Introduction
The tetraspanins (TM4SF) are a family of membrane
proteins that are charaterized by the presence of four highly
conserved transmembrane domains (1). They have a small intracellular loop,
intracellular N- and C-termini with short cytoplasmic tails and two
extracellular loops, the larger of which contains a distinctive
pattern of cysteine residues that helps to define the family
(2). The tetraspanin superfamily
member CD151 (also named PETA-3/SFA-1), is implicated in various
biological processes, including cell adhesion, signal tranduction,
cell proliferation, cell differentiation, pathological angiogenesis
and metastasis (3,4). CD151 is found in a large variety of
cell types, including the epithelia, endothelia, platelets and
smooth and striated muscles (5,6).
CD151 forms complexes by interacting with other tetraspanins or
non-tetraspanins to exert its effects; the most prominent
non-tetraspanin partners are integrins, particularly α3β1 and α6β1
(7). A previous study revealed
that an extracellular CD151 site, QRD194-196, in the larger of the
extracellular loops, is required for strong α3β1 association
(8).
A number of studies have shown that CD151 is related
to numerous different types of carcinoma, including prostate
cancer, pancreatic cancer, breast cancer, colorectal cancer and
non-small cell lung cancer; in all of them, high CD151 expression
is associated with a poor prognosis (9,10).
Most of these studies emphasize clinical study. As previously
mentioned, CD151 forms complexes with integrins, so we formed a
mutant of CD151 QRD194-196, AAA194-196, which may impair the
association between CD151 and integrins. We made use of this
mutant, taking HepG2 as our target cell line, to examine the role
of the CD151-AAA mutant in cell proliferation, migration and
invasion, which are the main processes involved in carcinoma
metastasis. The purpose of this study was to investigate the
function of CD151-integrin complexes in carcinoma metastasis and
the mechanism involved.
Materials and methods
Materials
The pZeoSV-CD151 plasmid and anti-CD151 monoclonal
antibody (mAb) 5C11 were provided by Professor Xin Zhang,
Department of Molecular Science, University of Tennessee Health
Science Center (Memphis, TN, USA). Dulbecco’s modified Eagle’s
medium (DMEM) culture medium was purchased from Gibco-BRL (Los
Angeles, CA, USA). The cell counting kit-8 (CCK-8) was purchased
from Beyotime (Haimen, China). The Attractene transfection reagent
was from Qiagen (Hilden, Germany). The enhanced chemiluminescent
(ECL) substrate was from Thermo Scientific Pierce (Rockford, IL,
USA). The polyvinylidene difluoride (PVDF) membrane was from Roche
Diagnostics (Basel, Switzerland). Matrigel was purchased from BD
Biosciences (Heidelberg, Germany). Mouse anti-human β-actin
antibody and antibodies against α3, α6 and β1 were purchased from
Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Antibodies
against Rac, cdc42 and phospho-Rac1/cdc42 (P-Rac1/cdc42) were from
Cell Signaling Technology, Inc. (Beverly, MA, USA). The protein
A/G-agarose kit was from Abmart (Shanghai, China). The study was
approved by the ethics committee of Huazhong University of Science
and Technology.
Cell culture and plasmid
transfection
The HepG2 cell line was bought from the China Center
for Type Culture Collection and cultured in DMEM supplemented with
10% fetal bovine serum (FBS), penicillin and streptomycin. The
HepG2 cells were grown on 6-well plates (60–80% confluent). The
pAAV-CD151 (1.2 μg), pAAV-CD151-AAA (1.2 μg) or pAAV-GFP plasmid
(1.2 μg) was mixed with the Attractene transfection reagent (4.5
μl/well) and incubated for 15 min at room temperature. The
lipid-coated DNA was then added to each well containing 2 ml DMEM
medium and incubated for 6 h. At the end of this period, the medium
was removed and replaced with complete medium, after which the
HepG2 cells were lysed or used in proliferation, migration and
chemotaxis assays.
Western blot analysis
HepG2 cell protein was extracted as follows.
Briefly, the medium in the 6-well plates was discarded, and the
cells were gently washed three times with cooled PBS. A
radioimmunoprecipitation assay (RIPA) buffer [50 mmol/l Tris-HCl
(pH 8.0), 150 mmol/l NaCl, 1% Nonidet-P40, 0.5% deoxycholic acid
and 0.1% sodium dodecyl sulfate] 150 μl] was then added to each
well. Following incubation on ice for 30 min, the lysate was
centrifuged at 12,000 × g at 4°C for 20 min. The protein
concentration of the supernatant was determined using the
bicinchoninic acid (BCA) method. Lysates (25 μg protein/lane) were
resolved by SDS-PAGE gel and transferred to PVDF membranes which
were then blocked with 20% non-fat dry milk in 10 mmol/l Tris-HCl,
pH 7.5, 100 mmol/l NaCl and 0.1% Tween-20. The membranes were then
incubated with primary antibodies against CD151, β-actin, Rac,
cdc42 and P-Rac/cdc42 overnight at 4°C. Peroxidase-conjugated
secondary antibodies were applied for 2–3 h. An ECL system was used
to visualize the separated proteins. The intensities of the various
protein bands were quantified by densitometry (using GeneTools
analysis software).
Proliferation assay
The HepG2 cells were transfected with pAAV-CD151,
pAAV-CD151-AAA or pAAV-GFP using the Attractene transfection
reagent in 6-well plates in triplicate, and 24 h later the cells
were trypsinized and seeded in 96-well plates in triplicate (2,000
cells/well). After attachment, CCK-8 solution (10 μl/well) was
added to the cells, which were then incubated for 1 h. The
absorbance was measured at 450 nm for each well using a microplate
reader (Bio-Tek Instruments, Inc., Winooski, VT, USA) according to
the manufacturer’s instructions.
Wound healing assay
Cell migration was assessed in a wound healing
assay. The HepG2 transfectant cells were cultured in 24-well
plates. When monolayers had formed, wounds were generated by
scraping the monolayers with the tips of sterile pipettes. The
detached cells were rinsed away with PBS and the wounded monolayers
were replenished with the complete medium. Following 12 h of
culturing at 37°C, the monolayers were fixed and images captured
under an inverted light microscope (Nikon TE2000; Nikon, Tokyo,
Japan).
Matrigel invasion assay
The HepG2 transfectant cells were detached from the
culture plates with EDTA, washed once with PBS and replated onto
the inserts of 8-μm pore sized Costar Transwell chambers (Corning
Inc., Corning, NY, USA). The undersides of the inserts were
pre-coated with matrigel (0.3 mg/ml) at 37°C overnight. The cell
number was between 1×105 and 5×105 cells/ml.
The media included DMEM containing 10% FBS in the lower wells and
DMEM containing 0.1% heat-inactivated bovine serum albumin in the
insert. Following incubation at 37°C for 12–18 h, cells that had
not migrated through the inserts were removed and cells that had
migrated to the undersides of the inserts were fixed and stained.
Cells on the lower surface of the membranes were counted. Data from
independent experiments were pooled and analyzed using a two-tailed
Student’s t-test.
Immunoprecipitation
The cells were lysed with ice-cold modified RIPA
buffer at 4°C for 30 min. Adherent cells were scraped off the dish
with a plastic cell scraper. Following the removal of the insoluble
material by 14,000 × g centrifugation, the pre-cleared lysates were
incubated with primary mAb (5C11) pre-absorbed onto protein A- and
G-agarose beads from 3 h to overnight at 4°C. The precipitates were
washed with the lysis buffer three times, dissolved in 1X SDS
loading buffer, heated at 100°C for 10 min, separated by SDS-PAGE
and then electrically transferred to PVDF membranes. After blocking
for 2.5 h in 20% non-fat dry milk in TBST, the membrane was
incubated with anti-α3, anti-α6 or anti-β1 antibody overnight at
4°C. After washing with TBST, the membrane was incubated with
horseradish peroxidase-conjugated secondary antibodies for 2–3 h at
room temperature and was then revealed by chemiluminescence.
Statistical analysis
All data are expressed as the mean ± standard
deviation (x±s). Differences between two groups were compared by
t-test and comparisons of groups were performed via one-way
analysis of variance (ANOVA) and the Student-Newman-Keuls test.
Statistical analysis was performed using SPSS 13.0 software.
P<0.05 was considered to indicate a statistically significant
result.
Results
Expression of CD151 protein in different
groups
The expression levels of CD151 in the cells
transfected with the pAAV-CD151 plasmid were significantly
increased (P<0.05) compared with the control and pAAV-GFP groups
48 h after transfection. In addition, the cells transfected with
pAAV-CD151-AAA demonstrated no significant difference in protein
expression levels from the pAAV-CD151 group (P>0.05) (Fig. 1)
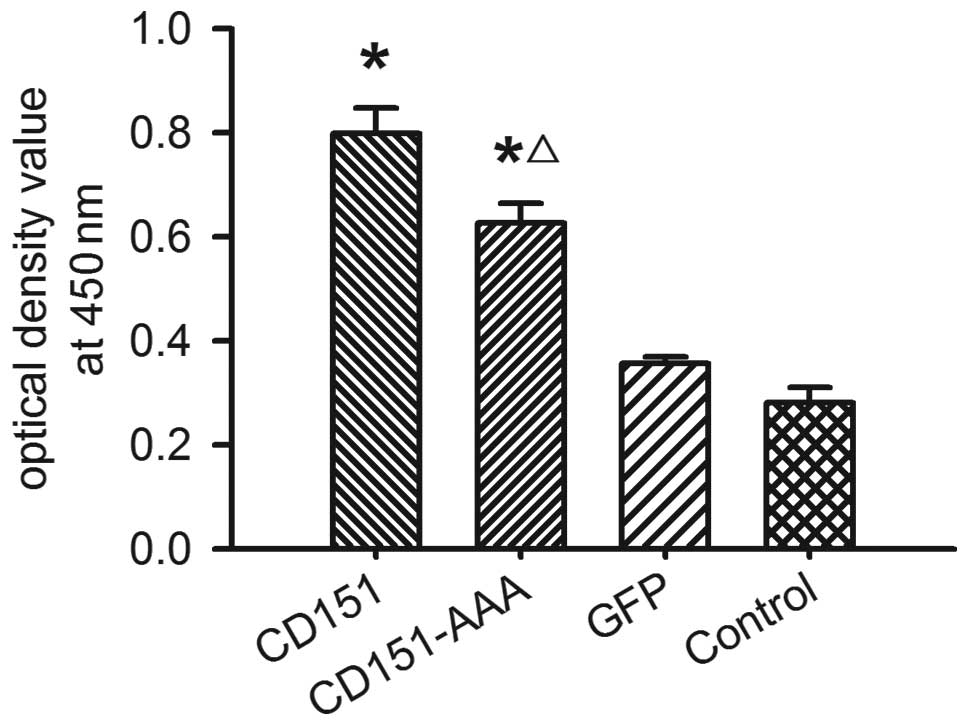
Effect of CD151 and CD151-AAA on HepG2
proliferation
To determine the effect of CD151 on HepG2
proliferation, we used a CCK-8 assay. The transfection of the HepG2
cells with the pAAV-CD151 plasmid significantly enhanced the HepG2
proliferation (P<0.05) compared with the pAAV-GFP-transfected
and control groups. Transfection with pAAV-CD151-AAA also
significantly enhanced the proliferation of the cells as compared
with the groups transfected with pAAV-GFP and the control (Fig. 2). The results indicated that
pAAV-CD151 was able to promote cell proliferation, while the
ability of pAAV-CD151-AAA to promote cell proliferation was
decreased compared with the pAAV-CD151 group.
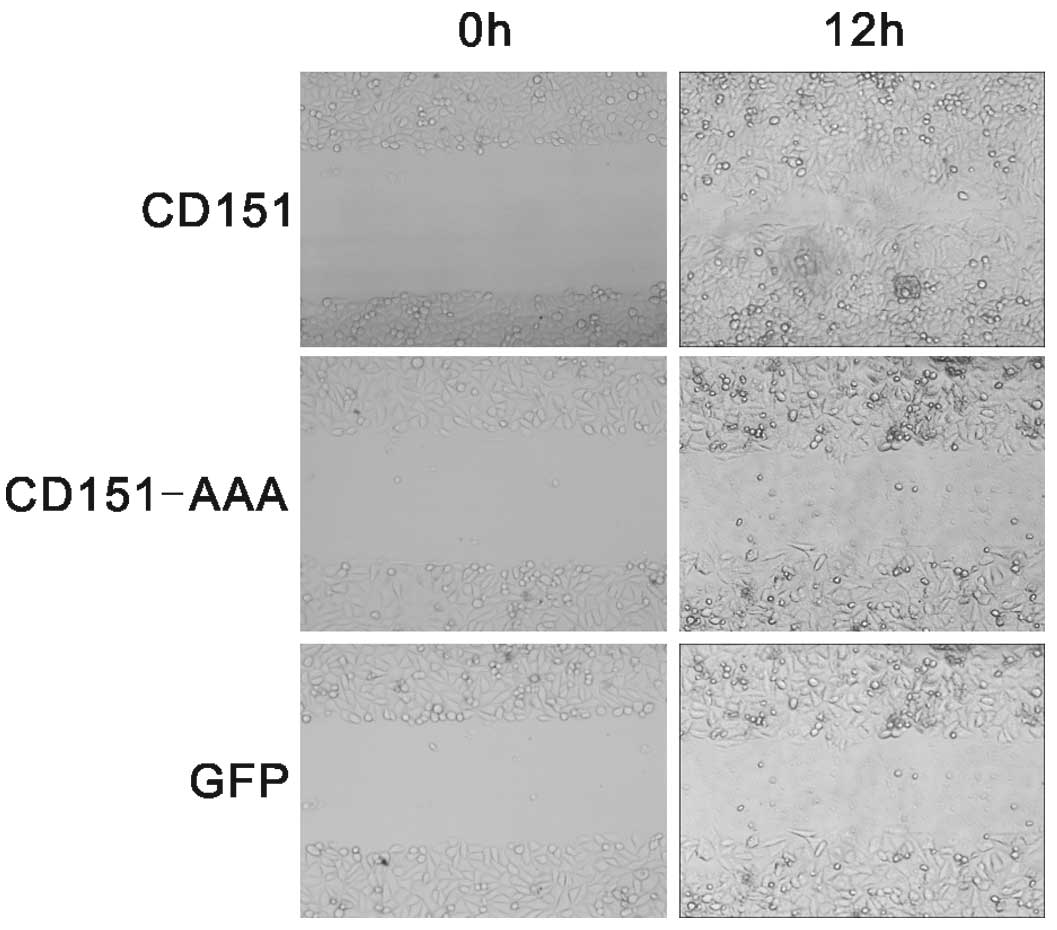
Effect of CD151 and CD151-AAA on HepG2
migration
In the wound healing assay (Fig. 3), the cells transfected with
pAAV-CD151 demonstrated significantly enhanced wound healing, The
ability of the pAAV-CD151-AAA mutant cells to undergo wound healing
was markedly delayed compared with the pAAV-CD151 transfectant
cells. The wound healing assay demonstrated that pAAV-CD151 gene
transfer was able to promote HepG2 migration and that this effect
was diminished in the pAAV-CD151-AAA group.
Effect of CD151 and CD151-AAA on HepG2
invasion
In the matrigel invasion assay (Fig. 4), pAAV-CD151 significantly promoted
HepG2 invasion compared with the control and pAAV-GFP groups.
pAAV-CD151-AAA was not able to promote cell invasion, which
suggests that this ability was damaged in the pAAV-CD151-AAA
group.
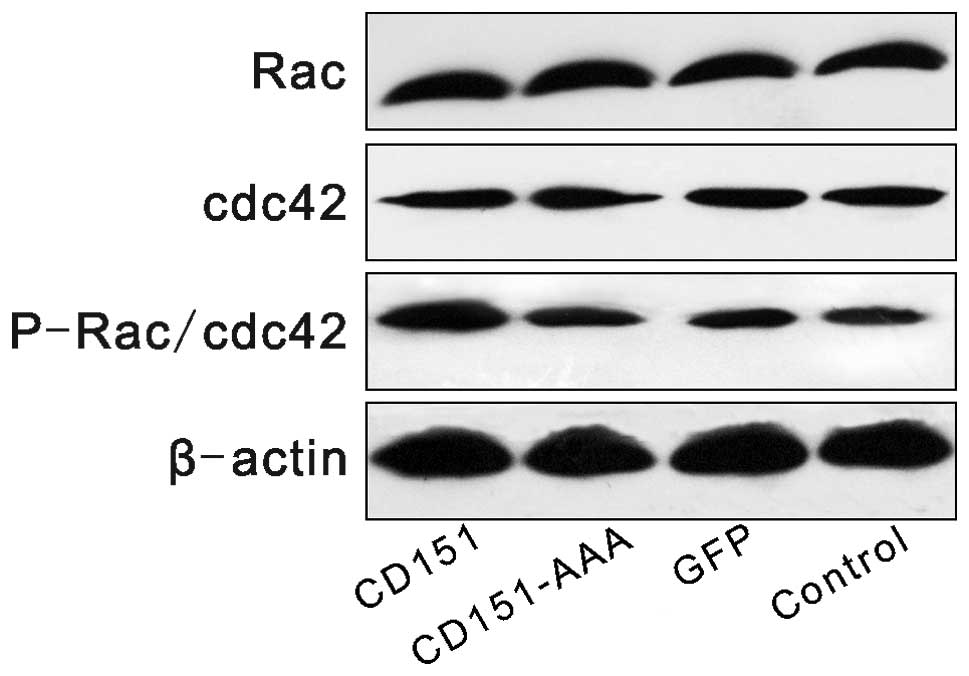
Effect of CD151 and CD151-AAA on the Rac,
cdc42 and P-Rac/cdc42 pathways
The expression levels of Rac, cdc42 and P-Rac/cdc42
were investigated following transfection. Western blot analysis
revealed that transfection with pAAV-CD151 increased the expression
level of phosphorylated P-Rac/cdc42 compared with the control group
and the GFP group, whereas transfection with pAAV-CD151-AAA had no
effect on the expression levels of P-Rac/cdc42 (Fig. 5).
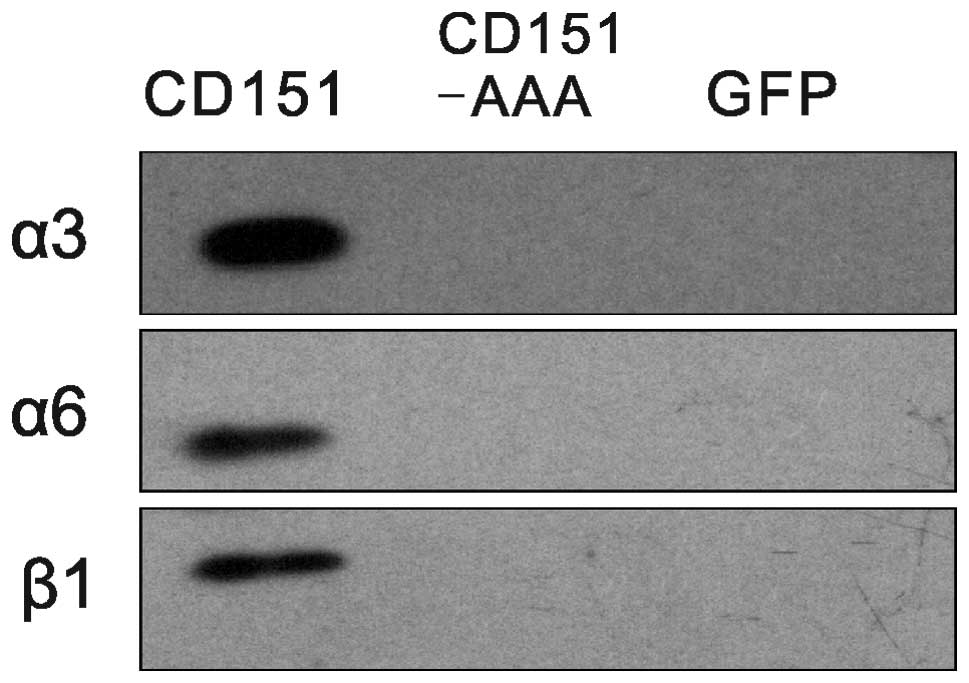
Effects of the CD151-AAA mutation on the
CD151-integrin association
To determine whether the pAAV-CD151-AAA mutant
altered the CD151-integrin complexation, we compared the
CD151-integrin associations between the CD151 and CD151-AAA
transfectants. Immunoblotting analysis indicated that the integrins
that were coprecipitated with CD151 (5C11) under the stringent
lysis conditions were endogenous α3, α6 and β1 (Fig. 6).
Discussion
In this study, we used the CD151-AAA mutant to
investigate the mechanism that governs the effects of CD151 on
carcinoma metastasis. Our results revealed that the CD151-AAA
mutant strongly inhibited the proliferation, migration and invasion
of HepG2 cells. These results demonstrated that the CD151-integrin
complex is functionally significant. In our previous study, this
mutant was shown to have the ability to impair angiogenesis due to
abrogation of the relationship between CD151 and integrins
(11).
CD151 has a function in a number of carcinomas. In
liver cancer, hepatocellular carcinoma patients with high
expression levels of the CD151/integrin β1 complex have been
reported to have the poorest prognosis (2). The expression levels of CD151 have
been found to correlate positively with clinical classification in
clear cell renal cell carcinoma patients; the patients with high
CD151 expression levels had significantly shorter survival times
(3). For tumors of the
gastrointestinal tract, CD151 is indicated to promote metastasis
formation (10). In endometrial
cancer, CD151 has been identified as a novel marker that may be
used to guide therapeutic decisions (12). It has also been reported that CD151
forms a structural and functional complex with integrin α3/α6 that
exerts oncogenic functions in human salivary gland cancer cells
(13). The expression levels of
the CD151-α6 integrin complex have been reported to be elevated in
31% of human breast cancers (14).
CD151 expression levels have also been found to be significantly
higher in prostate cancer, with poorly differentiated cancers
demonstrating the strongest staining (15). Patients with pancreatic
adenocarcinoma also revealed high expression levels of CD151, and
α6β4 was found to be selectively upregulated (16). In all, CD151 is the most harmful
molecule that may be detected in patients with carcinoma and many
of its functions are associated with integrins.
CD151 closely associates with laminin-binding
integrins (α3β1 and α6β1) and affects their functions (17,18).
The CD151 site QRD194-196 is the key site of complex formation
between CD151 and integrins. Therefore, in this study we used
pAAV-CD151-AAA, which we constructed in our former study by
applying the technology of oriented direction mutation, to
investigate whether the function of CD151 is weakened without the
aid of integrins. The results confirm our hypothesis. This is the
first time that we have used the mutant CD151-AAA to study tumor
cells. In our former study it was used in the investigation of
angiogenesis (11). The mutant
CD151-AAA abrogated cell proliferation, migration and invasion
ability. Its mechanism may involve the downregulation of
P-Rac1/cdc42 activity.
From these findings, it appears that the enhancement
of HepG2 cell proliferation, migration and invasion by CD151 is due
to P-Rac1/cdc42-mediated events activated by CD151-integrin
complexes, but not by CD151 alone. Our study suggests that, since
the relationship between CD151 and integrins may be disrupted to
result in marked decreases in tumor cell quantity and other
effects, interrupting their connection may be a new method to
prevent malignant cell metastasis.
Acknowledgements
We thank the Laboratory of Cardiology of Tongji
Hospital, Wuhan, China. This study was financially supported by
grants from the National Science Foundation for Young Scientists of
China (Project No. 81000047 and No. 81000139).
References
|
1
|
Wright MD and Tomlinson MG: The ins and
outs of the transmembrane 4 superfamily (Review). Immunol Today.
15:588–594. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Devbhandari RP, Shi GM, Ke AW, et al:
Profiling of the tetraspanin CD151 web and conspiracy of
CD151/integrin β1 complex in the progression of hepatocellular
carcinoma. PLoS One. 6:e249012011.PubMed/NCBI
|
|
3
|
Yoo SH, Lee K, Chae JY and Moon KC: CD151
expression can predict cancer progression in clear cell renal cell
carcinoma. Histopathology. 58:191–197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takeda Y, Kazarov AR, Butterfield CE, et
al: Deletion of tetraspanin Cd151 results in decreased pathologic
angiogenesis in vivo and in vitro. Blood. 109:1524–1532. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Caplan MJ, Kamsteeg EJ and Duffield A:
Tetraspan proteins: regulators of renal structure and function
(Review). Curr Opin Nephrol Hypertens. 16:353–358. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fitter S, Tetaz TJ, Berndt MC and Ashman
LK: Molecular cloning of cDNA encoding a novel platelet-endothelial
cell tetra-span antigen, PETA-3. Blood. 86:1348–1355.
1995.PubMed/NCBI
|
|
7
|
Zöller M: Tetraspanins: push and pull in
suppressing and promoting metastasis (Review). Nat Rev Cancer.
9:40–55. 2009.PubMed/NCBI
|
|
8
|
Kazarov AR, Yang X, Stipp CS, Sehgal B and
Hemler ME: An extracellular site on tetraspanin CD151 determines
alpha 3 and alpha 6 integrin-dependent cellular morphology. J Cell
Biol. 158:1299–1309. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lazo PA: Functional implications of
tetraspanin proteins in cancer biology (Review). Cancer Sci.
98:1666–1677. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zöller M: Gastrointestinal tumors:
metastasis and tetraspanins (Review). Z Gastroenterol. 44:573–586.
2006.PubMed/NCBI
|
|
11
|
Liu WF, Zuo HJ, Chai BL, et al: Role of
tetraspanin CD151-α3/α6 integrin complex: Implication in
angiogenesis CD151-integrin complex in angiogenesis. Int J Biochem
Cell Biol. 43:642–650. 2011.
|
|
12
|
Voss MA, Gordon N, Maloney S, et al:
Tetraspanin CD151 is a novel prognostic marker in poor outcome
endometrial cancer. Br J Cancer. 104:1611–1618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Klosek SK, Nakashiro K, Hara S, Shintani
S, Hasegawa H and Hamakawa H: CD151 forms a functional complex with
c-Met in human salivary gland cancer cells. Biochem Biophys Res
Commun. 336:408–416. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang XH, Richardson AL, Torres-Arzayus MI,
et al: CD151 accelerates breast cancer by regulating alpha 6
integrin function, signaling, and molecular organization. Cancer
Res. 68:3204–3213. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ang J, Lijovic M, Ashman LK, Kan K and
Frauman AG: CD151 protein expression predicts the clinical outcome
of low-grade primary prostate cancer better than histologic
grading: a new prognostic indicator? Cancer Epidemiol Biomarkers
Prev. 13:1717–1721. 2004.PubMed/NCBI
|
|
16
|
Gesierich S, Paret C, Hildebrand D, et al:
Colocalization of the tetraspanins, CO-029 and CD151, with
integrins in human pancreatic adenocarcinoma: impact on cell
motility. Clin Cancer Res. 11:2840–2852. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sincock PM, Fitter S, Parton RG, Berndt
MC, Gamble JR and Ashman LK: PETA-3/CD151, a member of the
transmembrane 4 superfamily, is localised to the plasma membrane
and endocytic system of endothelial cells, associates with multiple
integrins and modulates cell function. J Cell Sci. 112:833–844.
1999.PubMed/NCBI
|
|
18
|
Yáñez-Mó M, Alfranca A, Cabañas C, et al:
Regulation of endothelial cell motility by complexes of tetraspan
molecules CD81/TAPA-1 and CD151/PETA-3 with alpha3 beta1 integrin
localized at endothelial lateral junctions. J Cell Biol.
141:791–804. 1998.PubMed/NCBI
|