Introduction
The regulation of mRNA post-transcriptional
processes plays a central role in the control of eukaryotic gene
expression. RNA-binding proteins (RBPs) are key players in the
determination of mRNA fate (1).
Certain RBPs recognize common mRNA features, such as the 5′ cap or
the 3′ poly (A) tail, but the majority of RBPs contain RNA-binding
domains (RBDs) for recognition of secondary structures or specific
sequence motifs. More than 40 RBDs have been identified thus far,
such as well-characterized RNA recognition motifs (RRMs), the
K-homology (KH) domain, double-stranded RNA binding motif (dsRBM),
the Arg-Gly-Gly (RGG) box, DEAD/DEAH box and Piwi/Argonaute/Zwille
(PAZ) domains (2). The aberrant
expression of RBPs, which affect and alter many steps of RNA
metabolism, are implicated in the development of various diseases,
including cancer. In humans, more than 500 RBPs are known, but only
a few have been identified to exert an oncogenic or
tumor-suppressive function (3).
These RBPs include Src-associated in mitosis, 68 kDa (SAM68),
β-catenin, serine/arginine-rich splicing factor 1 (SRSF1), KH-type
splicing regulatory protein (KSRP) and human antigen R (HuR)
(4–8). The role of other RBPs in
tumorigenesis remains to be elucidated.
Human Mex-3 is a novel family of evolutionarily
conserved RBPs (hMex-3A to 3D) with two KH RBDs (9). They share the highest identity with
Caenorhabditis elegans Mex-3, which is involved in the
establishment of anterior-posterior embryonic asymmetry and in the
maintenance of germline pluripotency. hMex-3D is ubiquitously
expressed in various tissues; however, the other three hMex-3
proteins are expressed at varying levels in different tissues. All
four proteins predominantly accumulate in the cytoplasm, and
shuttle between the cytoplasm and the nucleus via the
CRM1-dependent export pathway (10). A previous report has demonstrated
that hMex-3A and 3B are two novel components of processing bodies
(P bodies), in which they co-localize and interact with the hDcp1a
decapping factor (9). P bodies are
the sites of mRNA decay and storage of non-translated transcripts,
indicating that hMex-3 may be involved in the regulation of a
variety of mRNA decay. In addition, hMex-3A and 3B are also
associated with Argonaute (Ago) proteins. Ago1 and Ago2, the key
components of the RNA-induced silencing complex, have been
implicated in tumor development (11). However, it remains unknown whether
the hMex-3 proteins play a role in human tumorigenesis.
Gastric cancer is one of the most common
malignancies worldwide, particularly in Eastern Asian countries
(12). In this study, we focused
on hMex-3A, one of the hMex-3 proteins, and explored its role in
gastric cancer development and metastasis. The results revealed
that the hMex-3A knockdown inhibited cell proliferation, reduced
the ability of cell transformation and suppressed cell migration.
In addition, it was also found that the expression of hMex-3A was
upregulated in gastric cancer tissues compared with matched
adjacent non-cancerous tissues. Thus, these data provide new
insight into the role of hMex-3 proteins in tumorigenesis, as well
as a novel promising therapeutic target for gastric cancer.
Materials and methods
Tissue specimens and cell culture
Paired gastric cancer and adjacent paracancer
tissues from 22 patients who underwent surgical resection were
obtained with informed consent. The use of these tissue materials
for research was approved by the Ethics Committee of the Shanghai
East Hospital. Clinical and pathological information was extracted
from the medical charts and pathology reports of these patients.
The diagnoses of these gastric cancer samples were verified by
pathologists. The three gastric cancer cell lines, SNU-16, AGS and
BCG823, were obtained from the American Type Culture Collection
(Manassas, VA, USA). Cells were grown in Dulbecco’s modified
Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum
(FBS; Gibco) and antibiotics (50 U/ml penicillin and 50 μg/ml
streptomycin) (Gibco) at 37°C in a 5% CO2-humidified
incubator.
RNA extraction and quantitative real-time
PCR
Total RNA was extracted from the tissue samples or
the cultured cell lines using TRIzol reagent (Invitrogen) and
reverse-transcribed into cDNA using the M-MLV reverse transcriptase
kit (Promega). The following primers were used to amplify the
Mex-3A and β-actin genes: Mex-3A forward,
5′-ATCGTGGGCAGG CAAGGCT-3′ and reverse, 5′-GCTGCTGAGATGATTT CCC-3′;
β-actin forward, 5′-AGAGCCTCGCCTTTGCCGA TCC-3′ and reverse,
5′-CTGGGCCTCGTCGCCCACATA-3′. Quantitative real-time PCR was
performed with a Thermal Cycler Dice Real Time System (Takara) and
SYBR-Green I reagent (Takara). The mRNA level of each sample was
normalized to that of β-actin prior to comparative analysis using
the 2-ΔCt method.
RNA interference and cell
transfection
In order to suppress the hMex-3A expression
in gastric cancer cells, hMex-3A-specific small interference RNAs
(siRNAs) were chemically synthesized (GenePharma). The siRNA
sequences used were as follows: si-2270 (sense, 5′-GUGUUUCCCUUCACU
CUCUdTdT-3′ and antisense, 5′-AGAGAGUGAAGGG AAACACdTdT-3′); si-5932
(sense, 5′-CUAGUGAAGAC ACGUACAAdTdT-3′ and antisense,
5′-UUGUACGUGUCUU CACUAGdTdT-3′). Irrelevant nucleotides not
targeting any annotated human genes were used as the negative
control as follows: si-NC (sense, 5′-UUCUCCGAACGUGUCAC GUdTdT-3′
and antisense, 5′-ACGUGACACGUUCGGAG AAdTdT-3′). The siRNA was
subsequently transfected into the indicated cells at a final
concentration of 50 nM using Lipofectamine 2000 reagent
(Invitrogen) according to the manufacturer’s instructions.
Cell viability assay
Gastric cancer cells transfected with siRNAs were
seeded into 96-well plates at a density of 3×103 cells
per well in 100 μl medium. Cell viability was measured using the
Cell Counting Kit-8 (Dojindo Laboratories) according to the
manufacturer’s instructions. Each assay was independently repeated
at least three times.
Cell cycle analysis
Following transfection with siRNAs at 48 h, SNU-16
cells were harvested, fixed in cold 70% ethanol, washed and
rehydrated in PBS. DNA staining was achieved by treating the cells
with RNase A (10 mg/ml) for 30 min and with propidium iodide (10
μg/ml) (Sigma) for 5 min. Flow cytometry analysis was performed
using a FACSCalibur instrument (Becton-Dickinson, Mountain View,
CA, USA) and CellQuest software (Becton-Dickinson).
Soft agar colony formation assay
hMex-3A knockdown cells or control cells were
suspended in medium containing 0.4% agar and overlaid on 1% agar in
24-well plates (500 cells/well), respectively. After two-three
weeks, colonies were counted and photographed. The results were
expressed as the means ± SD of triplicate counts within the same
experiment.
Cell migration assay
Cell migration assays were performed using 24-well
transwells (8-μm pore size, BD Biosciences) according to the
manufacturer’s instructions. Briefly, BCG-823 cells were
trypsinized and washed three times in DMEM without FBS. A total of
5×104 cells was then suspended in 500 μl DMEM without
FBS and added to the upper chamber, while 750 μl DMEM containing
10% FBS was placed in the lower chamber. The cells were incubated
for 24 to 48 h at 37°C in a 5% CO2-humidified incubator.
The non-migrating cells were removed with cotton swabs, and the
migrated cells were fixed in 4% paraformaldehyde and stained with
0.5% crystal violet. Cells in at least five random microscopic
fields (magnification, ×200) were counted and photographed. All
experiments were performed in duplicate and repeated three
times.
Wound-healing assay
The cells transfected with siRNAs were grown to
90–100% confluence in six-well plates. Cell monolayers were wounded
with a sterile pipette tip and then rinsed with PBS to remove
cellular debris. The phase contrast images of the wounds were
recorded at 37°C for incubations of 0, 24 and 48 h, and three
separate experiments were performed.
Statistical analysis
All statistical analyses were evaluated using the
Student’s t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
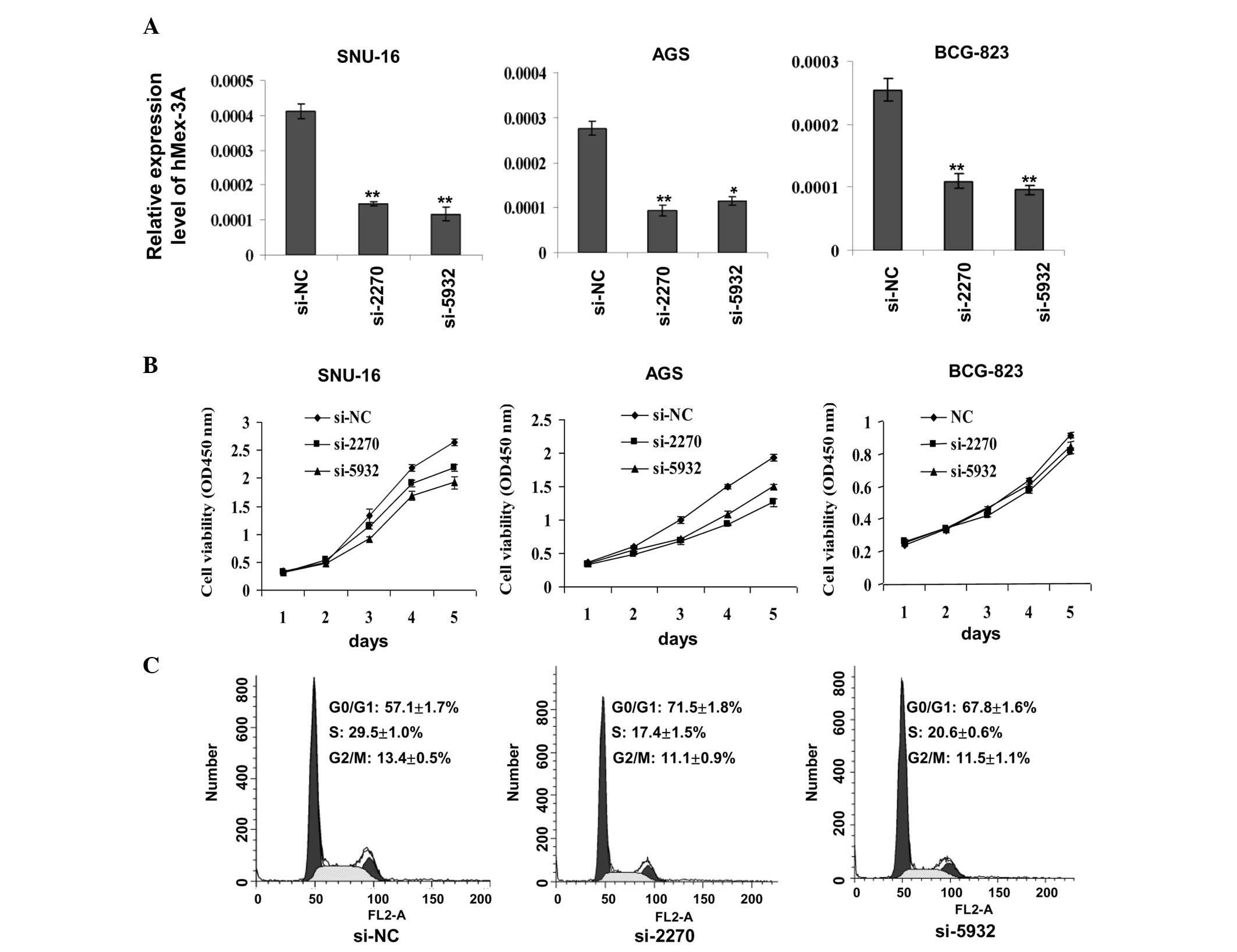
Silencing of hMex-3A by RNA interference
inhibits gastric cancer cell growth
To assess whether hMex-3A has an effect on cell
growth, RNA interference against hMex-3A was carried out in the
three gastric cancer cell lines, SNU-16, AGS and BCG-823. As shown
in Fig. 1A, the chemically
synthesized siRNAs, si-2270 and si-5932, exhibited an efficient
knockdown of hMex-3A relative to the control, si-NC. Cell viability
assays using the Cell Counting Kit-8 indicated that cells in which
hMex-3A was knocked down grew more slowly than in the controls
(shown for SNU-16 and AGS cells) (Fig.
1B). However, there was no marked difference observed in the
BCG-823 cells, implying that the effect of hMex-3A on cell growth
is dependent on cell type. The effect of hMex-3A on the cell cycle
of SNU-16 cells was then evaluated by flow cytometry. As observed
in Fig. 1C, the percentages of
cells in the S phase were 29.5% in those transfected with si-NC,
17.4% in those transfected with si-2270 and 20.6% in those
transfected with si-5932. These results suggest that the silencing
of hMex-3A inhibits cell proliferation by lengthening the cell
cycle.
Knockdown of hMex-3A affects
anchorage-independent growth of SNU-16 and AGS cells
An important hallmark of cellular transformation is
cell anchorage-independent growth. Normal cells often require a
solid substratum on which to proliferate, whereas malignant cells
gain the ability to grow regardless of their attachment status.
Previous studies have reported that SNU-16 and AGS cells have such
an ability of growth (13,14). To examine whether hMex-3A is
capable of contributing to anchorage-independent growth, a soft
agar colony formation assay was performed in these two cell lines.
Compared to those cells transfected with si-NC, the cells
transfected with si-2270 and si-5932 formed smaller and fewer
colonies in the two cell lines (Fig.
2), indicating that hMex-3A knockdown reduced the ability of
anchorage-independent growth of gastric cancer cells.
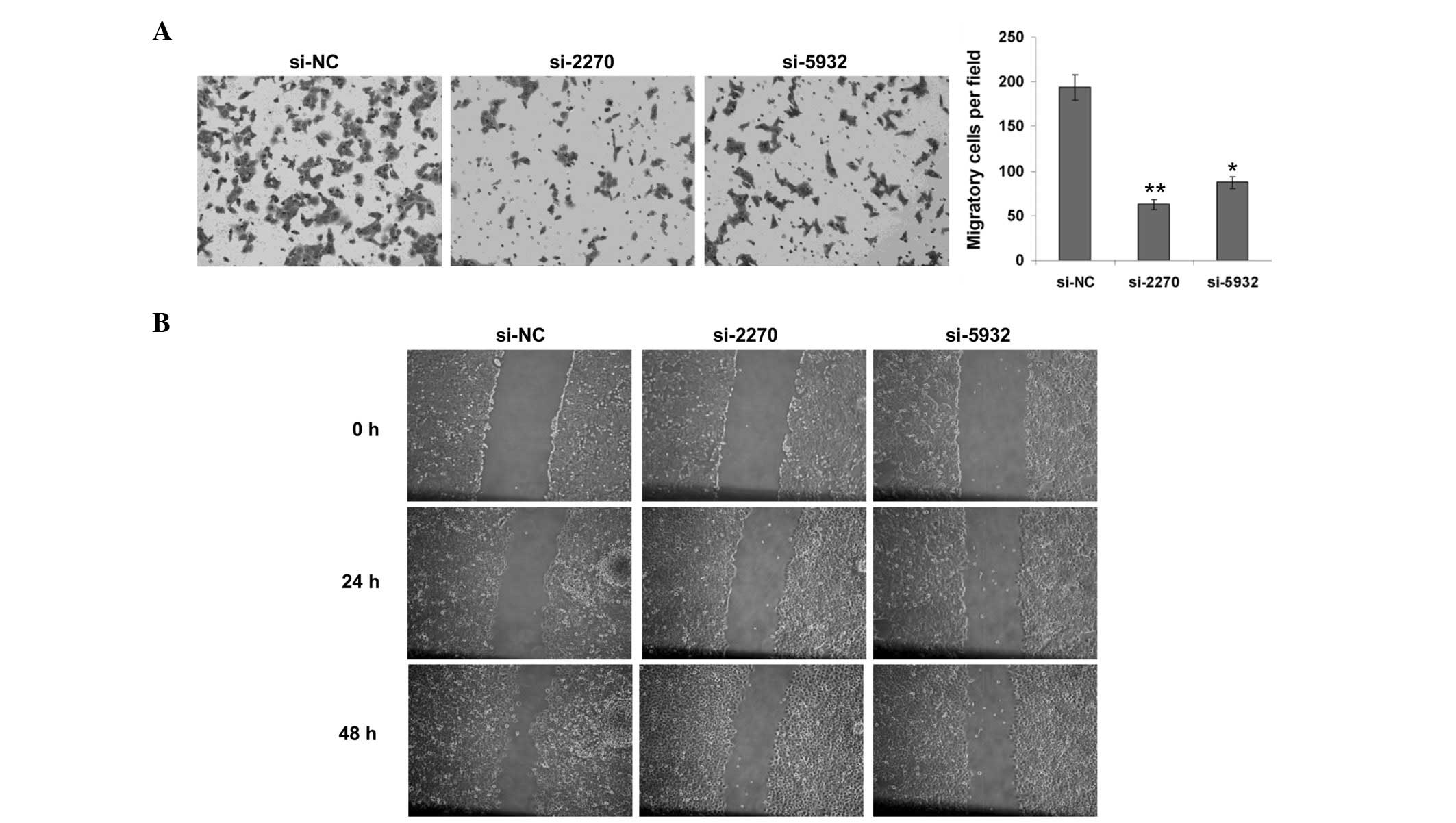
Knockdown of hMex-3A suppresses cell
migration of BCG-823 cells
In order to further characterize the function of
hMex-3A in cancer progression, a Transwell chamber assay was
performed for cell migration in BCG-823 cells. Since the knockdown
of hMex-3A in BCG-823 cells only had a slight effect on cell
growth, the possibility that the impact on cell migration was due
to cell proliferation was excluded. After 24 h of suspending the
cells in the upper chamber, it was found that the cells transiently
transfected with si-2270 and si-5932 migrated to the lower chamber
less than those transiently transfected with si-NC (Fig. 3A). Furthermore, a wound healing
assay was also performed in BGC823 cells. Consistent with the
results from the Transwell assays, hMex-3A knockdown cells
displayed a significant decrease in the cell migration ability
compared with the control (Fig.
3B). Taken together, these data demonstrate that a decrease in
the hMex-3A level markedly influences cell migration, implying that
hMex-3A contributes to the metastasis of gastric cancer.
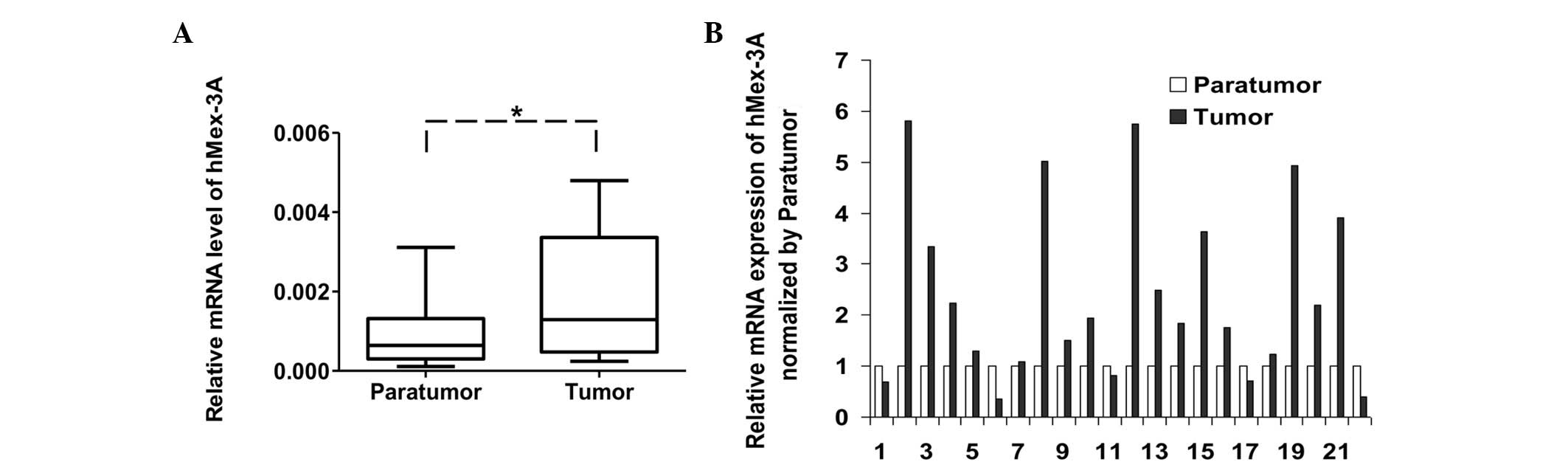
hMex-3A is frequently upregulated in
gastric cancer tissues
To investigate the clinical relevance of hMex-3A in
gastric cancer, the mRNA levels of hMex-3A in 22 pairs of human
gastric cancer samples were measured by quantitative real- time
PCR. The resulting data revealed that hMex-3A was significantly
upregulated in tumor tissues in comparison with adjacent
non-tumorous tissues (Fig. 4A).
Among the samples, 14 of 22 (63.6%) displayed at least a 1.5-fold
increase in tumor tissues compared to paratumor tissues (Fig. 4B). The expression pattern of
hMex-3A in clinical samples suggests its involvement in the
pathogenesis of gastric cancer.
Discussion
Human Mex-3A proteins, including hMex-3A, -3B, -3C
and -3D, are located on distinct chromosomes at 1q22,
15q25.2, 18q21.1 and 19p13.3, respectively
(9). All the four proteins contain
two highly conserved KH domains specifically for RNA binding and a
RING finger domain in their carboxy-terminal region probably
involved in mediating protein-protein interactions (15–17).
The similarity in structure of the hMex-3 proteins implies that
they may share certain similar biological functions. In the present
study, one of the four proteins, hMex-3A, was selected to
investigate its function in gastric cancer development and
progression for the first time. The final data demonstrated that a
reduction in the hMex-3A expression level led to a suppression of
cell growth in an anchorage-dependent and -independent manner.
Furthermore, the silencing of hMex-3A also resulted in a decrease
in the migratory ability of gastric cancer cells. These findings
shed light on the role of hMex-3 proteins, particularly for hMex-3A
in tumorigenesis.
In recent years, RNA interference has been employed
as a mature and powerful strategy for exploring gene functions by
downregulating the expression of targeted genes. We applied this
technique to specifically silence hMex-3A expression in the gastric
cancer cell lines, SNU-16, AGS and BCG-823. Notably, the
significant inhibitory effect on cell growth was present in SNU-16
and AGS cells, but not in BCG-823 cells, indicating that the effect
of hMex-3A on cell growth is dependent on cell type. More
importantly, flow cytometry analysis confirmed the inhibitory
effect on the cell growth of SNU-16 cells. These data imply that
hMex-3A may be essential for maintaining efficient cell division in
gastric cancer cells. Moreover, we also show that the hMex-3A
expression level severely influences cell growth ability in soft
agar. This ability of cell growth in soft agar is assumed to
closely reflect the ability of cell transformation in in
vivo carcinogenesis. Another hallmark of tumors is metastasis.
To date, tumor recurrence and metastasis remain the main obstacles
in treatment. The results of cell metastasis assays revealed that
hMex-3A may be involved in this process. In addition, the role of
hMex-3A involvement in the gastric cancer development was supported
by clinical data. Quantitative real-time PCR results indicated that
the transcription level of hMex-3A was significantly upregulated in
tumor tissues compared to adjacent non-tumorous tissues; however,
this upregulation of hMex-3A did not statistically correlate with
gender, age, tumor size or metastasis. One possible reason for this
may be that we did not have not enough clinical samples for
statistical analysis.
hMex-3 proteins were initially identified in a
screening for genes with similarity to the Caenorhabditis
elegans Mex-3. In the nematode, the Mex-3 protein acts in the
cytoplasm during the early cleavage stages of the embryonic cell
and is localized in P-granules (18), which are cytoplasmic structures
containing RNA and RBPs playing a role in mRNA processing or
packaging. By contrast, hMex-3 proteins are mostly distributed in
the nuclear-perinuclear cell compartment and may play a similar
role in RNA metabolism as RBPs. Deregulation of RBP expression or
activity has been reported in a number of malignancies (4,19).
RBPs are also key components in the determination of microRNA
(miRNA) function, as they control different stages of miRNA
biogenesis and their localization, degradation and activity. The
global downregulation of miRNA expression is an emerging feature in
cancer, and the specific deregulation of certain miRNAs has been
observed in specific tumor types (20,21).
Most notably, hMex-3A has been reported to be associated with Ago
proteins. Ago proteins are components of the RNA-induced silencing
complex and are crucial effectors of RNA silencing (22,23).
Recent studies have revealed that Ago1 and Ago2 play an important
oncogenic role in breast and colon cancer. These insights uncover a
probable mechanism for hMex-3A in RNA regulation, which could have
relevance for cancer development and progression. Taken together,
tumorigenesis is a complicated process and the exact mechanism by
which hMex-3A contributes to tumorigenesis should be explored in
the future.
In conclusion, our present study is the first to
show that the knockdown of hMex-3A using RNA interference can
effectively inhibit cell proliferation and migration in gastric
cancer cells. These results further indicate that hMex-3A may serve
as a potential target for the therapy of gastric cancer, although
additional studies in vivo are necessary.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (81070343) and Shanghai
Excellent Academic leaders Program (08xD14045).
References
|
1
|
Kim MY, Hur J and Jeong S: Emerging roles
of RNA and RNA-binding protein network in cancer cells. BMB Rep.
42:125–130. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lunde BM, Moore C and Varani G:
RNA-binding proteins: modular design for efficient function. Nat
Rev Mol Cell Biol. 8:479–490. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lukong KE, Chang KW, Khandjian EW and
Richard S: RNA-binding proteins in human genetic disease. Trends
Genet. 24:416–425. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bielli P, Busa R, Paronetto MP and Sette
C: The RNA-binding protein Sam68 is a multifunctional player in
human cancer. Endocr Relat Cancer. 18:R91–R102. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shultz JC, Goehe RW, Murudkar CS, et al:
SRSF1 regulates the alternative splicing of caspase 9 via a novel
intronic splicing enhancer affecting the chemotherapeutic
sensitivity of non-small cell lung cancer cells. Mol Cancer Res.
9:889–900. 2011. View Article : Google Scholar
|
|
6
|
Denkert C, Koch I, von Keyserlingk N, et
al: Expression of the ELAV-like protein HuR in human colon cancer:
association with tumor stage and cyclooxygenase-2. Mod Pathol.
19:1261–1269. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gherzi R, Trabucchi M, Ponassi M, et al:
The RNA-binding protein KSRP promotes decay of beta-catenin mRNA
and is inactivated by PI3K-AKT signaling. PLoS Biol. 5:e52006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee HK and Jeong S: Beta-catenin
stabilizes cyclooxygenase-2 mRNA by interacting with AU-rich
elements of 3′-UTR. Nucleic Acids Res. 34:5705–5714.
2006.PubMed/NCBI
|
|
9
|
Buchet-Poyau K, Courchet J, Le Hir H, et
al: Identification and characterization of human Mex-3 proteins, a
novel family of evolutionarily conserved RNA-binding proteins
differentially localized to processing bodies. Nucleic Acids Res.
35:1289–1300. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fornerod M, Ohno M, Yoshida M and Mattaj
IW: CRM1 is an export receptor for leucine-rich nuclear export
signals. Cell. 90:1051–1060. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Parisi C, Giorgi C, Batassa EM, et al:
Ago1 and Ago2 differentially affect cell proliferation, motility
and apoptosis when overexpressed in SH-SY5Y neuroblastoma cells.
FEBS Lett. 585:2965–2971. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Alberts SR, Cervantes A and van de Velde
CJ: Gastric cancer: epidemiology, pathology and treatment. Ann
Oncol. 14(Suppl 2): ii31–36. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim YB, Lee SY, Ye SK and Lee JW:
Epigenetic regulation of integrin-linked kinase expression
depending on adhesion of gastric carcinoma cells. Am J Physiol Cell
Physiol. 292:C857–866. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu X, Li W, Fan X, et al: Identification
and characterization of a novel p42.3 gene as tumor-specific and
mitosis phase-dependent expression in gastric cancer. Oncogene.
26:7371–7379. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Joazeiro CA and Weissman AM: RING finger
proteins: mediators of ubiquitin ligase activity. Cell.
102:549–552. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fang S, Lorick KL, Jensen JP and Weissman
AM: RING finger ubiquitin protein ligases: implications for
tumorigenesis, metastasis and for molecular targets in cancer.
Semin Cancer Biol. 13:5–14. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Draper BW, Mello CC, Bowerman B, Hardin J
and Priess JR: MEX-3 is a KH domain protein that regulates
blastomere identity in early C. elegans embryos. Cell.
87:205–216. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hunter CP and Kenyon C: Spatial and
temporal controls target pal-1 blastomere-specification activity to
a single blastomere lineage in C. elegans embryos. Cell.
87:217–226. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gubin MM, Calaluce R, Davis JW, et al:
Overexpression of the RNA binding protein HuR impairs tumor growth
in triple negative breast cancer associated with deficient
angiogenesis. Cell Cycle. 9:3337–3346
|
|
20
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Janowski BA, Huffman KE, Schwartz JC, et
al: Involvement of AGO1 and AGO2 in mammalian transcriptional
silencing. Nat Struct Mol Biol. 13:787–792. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adams BD, Claffey KP and White BA:
Argonaute-2 expression is regulated by epidermal growth factor
receptor and mitogen-activated protein kinase signaling and
correlates with a transformed phenotype in breast cancer cells.
Endocrinology. 150:14–23. 2009. View Article : Google Scholar
|