Introduction
Yigit et al first reported Klebsiella
pneumoniae strains with a medium to high degree of resistance
to imipenem and meropenem in 2001 (1). The authors isolated a new
carbapenemase which they named Klebsiella pneumoniae
carbapenemase (KPC)-1. From then on, KPC-producing bacteria have
been reported successively in various countries and regions and are
now spreading globally (2–5).
An imipenem-resistant strain of Klebsiella
pneumoniae was identified in our hospital for the first time by
the ICU department on 5th July, 2008. It was detected in the blood
of a patient who was transferred to our hospital following
prolonged therapy in another hospital in Hangzhou. Investigations
were immediately carried out to determine the source and path of
the infection, and appropriate measures were taken to get the
infection under control in a timely and effective manner. The aim
of the present study was to analyze the resistance mechanisms and
epidemiology of the imipenem-resistant Klebsiella pneumoniae
strains isolated from the patients and hygienic surveillance
samples obtained at the hospital.
Materials and methods
Strains
Bacteriological specimens were collected from the
ICU environment, medical items, hands of the medical staff and
sputum and blood specimens of other patients in the ward at the
Central Hospital of Taizhou City. Among the 57 collected specimens,
four were identified to carry imipenem-resistant Klebsiella
pneumoniae. The four imipenem-resistant Klebsiella
pneumoniae clinical isolates 2011, 2163, 2193 and 2285 were
investigated in this study. One was isolated from a patient’s blood
specimen, two were from sputum specimens and one was from an
inspection specimen which was collected from a mop used in the
ward.
Apparatus and reagents
The automatic bacteria calibrator VITEK-60 was
acquired from bioMérieux SA (Marcy l’Etoile, France). Pulsed-field
gel electrophoresis (PFGE) typing was performed using the CHEF
Mapper® XA system (Bio-Rad Laboratories, Hercules, CA,
USA). Gel images were captured using a Bio-Rad GelDoc 2000 gel
documentation system (Bio-Rad Laboratories). PCR was performed
using a Bio-Rad PTC-200 thermal cycler (Bio-Rad Laboratories).
Primers were purchased from Yingwei Jieji (Shanghai, China).
Restriction endonuclease XbaI was purchased from Dalian
Baosheng Biological Engineering Co., Ltd. (Dalian, China). SeaKem
Gold Agarose was acquired from Cambrex Bioscience Rockland, Inc.
(Rockland, ME, USA). Protease K was from Merck & Co., Inc.
(Whitehouse Station, NJ, USA). The standard control strain H9812,
susceptibility paper, meropenem, cefoperazone/sulperazon and
polymyxin B were all from Oxoid (Basingstoke, UK).
Drug susceptibility test
The minimum inhibitory concentration (MIC) was
determined using a VITEK-60 automatic bacteria calibrator by the
microplate dilution method. Fifteen antimicrobial agents were
examined as follows: imipenem, ertapenem, cefoperazone/sulbactam,
cefepime, ciprofloxacin, tobramycin, gentamicin, aztreonam,
ceftazidime, ceftriaxone, piperacillin/tazobactam, cefazolin,
ampicillin, ampicillin/sulbactam and cefoxitin. The agar disc
diffusion method (KB method) was also used to evaluate the
susceptibility of the bacteria to cefoperazone/sulperazon,
polymyxin B and meropenem.
Modified Hodge test for detection of
carbapenemases
An overnight culture suspension of Escherichia
coli ATCC 25922 adjusted to 0.5 McFarland standard was
inoculated on the surface of a Mueller-Hinton agar (MHA) plate. A
paper disk impregnated with 10 μg imipenem was placed at the center
of the plate. The test strain was streaked from the edge of the
paper to the periphery of the plate in four different directions.
The plate was incubated for 16–18 h at 35°C. The presence of
growing bacteria within the imipenem antibacterial circle due to
carbapenemase production by the test strain was considered as
positive.
PCR amplification of extended-spectrum
β-lactamases (ESBL), cephalosporinase (AmpC) and KPC genes and DNA
sequence analysis
The primers used for β-lactamases (Table I) were as used in a previous study
by Wang et al(6). The PCR
conditions were: preparation of the template by boiling;
denaturation at 94°C for 5 min, then 94°C for 60 sec, 56°C for 30
sec and 72°C for 45 sec for 30 cycles; followed by a final step at
72°C for 10 min.
 | Table IPrimers for β-lactamases. |
Table I
Primers for β-lactamases.
| Target gene | Primer sequence
(3′-5′) | Size (bp) |
|---|
| CTX-M |
ACGCTTTCCAATGTGCAGTA | |
|
ACGTCACCAACTGCGCCC | 436 |
| CTX-M-1 |
TTAATTCGTCTCTTCCAGA | |
|
CAGCGCTTTTGCCGTCTAAG | 971 |
| CTX-M-2 |
CTCAGAGCATTCGCCGCTCA | |
|
GCGCCGCAGCCAGAATATCC | 842 |
| CTX-M-9 |
GTGACAAAGAGAGTGCAACGG | |
|
ATGATTCTCCCCCCTGAACCC | 857 |
| SHV |
GCCTTTATCGGCCCTCACTCAA | |
|
TTAGCGTTGCCAGTGCTCGATCA | 928 |
| TEM |
ATAAAATTCTTGAAGACGAAA | |
|
GACAGTTAGCAATGCTTAATCA | 1079 |
We used the primers reported by Jiang et
al(7) for AmpC (Table II). The PCR conditions were:
preparation of the template by boiling; denaturation at 94°C for 5
min, then 94°C for 30 sec, 52°C for 30 sec and 72°C for 1 min for
30 cycles; followed by a final step at 72°C for 10 min.
 | Table IIPrimers for cephalosporinase
(AmpC). |
Table II
Primers for cephalosporinase
(AmpC).
| Target genes | Primer | Primer sequence
(3′-5′) | Size (bp) |
|---|
| MOX-1, 2; CMY-1, 8,
9, 10, 11 | MOXMF |
GCTGCTCAAGGAGCACAGGAT | 520 |
| MOXMR |
CACATTGACATAGGTGTGGTGC | |
| LAT-1, 2, 3, 4;
BIL-1; CMY-2, 3, 4, 5, 6, 7 | CITMF |
GGCCAGAACTGACAGGCAAA | 462 |
| CITMR |
TTTCTCCTGAACGTGGCTGGC | |
| DHA-1; DHA-2 | DHAMF |
AACTTTCACAGGTGTGCTGGGT | 405 |
| DHAMR |
CCGTACGCATACTGGCTTTGC | |
| ACC | ACCMF |
AACAGCCTCAGCAGCCGGTTA | 346 |
| ACCMR |
TTCGCCGCAATCATCCCTAGC | |
| MIR-1; ACT-1 | EBCMF |
TCGGTAAAGCCGATGTTGCGG | 302 |
| EBCMR |
CTTCCACTGCGGCTGCCAGTT | |
| FOX-1, 2, 3, 4,
5 | FOXMF |
AACATGGGGTATCAGGGAGATG | 190 |
| FOXMR |
CAAAGCGCGTAACCGGATTGG | |
| CMY-4 | CMY-4-F |
ATGATGAAAAAATCGTTATGC | 1100 |
| CMY-4-R |
TTGCAGCTTTTCAAGAATGCGC | |
For KPC, in accordance with a previous study
(8), we used the following
primers: KPC-1 forward, GCTACACCTAGC TCCACCTTC, and reverse
ACAGTGGTTGGTAATCCATGC. The PCR conditions were: preparation of the
template by boiling; denaturation at 94°C for 5 min, then 94°C for
25 sec, 55°C for 45 sec and 72°C for 60 sec for 35 cycles; followed
by a final step at 72°C for 10 min. The purified PCR amplification
products were sequenced by Yingwei Jieji (Invitrogen, Shanghai,
China) and compared with sequences available in the GenBank
database.
PFGE typing
We used a previously reported method (9) which involved digestion with
endonuclease XbaI at 37°C for 4 h and electrophoresis at
120°C, 6 V/cm, 5–40 sec for 24 h. The XbaI restriction
patterns of the genomic DNA of the isolates were analyzed and
interpreted according to the criteria of Tenover et
al(10).
Results
Modified Hodge test results
The four imipenem-resistant Klebsiella
pneumoniae isolates 2011, 2163, 2193 and 2285 revealed an area
of inhibition due to carbapenemase production. Fig. 1 presents the result for isolate
2011.
Analysis of the susceptibility of four
Klebsiella pneumoniae isolates to 18 antimicrobial agents
The four isolates were sensitive to polymyxin B and
tobramycin, had a moderate degree of resistance to meropenem and a
high degree of resistance to the other antibiotics (Table III).
 | Table IIIAntimicrobial susceptibility patterns
of Klebsiella pneumoniae. |
Table III
Antimicrobial susceptibility patterns
of Klebsiella pneumoniae.
| A, Minimal inhibitory
concentration (MIC) |
|---|
|
|---|
| MIC (μg/ml) of
isolates |
|---|
|
|
|---|
| Antimicrobial
agents | 2163 | 2193 | 2285 | 2011 |
|---|
| Imipenem | >16 | >16 | >16 | >16 |
| Ertapenem | >16 | >16 | >16 | >16 |
| Cefoxitin | ≥64 | ≥64 | ≥64 | ≥64 |
| Cefepime | ≥64 | ≥64 | ≥64 | ≥64 |
| Ciprofloxacin | ≥4 | ≥4 | ≥4 | ≥4 |
| Tobramycin | 4 | 4 | 4 | 4 |
| Gentamicin | ≥16 | ≥16 | ≥16 | ≥16 |
| Aztreonam | ≥64 | ≥64 | ≥64 | ≥64 |
| Ceftazidime | ≥64 | ≥64 | ≥64 | ≥64 |
| Ceftriaxone | ≥64 | ≥64 | ≥64 | ≥64 |
|
Piperacillin-tazobactam | ≥128 | ≥128 | ≥128 | ≥128 |
| Cefazolin | ≥64 | ≥64 | ≥64 | ≥64 |
| Ampicillin | ≥32 | ≥32 | ≥32 | ≥32 |
|
Ampicillin-sulbactam | ≥32 | ≥32 | ≥32 | ≥32 |
|
| B, Agar disc
diffusion (KB testing) |
|
| KB (mm) of
isolates |
|
|
| Antimicrobial
agents | 2163 | 2193 | 2285 | 2011 |
|
| Polymyxin B | 20 | 20 | 21 | 20 |
| Meropenem | 16 | 16 | 16 | 16 |
|
Cefoperazone-sulbactam | 6 | 6 | 6 | 6 |
Screening of extended-spectrum
β-lactamases
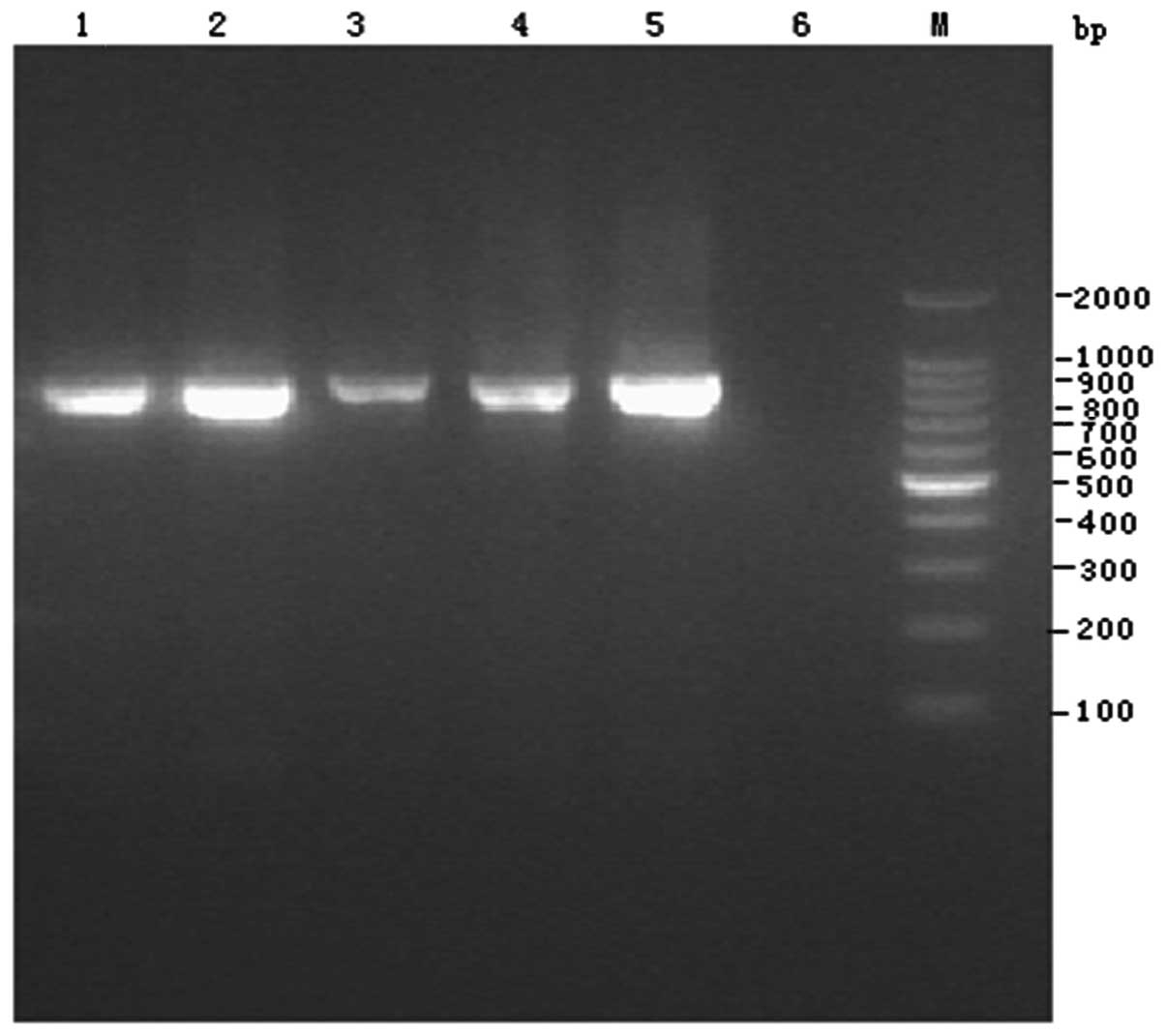
The four isolates all produced extended-spectrum
β-lactamases and all had an 857-bp positive band (Fig. 2), which was verified to be CTX-M-9
β-lactamase by sequencing.
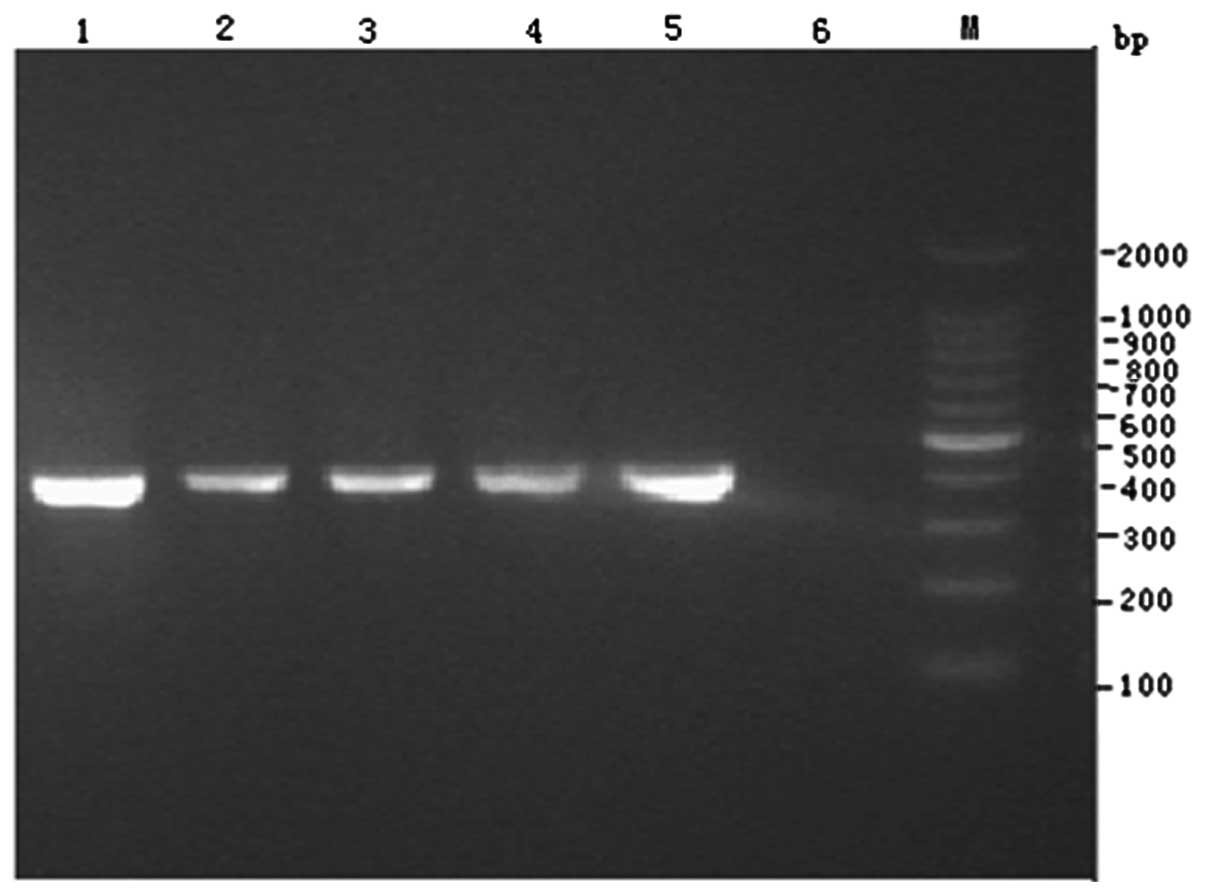
Screening of cephalosporinase
The four isolates all produced cephalosporinase and
all had a 405-bp positive band (Fig.
3), which was verified to be DHA-1 cephalosporinase by
sequencing.
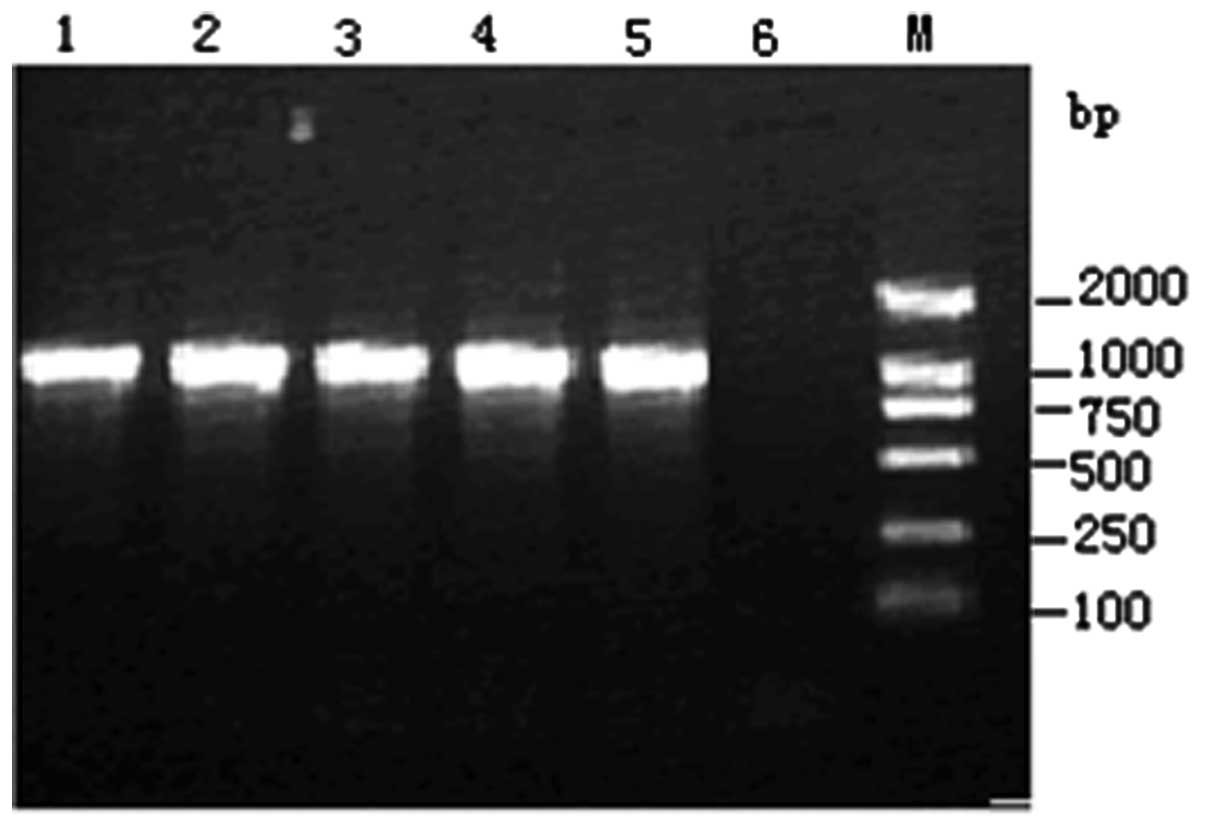
Screening of carbapenemases
The four isolates all produced carbapenemases and
all had a 989-bp positive band (Fig.
4), which was verified to be KPC-2 carbapenemase by
sequencing.
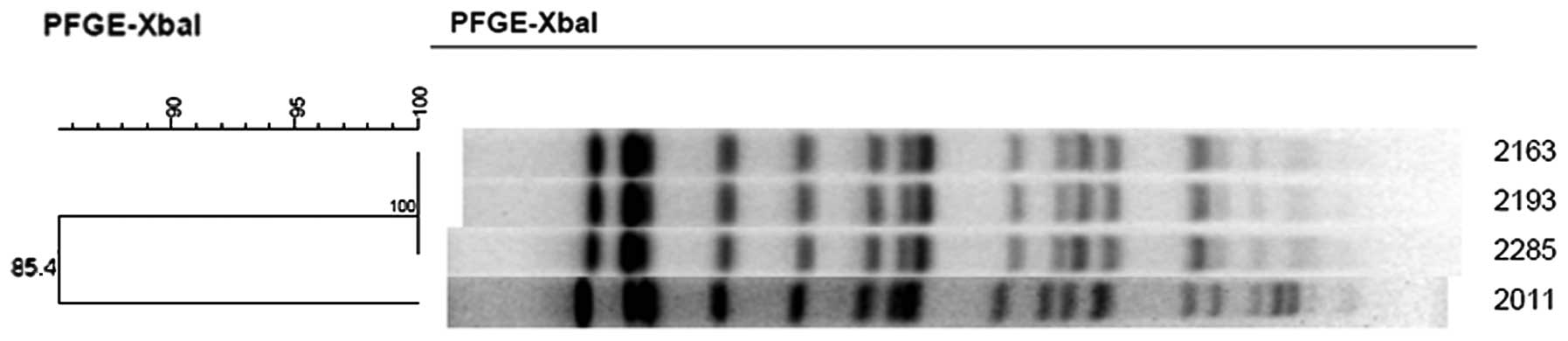
PFGE typing
Fig. 5 presents the
PFGE typing of the four isolates. These four isolates were
homologous with a difference of 1–3 bands, so they may be
considered as four subclones of one pulsed-field-type clone.
Discussion
Carbapenem antibiotics are highly stable to most
β-lactamases, show a high affinity for penicillin-binding proteins
and are easily able to enter the periplasmic space by effectively
penetrating the bacterial outer membrane. Therefore, they are among
the most potent and rapidly acting antibiotics and, in particular,
they have unrivalled effects in the treatment of severe infections
caused by Enterobacteriaceae. However, reports concerning
Enterobacteriaceae which are not sensitive to carbapenem
antibiotics have been rapidly increasing in number over the last
ten years, the main mechanism of drug resistance being the
production of carbapenemases. These enzymes are a class of
β-lactamase which are able to significantly hydrolyze imipenem or
meropenem. Among the three classes of enzymes A, B and D in the
Ambler molecular classification (11), the KPC enzyme is an A type with its
gene on a transferable plasmid. It may be horizontally transmitted
through plasmids, integrons and gene elements which insert the
sequence, and readily leads to an outbreak. However, this plasmid
often carries a number of other resistance-determining factors,
which is one of the causes of the multidrug resistance of the
strains containing it. There are numerous bacteria which produce
the KPC enzyme: Escherichia coli, Salmonella,
Enterobacter cloacae, acid bacteria isolated from the oak
tree (Freund), Serratia marcescens, Proteus,
Klebsiella oxytoca, bacteria isolated from the oak tree
(Rostock Reber), citric acid bacteria (Freund), Pseudomonas
aeruginosa and Pseudomonas putida, Acinetobacter
spp and particularly Klebsiella pneumoniae. There are 11
subtypes of the KPC enzyme; the KPC subtypes from Klebsiella
pneumoniae are KPC-1/2, KPC-3, KPC-4, KPC-6, KPC-7, KPC-8 and
KPC-11, and the other subtypes have been identified in other
bacteria (12). In view of the
harmfulness of KPC-producing bacteria, the American Clinical and
Laboratory Standards Institute (CLSI) suggested that clinical
laboratories should detect the KPC enzyme, test the susceptibility
of the Enterobacter towards ertapenem, and carry out the
adjusted Hodge test for all Enterobacter whose MIC values
for meropenem and imipenem are greater than 2–4 μg/ml, in order to
strengthen the monitoring of the KPC enzyme on a global scale.
In 2008, we identified KPC-2-producing Klebsiella
pneumoniae in the sputum and blood of a patient who was
transferred to our hospital following prolonged therapy in other
hospital in Hangzhou. The patient succumbed on the second day after
the bacterial infection was detected. Bacteriological specimens
were then collected from the ICU environment, medical items, hands
of the medical staff and sputum and blood specimens from other
patients in the ward. Among the 57 collected specimens, the four
isolates 2011, 2163, 2193 and 2285 were shown to be homologous by
PFGE. According to the definition of hospital infection outbreaks,
which is more than three cases of same type of homologous infection
in a medical institution or its patients in a short time, we may
conclude that there was a small-scale outbreak of KPC-2-producing
Klebsiella pneumoniae in our hospital and the source of this
medicine-resistant pathogen was another hospital.
The enzyme KPC-2 was detected in all four isolates
and was the main cause of their imipenem resistance. In addition,
these four isolates also contained the extended-spectrum
β-lactamase (ESBL) gene blaCTX-M-9 and the
cephalosporinase (AmpC) gene blaDHA-1, which resulted in
multidrug resistance.
Investigation of the epidemic started immediately
following the hospital infection outbreak. At the same time, the
hospital infection control committee developed infection control
programs after investigating the infection source, transmission
route and susceptible populations. The detailed measures undertaken
were as follows: evacuation of the patients and thorough
disinfection of the ICU ward; air disinfection by steaming with 2
g/m3 15% peracetic acid and ozone, mopping the ground
and wiping the surfaces of objects with 0.5% peracetic acid;
reopening the ICU when the monitoring results were qualified after
disinfection; high-pressure steaming or immersion in 0.5% peracetic
acid for other medical supplies according to the nature of the
materials; and isolation and special care of the infected patient.
Following all these measures, no Klebsiella pneumoniae with
decreased susceptibility to imipenem was identified during tests of
air, article surfaces and medical supplies in the ICU ward. Due to
the timely detection of the hospital infection outbreak, the
control measures effectively prevented the infection from spreading
on a larger scale, demonstrating that significance of molecular
biology techniques in hospital infection control.
As carbapenem antibiotics are developed and new oral
drugs emerge, the quantity of this type of antibiotic used will
increase. However, an increased number of drug-resistant strains
are likely to arise with an unlimited and extensive use of
antibiotics, so this type of efficient and broad-spectrum
antibiotic must be used reasonably and prudently. The timely
reporting of KPC-producing bacteria in the clinical laboratory is
of great significance for guiding the reasonable use of antibiotic,
slowing down the emergence of drug-resistant strains and
controlling the transmission of and infection with drug-resistant
strains.
Acknowledgements
This study was supported by a grant from the
Education Department of Zhejiang Provincial Government:
Y200805658.
References
|
1
|
Yigit H, Queenan AM, Anderson GJ,
Domenech-Sanchez A, Biddle JW, Steward CD, et al: Novel
carbapenem-hydrolyzing beta-lactamase, KPC-1, from a
carbapenem-resistant strain of Klebsiella pneumoniae.
Antimicrob Agents Chemother. 45:1151–1161. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mavroidi A, Miriagou V, Malli E, Stefos A,
et al: Emergence of Escherichia coli sequence type 410
(ST410) with KPC-2 β-lactamase. Int J Antimicrob Agents.
39:247–250. 2012.
|
|
3
|
Brink AJ, Coetzee J, Clay CG, et al:
Emergence of New Delhi metallo-beta-lactamase (NDM-1) and
Klebsiella pneumoniae carbapenemase (KPC-2) in South Africa.
J Clin Microbiol. 50:525–527. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nordmann P, Cuzon G and Naas T: The real
threat of Klebsiella pneumoniae carbapenemase 2-producing
bacteria. Lancet Infect Dis. 9:228–236. 2009.
|
|
5
|
Wei ZQ, Du XX, Yu YS, et al:
Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate
from China. Antimicrob Agents Chemother. 51:763–765. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Y, Jiang X, Sun J, et al: Detection
of plasmid-mediated ampC gene, blaDHA-1 from clinical
isolates of Klebsiella pneumoniae. Shanghai J Med Lab Sci.
18:331–335. 2003.(In Chinese).
|
|
7
|
Jiang Y, Cao N, Chen T, et al: Detection
and analysis of plasmid-mediated ampC genes from gram-negative
clinical strains. Shanghai J Med Lab Sci. 18:328–330. 2003.(In
Chinese).
|
|
8
|
Smith Moland E, Hanson ND, Herrera VL, et
al: Plasmid-mediated, carbapenem-hydrolysing beta-lactamase, KPC-2,
in Klebsiella pneumoniae isolates. J Antimicrob Chemother.
51:711–714. 2003.
|
|
9
|
Piekarska K, Zacharczuk K, Szych J, et al:
Dissemination of the KPC carbapenemase producing Klebsiella
pneumoniae in a hospital in Warsaw, Poland. Med Dosw Mikrobiol.
62:9–20. 2010.(In Polish).
|
|
10
|
Tenover FC, Arbeit RD, Goering RV, et al:
Interpreting chromosomal DNA restriction patterns produced by
pulsed-field gel electrophoresis: criteria for bacterial strain
typing. J Clin Microbiol. 33:2233–2239. 1995.
|
|
11
|
Endimiani A, Hujer AM, Perez F, et al:
Characterization of blaKPC-containing Klebsiella pneumoniae
isolates detected in different institutions in the Eastern USA. J
Antimicrob Chemother. 63:427–437. 2009.
|
|
12
|
Fu W, Chen D, et al: Research advances in
KPC-type carbapenemases. Chin J Nosocomiology. 21:2159–2160.
2011.
|