li W,
Yu G and Gao Y: The prognostic potential and oncogenic effects of
PRR11 expression in hilar cholangiocarcinoma. Oncotarget.
6:20419–20433. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ji Y, Xie M, Lan H, Zhang Y, Long Y, Weng
H, Li D, Cai W, Zhu H, Niu Y, et al: PRR11 is a novel gene
implicated in cell cycle progression and lung cancer. Int J Biochem
Cell Biol. 45:645–656. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Wang Y, Zhang Y, Zhang C, Weng H, Li Y,
Cai W, Xie M, Long Y, Ai Q, Liu Z, et al: The gene pair PRR11 and
SKA2 shares a NF-Y-regulated bidirectional promoter and contributes
to lung cancer development. Biochim Biophys Acta. 1849:1133–1144.
2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Tan S, Jiang Z, Hou A, Wang J, Zhang J and
Dai L: Expression of PRR11 protein and its correlation with
pancreatic cancer and effect on survival. Oncol Lett. 13:4117–4122.
2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Li K, Yu H, Zhao C, Li J, Tan R and Chen
L: Down-regulation of PRR11 affects the proliferation, migration
and invasion of osteosarcoma by inhibiting the Wnt/β-catenin
pathway. J Cancer. 12:6656–6664. 2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hu H, Song Z, Yao Q, Geng X, Jiang L, Guo
C and Li H: Proline-rich protein 11 regulates self-renewal and
tumorigenicity of gastric cancer stem cells. Cell Physiol Biochem.
47:1721–1728. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhou F, Liu H, Zhang X, Shen Y, Zheng D,
Zhang A, Lai Y and Li H: Proline-rich protein 11 regulates
epithelial-to-mesenchymal transition to promote breast cancer cell
invasion. Int J Clin Exp Pathol. 7:8692–8699. 2014.PubMed/NCBI
|
|
11
|
Anouar Tadlaoui K, Alaoui Sosse S,
Benhessou M, El Karroumi M and Ennaji MM: Proline-Rich Protein 11
Overexpression in Invasive Breast Carcinoma: A Potential Diagnosis
Biomarker. Indian J Gynecol Oncolog. 21(35)2023.
|
|
12
|
Zheng W, Zhu G, Huang Y, Hua J, Yang S,
Zhuang J, Wang J, Huang Q, Xu J and Ye J: PRR11 promotes growth and
progress of colorectal cancer via epithelial-mesenchymal
transition. Int J Clin Exp Med. 10:13109–13122. 2017.
|
|
13
|
Zhu J, Hu H, Wang J, Yang Y and Yi P:
PRR11 overexpression facilitates ovarian carcinoma cell
proliferation, migration, and invasion through activation of the
PI3K/AKT/β-catenin pathway. Cell Physiol Biochem. 49:696–705.
2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhao Z, Pang Z, Xu L, Chen Y and Yang Y:
Expression of PRR11 and SKA2 in cervical cancer tissues and its
relationship with prognosis. Chin J Cancer Prev Control.
12:206–211. 2020.
|
|
15
|
Agha R, Abdall-Razak A, Crossley E, Dowlut
N, Iosifidis C and Mathew G: STROCSS Group. STROCSS 2019 Guideline:
Strengthening the reporting of cohort studies in surgery. Int J
Surg. 72:156–165. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lee EY and Muller WJ: Oncogenes and tumor
suppressor genes. Cold Spring Harb Perspect Biol.
2(a003236)2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang C, Yu L, Ren X, Wu T, Chen X, Huang Y
and Cheng B: The oncogenic potential of PRR11 gene in tongue
squamous cell carcinoma cells. J Cancer. 10:2541–2551.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhang L, Lei Y, Zhang Y, Li Y, Bu Y, Song
F and Zhang C: Silencing of PRR11 suppresses cell proliferation and
induces autophagy in NSCLC cells. Genes Dis. 5:158–166.
2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Tadlaoui KA and Ennaji MM: The molecular
mechanism of novel oncogenes dysregulating signaling pathways
associated with cervical carcinoma. Immunological Implications and
Molecular Diagnostics of Genitourinary Cancer. 19–31. 2023.
|
|
21
|
Xu M and Chang L: The Expression of PRR11
in Cervical Cancer and Its Effect on the Proliferation and
Apoptosis of Cervical Cancer Cell. Labeling Immunoassays and
Clinical Medicine. 27:349–355. 2020.DOI:
10.11748/bjmy.issn.1006-1703.2020.02.035.
|
|
22
|
Qiao W, Wang H, Zhang X and Luo K:
Proline-rich protein 11 silencing inhibits hepatocellular carcinoma
growth and epithelial-mesenchymal transition through β-catenin
signaling. Gene. 681:7–14. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Song Z, Liu W, Xiao Y, Zhang M, Luo Y,
Yuan W, Xu Y, Yu G and Hu Y: PRR11 is a prognostic marker and
potential oncogene in patients with gastric cancer. PLoS One.
10(e0128943)2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang C, Yu L, Hu F, Wang J, Chen X, Tai S
and Cheng B: Upregulation of proline rich 11 is an independent
unfavorable prognostic factor for survival of tongue squamous cell
carcinoma patients. Oncol Lett. 14:4527–4534. 2017.PubMed/NCBI View Article : Google Scholar
|