Introduction
Gastric cancer is one of the most common cancers
worldwide, accounting for the fourth most frequent malignancy and
the second cause of cancer-related mortality annually (1).Gastric cancer is usually diagnosed in
the later stages of the disease and has poor prognosis due to
insufficient therapeutic options for patients with unresectable,
metastatic, or recurrent gastric cancer. Therefore, investigation
of the molecular mechanisms of gastric carcinogenesis, and the
development of novel therapeutic strategies for the control of
gastric cancer is crucial. The hedgehog pathway is important in
embryonic development, tissue polarity and carcinogenesis (2–4).
Gastric cancer development has been associated with a variety of
genetic alterations, including genes in the Sonic hedgehog (Shh),
Notch, and Wnt signaling pathways (5–9).
Recently, aberrant activation of this pathway was described in
human gastric tumor biopsies, and a high expression of proteins in
the Shh pathway was found in gastric tumor cell lines (10). The canonical hedgehog pathway
includes three hedgehog ligands, sonic (SHH), Indian (IHH), and
desert (DHH) hedgehog, which bind to PTCH, a 12 transmembrane
domain protein that releases smoothened homolog (SMO). SMO
subsequently allows Gli family transcription factors to translocate
to the nucleus and affect expression of target genes. Human
hedgehog-interacting protein (HHIP) was identified by screening a
mouse cDNA expression library for proteins that bind to Shh
(11). HIP binds all three Hh
proteins with an affinity equal to that of Patched-1 (Ptch-1), and
functions to negatively regulate the hedgehog pathway.
Specifically, expression of the HHIP and PTCH genes, two negative
regulators of hedgehog signaling (12,13),
has been shown to be reduced in gastric cancer tissues, but
retained in normal gastric tissues or atypical hyperplasia
(14). However, the clinical
significance of their expression loss in gastric cancer, and the
underlying mechanism responsible remains to be determined.
DNA methylation is a stable albeit reversible
epigenetic change for effective silencing of gene expression
(15), and plays a crucial role in
normal organism development and cellular differentiation as it can
stably alter gene expression patterns in cells. However, it is also
thought that somatic alterations in DNA methylation patterns
contribute to cancer development and aging, particularly the
methylation of tumor suppressor gene promoters in the development
of human cancer (16). In gastric
cancer, DNA methylation of different genes has been studied,
including methylation of the PTCH gene transcriptional regulation
region (17–20). However, to the best of our
knowledge, no study has accurately analyzed the methylation of PTCH
and HHIP gene promoters in gastric cancer. In this study, we first
determined the expression of PTCH and HHIP mRNA and protein in
gastric cancer tissues and adjacent normal tissues, and then
detected methylation of these two genes to associate their
expression and gene promoter methylation in gastric cancer.
Materials and methods
Patient tissue samples
Surgical specimens from 30 patients with gastric
cancer and adjacent normal tissues were collected from the
Department of Surgery, Zhangjiagang First Hospital (Jiangsu, China)
between 2008 and 2010. Surgically resected tissue specimens were
snap-frozen in liquid nitrogen until use. The specimens were
examined by at least two experienced pathologists and tumors were
classified according to the tumor-node-metastasis (TNM)
classification. Patients included 17 males and 13 females with an
age range of 36–72 years (mean, 60.82 years). According to the TNM
staging system, 20 cases were stage II and 10 cases were stage III.
Sixteen tumors were well- and moderately differentiated, while 14
were poorly differentiated. Twelve tumors had lymph node
metastasis, but 18 cases were without lymph node metastasis. The
study was approved by the Ethical Committee of National Drug
Clinical Trial Institution of The Second Affiliated Hospital of
Soochow University, and written informed consent was obtained from
all participants.
RNA isolation and reverse
transcription-PCR (RT-PCR)
RNA was isolated from the frozen gastric cancer and
normal tissues. Briefly, tissue samples of ~100–300 μg were
pulverized over liquid nitrogen, placed in a denaturing buffer, and
disrupted using a Mixer Mill (Retsch) or rotor-stator homogenizer
(Omni International, Kennesaw, GA, USA). RNA was isolated from the
homogenate either by using phenol-chloroform extraction followed by
precipitation with isopropanol and washing with 70% ethanol, or
using an RNeasy kit (Qiagen, Dusseldorf, Germany). RNA
concentration was measured using a spectrophotometer (Hongji,
Shanghai, China) and the absorbance was read at 260 nm. cDNA was
then converted from RNA using a reverse transcription kit (Jingmei,
Shanghai, China) according to the manufacturer’s instructions, and
stored at −20°C. For PCR amplification of gene expression, a 25 μl
reaction mixture containing 2 μl of sample cDNA, 2.5 μl 10X PCR
reaction buffer, 0.5 μl of 10 mM/l dNTP intermixture, 0.2 μl of 10
mM/l of each primer, 0.5 μl of 5 U/μl Taq DNA polymerase, and 19.1
μl ddH2O. PCR amplification was set for an initial cycle
of 94°C for 3 min and 32 cycles of 94°C for 30 sec, 58°C for 45
sec, and 72°C for 45 sec and a final extension at 72°C for 10 min.
The amplification of each gene was in the linear range. RT-PCR
products were then separated on ethidium bromide-stained 2% agarose
gels. The primer sequences for PTCH, HHIP, and β-actin genes are
listed in Table I.
 | Table IPrimers and PCR product size. |
Table I
Primers and PCR product size.
| Methods | Primers | Sequence (5′-3′) | Length (bp) | Temperature (°C) |
|---|
| RT-PCR | PTCH |
CTGCTGGTATGCTCGGGACTG
TAAATCGCTCGGAGTTTCTGG | 175 | 60 |
| HHIP |
CTGCTTCTGTATTCAGGAGGTT
GGGATGGAATGCGAGGCTTA | 229 | 55 |
| β-actin |
AGAGCTACGAGCTGCCTGAC
AGCACTGTGTTGGCGTACAG | 184 | 60 |
| BSP | PTCH |
GGGAGTATTTGGGTGGTATATT
ATTNCTACAAAAAAACACCACCTTTC | 349 | 60 |
| HHIP |
GGGGAGGAGAGAGGAGTTTG
CCCRACRACCTCCCTACTA | 243 | 60 |
| β-actin |
AGAGCTACGAGCTGCCTGAC
AGCACTGTGTTGGCGTACAG | 184 | 60 |
| MSP (HHIP) | Methylation |
GTAGTAGTCGGGTAGTTTCGGAATTTTC
AAAAACGACTAACCGCGACG | 190 | 60 |
| Non-methylation |
AGTAGTTGGGTAGTTTTGGAATTTTTGG
AAAAACAACTAACCACAACA | 188 | 60 |
Quantitative RT-PCR using the ABI PRISM™ 7700
Sequence Detection System and TaqMan™ chemistry was also performed.
Reactions contained 0.3 μl of each primer, 0.1 μl cDNA, 0.25 U/ml
MultiScribe reverse transcriptase, 0.4 U/ml RNase inhibitor, and 1X
PCR master mix (Applied Biosystems, Foster City, CA, USA). qRT-PCR
data were quantified using the comparative CT method (i.e., CT
values were normalized to the housekeeping gene β-actin). The
melting curve was analyzed to ensure the specificity of the PCR
products. The RQ values of PTCH and HHIP expression in tumor
samples were compared to that of pooled adjacent normal tissue
samples. To ensure experimental accuracy, all reactions were
performed in triplicate. The primer sequences of PTCH, HHIP and the
β-actin genes are listed in Table
I.
Immunohistochemistry
Representative formalin-fixed and paraffin-embedded
tissue sections (5-μm) were prepared and used for
immunohistochemistry with specific antibodies to PTCH (sc-6149;
Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) and HHIP
(sc-25465; Santa Cruz, Biotechnology Inc.). Briefly, the tissue
sections were deparaffinized, followed by rehydration with serially
decreased concentrations of ethanol, and immersed in 3%
H2O2 (in distilled water) for 10 min to
inhibit endogenous peroxidase activity. Antigen retrieval was
performed in citrate buffer (pH 6.0) and the tissue sections were
incubated with a normal goat serum to block non-specific antibody
binding for 20 min at room temperature. The sections were then
incubated with primary antibodies against PTCH (diluted at 1:100)
and HHIP (diluted at 1:50) at 37°C in humid chambers for 2 h. After
washing 3 times with PBS, the sections were incubated with a
biotinylated secondary antibody (the goat anti-rabbit IgG) and
streptavidin conjugated to horseradish peroxidase for 20 min at
37°C, followed by PBS washing. The sections were incubated with DAB
substrate for 10 min and counterstained with haematoxylin. Negative
controls were performed in all the cases by omitting the primary
antibodies. The sections were reviewed under a microscope and
scored. Positive staining was reviewed as yellow or brown staining
in the stained sections. Each section was counted for 200 cancer
cells in five different high-power fields, and the section was
reviewed as positive staining if ≥5% tumor cells were stained
positively.
DNA isolation and methylation-specific
PCR (MSP) and bisulfite sequencing PCR (BSP)
DNA was extracted according to the standard
protocol. Briefly, 50 mg of tissue specimens were placed into a
mortar, (cells were repeatedly washed with PBS buffer via
centrifugation), and ground into powder. TE buffer (1 ml), 50 μl of
10% SDS, and 2 μl of 10 mg/ml RNase, were added followed by
incubation in a 37°C water bath for 1 h, and the addition of 5 μl
of 20 mg/ml proteinase K and a subsequent incubation in a 55°C
water bath overnight. The following day, an equal volume of
saturated phenol was added to the tissue mixture and centrifuged at
16,000 × g for 10 min at 4°C. The supernatant was transferred to a
new microtube and an equal volume of phenol/chloroform/isopropyl
alcohol mixture was then added followed by centrifugation at 16,000
× g for 10 min at 4°C. The supernatant was transferred to a new
microtube and the aforementioned protocol was repeated again. The
final supernatant was precipitated with two volumes of ethanol and
one tenth volume of 3 M sodium acetate at −20°C for 2 h, and
centrifuged at 16,000 × g for 10 min at 4°C. The supernatant was
discarded and then 70% ethanol was added followed by centrifugation
at 16,000 × g for 10 min at 4°C. The supernatant was then dissolved
in 80 μl TE buffer and stored at −20°C until use.
For MSP and BSP, DNA samples were treated with
bisulfite modification as previously described (21). One microgram of genomic DNA was
treated using a Beijing Science and Technology Development Co.
methylation kit (Beijing, China) according to the manufacturer’s
instructions. The primers of MSP and BSP used are shown in Table I. For MSP, 1 μl of the modified DNA
was amplified using MSP primers that specifically recognized the
methylated (M) or unmethylated (U) DNA after bisulfite conversion.
CpGnome Universal Methylated DNA (S7821) and CpGnome Universal
Unmethylated DNA (S7822) (Chemicon Co., Temecula, CA, USA) were
used as the controls for methylated and unmethylated detection,
respectively. Amplification products were resolved with 2% agarose
gel electrophoresis and visualized with ethidium bromide staining.
For BSP clone sequence analysis, the PCR products were submitted to
Shanghai Sangon Co. Ltd. (Shanghai, China) for subcloning of PCR
products and then amplified. Twenty clones were chosen for DNA
sequencing by Shanghai Sangon Co. Ltd.
Cell culture and 5-aza-dc treatment
The AGS gastric cancer cell line was purchased from
the Shanghai Institute Cell Bank (Shanghai, China) and cultured in
RPMI-1640 (Invitrogen, Carlsbad, CA, USA) supplemented with 10%
fetal bovine serum (FBS), streptomycin 100 μg/ml, and penicillin
100 U/ml at 37°C in a humidified atmosphere with 5% CO2.
For our experiments, AGS cells were then treated with 5-aza-dc
(Sigma, St. Louis, MO, USA) for 3 days and the control cells were
cultured with an equivalent volume of dimethyl sulfoxide
(DMSO).
Cell viability MTT assay
Exponentially growing cells (2×103/200
μl/well) were seeded in 96-well plates and were grown up to 4 days.
The surviving cells were evaluated by adding 50 μl of 2.5 mg/ml
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
Sigma) in DMSO into the culture medium and incubated for 4 h at
37°C in the dark. Subsequently, the culture medium was removed and
100 μl of DMSO were added into the plates and absorbance was
measured at 570 nm with a microplate reader (Bio-Rad, Hercules, CA,
USA).
Annexin V/PI-flow cytometry assay
AGS gastric cancer cells were grown and treated with
or without 5-aza-dc for 3 days. Approximately 1×106 AGS
cells were centrifuged at 400 × g for 10 min to remove the culture
medium. Buffer (20 ml) and 60 ml deionized water were added to the
cells, and then the cells were centrifuged at 400 × g for 5 min and
washed with PBS. Approximately 5–12.5×104 cells were
used for the assay. Briefly, the cells were re-suspended in 195 μl
of binding buffer to reach a final concentration of
2–5×105 cells/ml. Annexin V/FITC (5 μl; from BD
Biosciences, Franklin Lakes, NJ, USA) was added and incubated for
10 min at room temperature in the dark. After washing with binding
buffer, 10 μl of propidium iodide (PI, 20 μg) was added to the cell
solution. The apoptosis rate was measured with a flow cytometer
(Human Biotech, Shanghai, China), and the data were summarized as
mean ± SD compared to the control cells.
Statistical analysis
The significance of data was analyzed by the
Chi-square test, Student’s t-test, and correlation analysis using
SPSS software, version 16.0 (SPSS, Chicago, IL, USA). P<0.05 was
considered statistically significant.
Results
Expression of PTCH and HHIP mRNA in
gastric cancer tissues
In this study, we first detected the expression of
PTCH and HHIP mRNA in gastric cancer tissues and adjacent normal
tissues and AGS gastric cancer cells by RT-PCR. The data showed
that the positive rate of PTCH mRNA in gastric cancer tissues was
40% (12/30) compared to that of adjacent normal tissues (83.3%,
25/30), while the positive rate of HHIP mRNA was 30% (9/30) in
gastric cancer tissue compared to that of adjacent normal tissues
(66.7%, 20/30) (Fig. 1 and
Table II). Data from our qRT-PCR
analysis were consistent with the RT-PCR data (Fig. 1D). We then associated PTCH and HHIP
mRNA expression with clinicopathological data from the gastric
cancer patients, and found that PTCH expression was associated with
tumor differentiation (P<0.05, Table III), but not with other factors,
such as tumor stages or lymph node metastasis (P>0.05).
 | Table IIExpression of PTCH and HHIP mRNA and
protein. |
Table II
Expression of PTCH and HHIP mRNA and
protein.
| | PTCH expression | HHIP expression |
|---|
| |
|
|
|---|
| | mRNA | Protein | mRNA | Protein |
|---|
| |
|
|
|
|
|---|
| Tissue | N | Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative |
|---|
| Carcinoma | 30 | 12 | 18 | 10 | 20 | 9 | 21 | 9 | 21 |
| Normal tissue | 30 | 25 | 5 | 24 | 6 | 20 | 10 | 18 | 12 |
| χ2
test | | 1.2 | | 3.333 | | 4.8 | | 4.8 | |
| P-value | | 0.273 | | 0.068 | | 0.028 | | 0.028 | |
 | Table IIIAssociation of PTCH and HHIP mRNA
expression with clinicopathological data. |
Table III
Association of PTCH and HHIP mRNA
expression with clinicopathological data.
| | PTCH | HHIP |
|---|
| |
|
|
|---|
| Clinical
characteristics | Total | Mean ± SD | t-test | P-value | Mean ± SD | t-test | P-value |
|---|
| Gender |
| Male | 17 | 1.6792±0.04060 | −0.387 | 0.706 | 1.3562±0.02339 | 0.545 | 0.596 |
| Female | 13 | 1.6983±0.02637 | | | 1.3386±0.01571 | | |
| Age (years) |
| <50 | 11 | 1.6867±0.04767 | 0.114 | 0.912 | 1.3502±0.02738 | 0.012 | 0.991 |
| ≥50 | 19 | 1.6812±0.02312 | | | 1.3497±0.01534 | | |
| TNM stage |
| II | 20 | 1.6841±0.05262 | −0.482 | 0.641 | 1.3448±0.01786 | −0.031 | 0.976 |
| III | 10 | 1.7084±0.02553 | | | 1.3455±0.01975 | | |
|
Differentiation |
| Well and
moderate | 16 | 1.7589±0.01190 | 2.914 | 0.012 | 1.3543±0.02174 | 0.972 | 0.394 |
| Poor | 14 | 1.6410±0.02906 | | | 1.3366±0.01468 | | |
| Lymph node
metastasis |
| Yes | 12 | 1.6860±0.04352 | −0.370 | 0.718 | 1.3524±0.02509 | 0.162 | 0.875 |
| No | 18 | 1.7021±0.02329 | | | 1.3489±0.01705 | | |
Expression of PTCH and HHIP proteins in
gastric cancer tissues
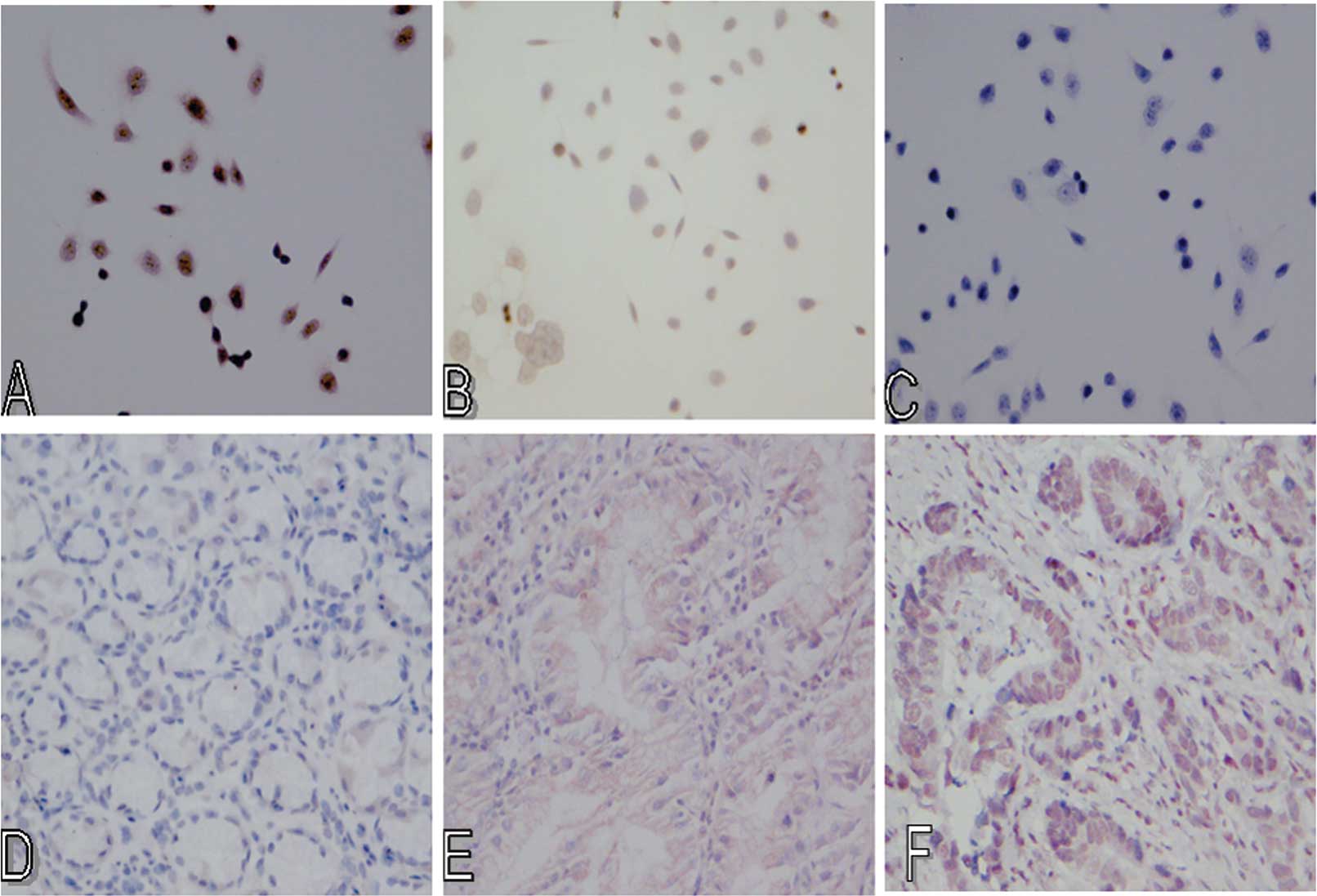
Immunohistochemistry was performed to analyze the
expression of PTCH and HHIP proteins in gastric cancer and adjacent
normal tissues. Results showed positive staining of PTCH and HHIP
proteins to be brown-yellow or brown granules, mainly expressed in
the cell membrane, and rare in the cytoplasm (Fig. 2). We then associated the expression
of PTCH and HHIP proteins with their mRNA levels and found a
positive association between them (Pearson’s test, r=0.902, P=0.000
and r=0.935, P=0.000, respectively; Table IV). Statistical analysis was
performed for the association between the expression of PTCH and
HHIP proteins and clinicopathological characteristics. However, we
did not find any statistical association, such as gender, age, TNM
stage, tumor differentiation, and lymph node metastasis (Table V).
 | Table IVAssociation of PTCH and HHIP mRNA
levels with their protein expression. |
Table IV
Association of PTCH and HHIP mRNA
levels with their protein expression.
| PTCH protein | | HHIP protein |
|---|
|
| |
|
|---|
| PTCH mRNA | Positive | Negative | HHIP mRNA | Positive | Negative |
|---|
| Positive | 32 | 5 | Positive | 26 | 3 |
| Negative | 2 | 21 | Negative | 1 | 30 |
| Pearson
correlation | r=0.902 | | Pearson’s
correlation | r=0.935 | |
| P=0.000 | | | P=0.000 | |
 | Table VImmunohistochemistry expression of
PTCH and HHIP in different clinical and pathological features. |
Table V
Immunohistochemistry expression of
PTCH and HHIP in different clinical and pathological features.
| | PTCH | HHIP |
|---|
| |
|
|
|---|
| Clinical
features | Total | Positive | Negative | χ2
test | P-value | Positive | Negative | χ2
test | P-value |
|---|
| Gender |
| Male | 17 | 5 | 12 | 0.533 | 0.144 | 4 | 13 | 0.533 | 0.465 |
| Female | 13 | 5 | 8 | | | 5 | 8 | | |
| Age (years) |
| <50 | 11 | 4 | 7 | 2.133 | 0.144 | 5 | 6 | 2.133 | 0.144 |
| ≥50 | 19 | 6 | 13 | | | 4 | 15 | | |
| TNM stage |
| II | 20 | 7 | 13 | 3.333 | 0.068 | 5 | 15 | 2.133 | 0.144 |
| III | 10 | 3 | 7 | | | 4 | 6 | | |
|
Differentiation |
| Well and
moderate | 16 | 6 | 10 | 0.133 | 0.715 | 6 | 10 | 0.133 | 0.715 |
| Poor | 14 | 4 | 10 | | | 3 | 11 | | |
| Lymph node
metastasis |
| Yes | 12 | 5 | 7 | 1.2 | 0.273 | 2 | 10 | 1.2 | 0.273 |
| No | 18 | 5 | 13 | | | 7 | 11 | | |
Detection of PTCH and HHIP gene promoter
methylation in gastric cancer tissues
To investigate the mechanism underlying the loss of
expression of PTCH and HHIP, we detected PTCH and HHIP gene
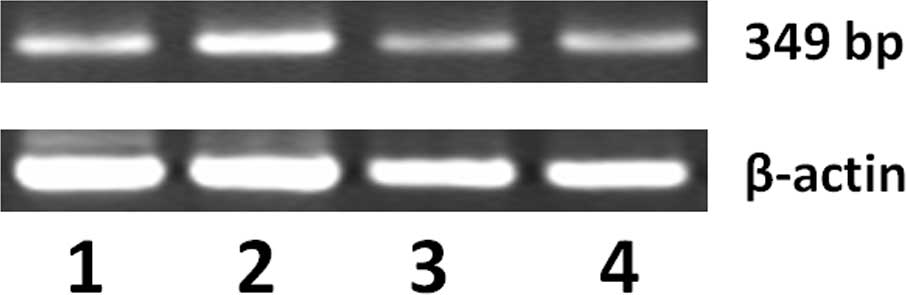
promoter methylation in gastric cancer tissues. We detected HHIP
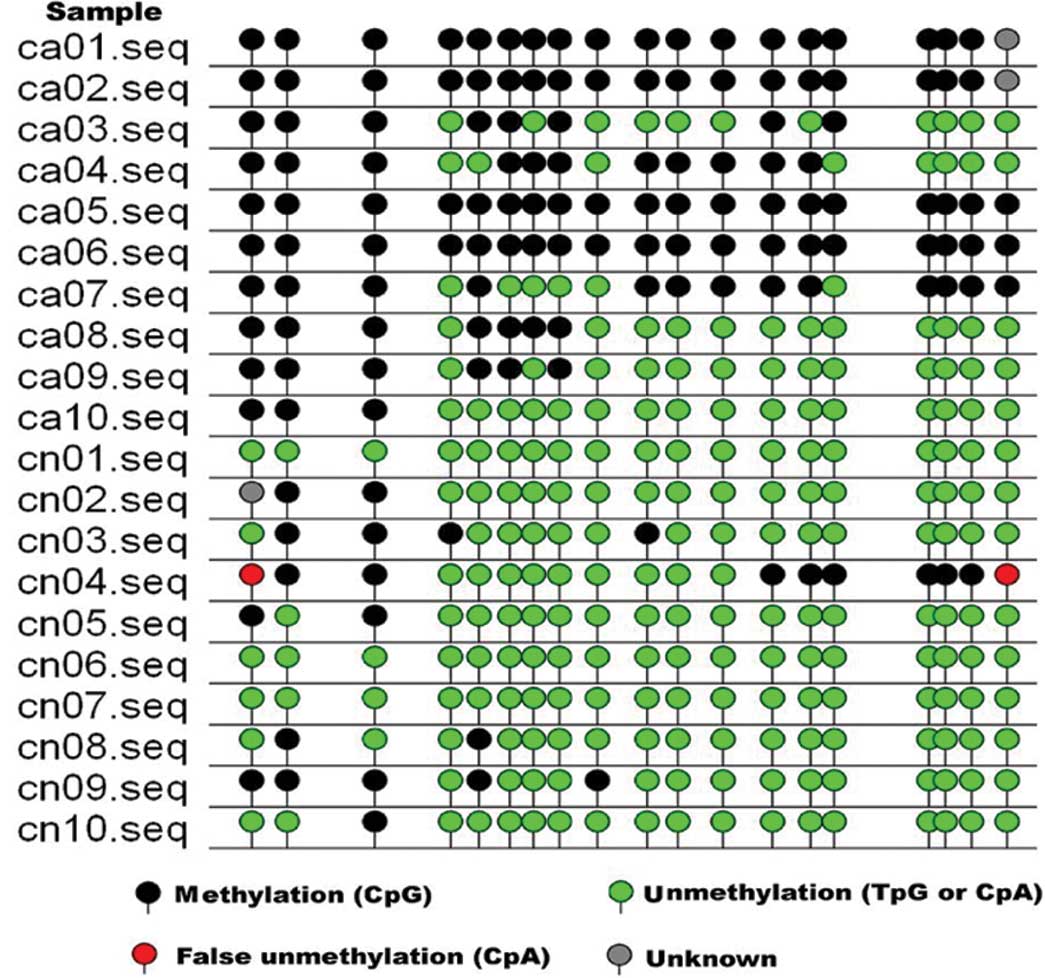
gene promoter methylation by MSP. As shown in Fig. 3, a number of gastric cancer tissues
had methylated HHIP gene promoters compared to adjacent normal
tissues. We then associated HHIP gene promoter methylation with
their mRNA expression and found that they were negatively
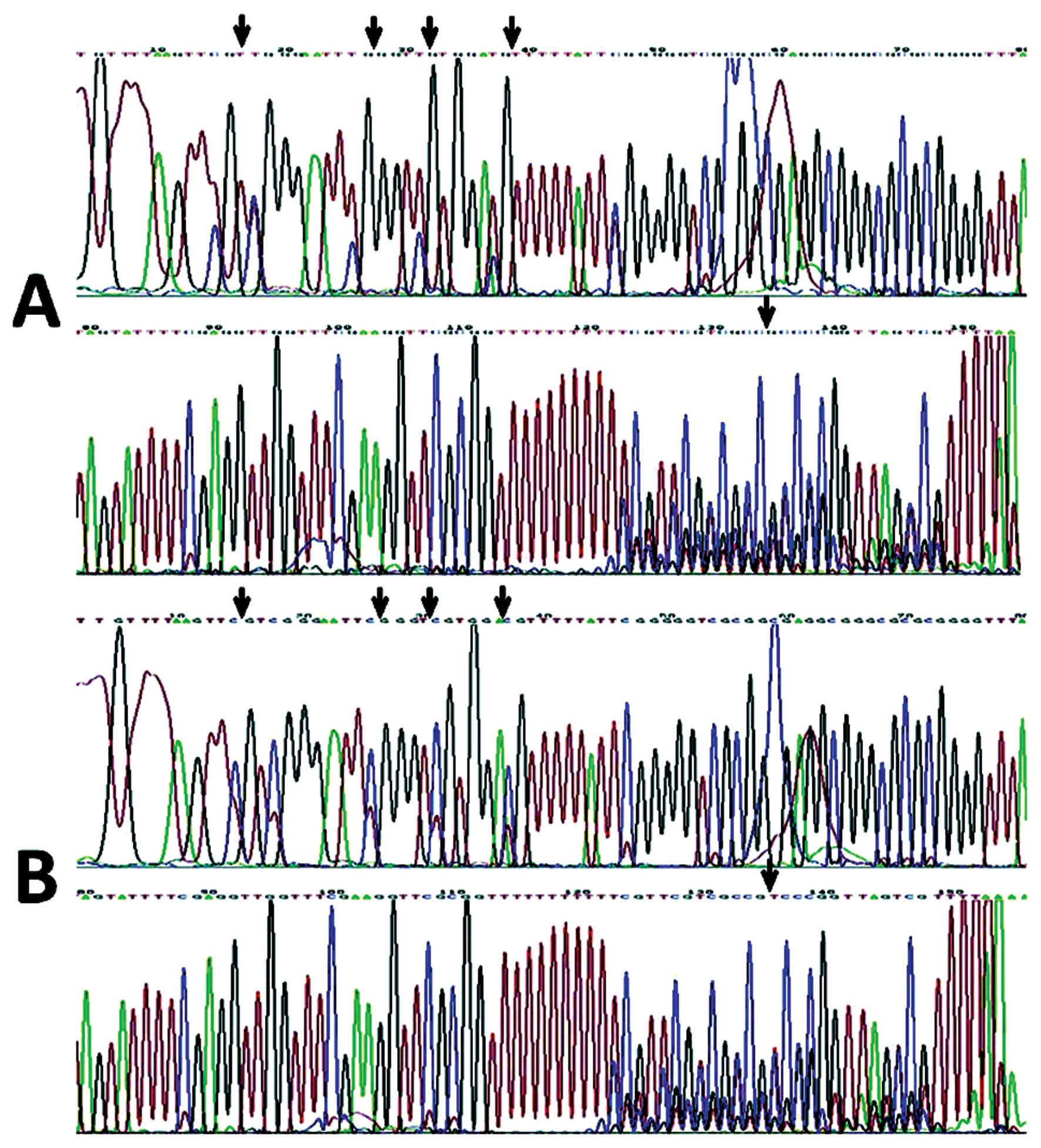
associated (Spearman’s test, r=−0.380, P=0.000). Fig. 4 shows the sequences of the
methylated and unmethylated clones of the MSP product of the HHIP
gene in gastric cancer; the arrows in Fig. 4A represent the methylated spots,
while the arrows in Fig. 4B
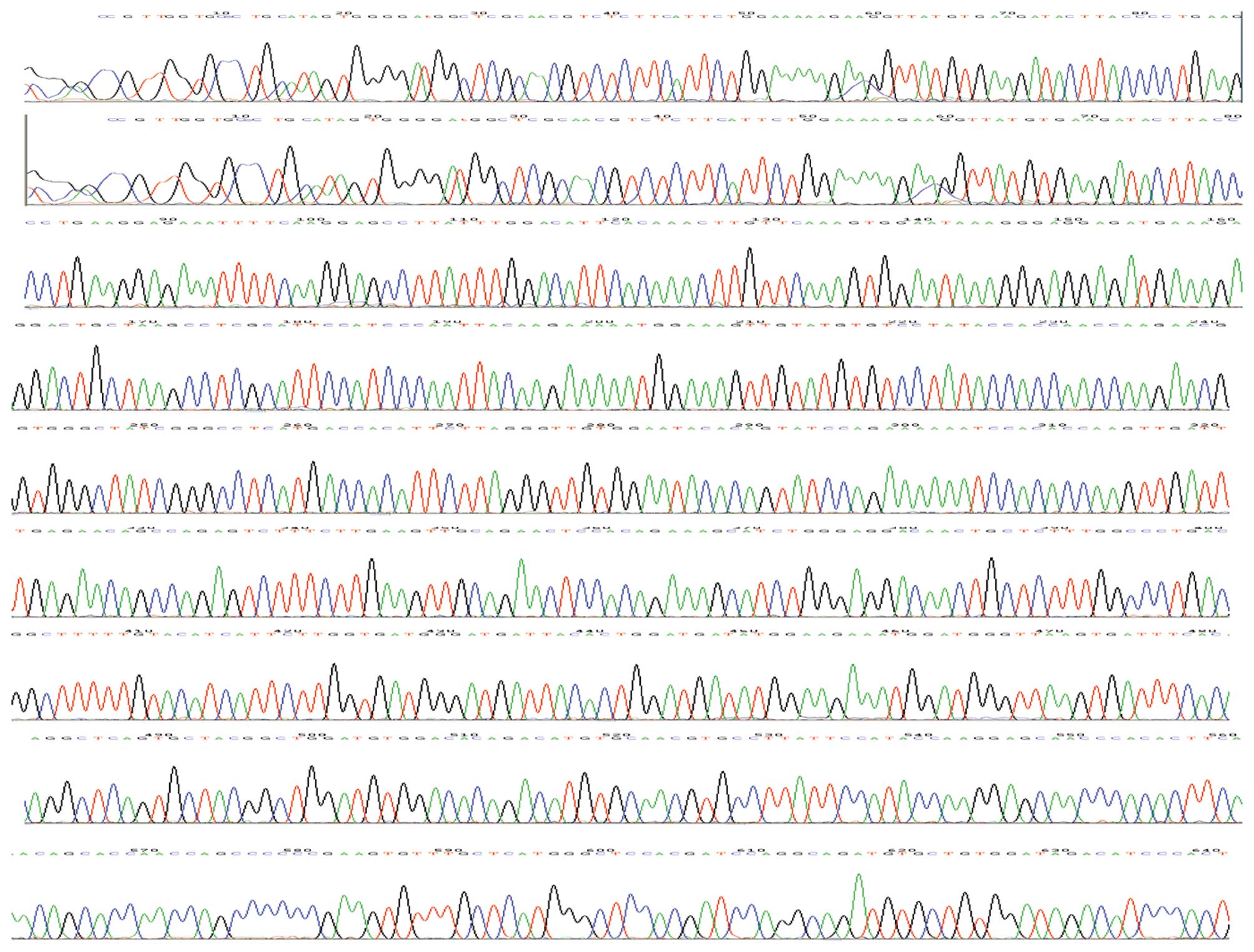
represent the unmethylated spots. As shown in Fig. 5 PTCH gene promoter methylation was
identified by BSP. Furthermore, we performed bisulfite sequencing
PCR analysis through Shanghai Sangon Co., and identified a negative
correlation between PTCH expression and methylation (Spearman’s
correlation r=−.693, P=0.000, Table
VI, Figs. 6 and 7).
 | Table VIAssociation of PTCH expression with
the gene promoter methylation. |
Table VI
Association of PTCH expression with
the gene promoter methylation.
| PTCH
expression | Methylation |
|---|
| Cancer | 1.7537±1.15046 | 0.5700±0.31724 |
| Normal tissues | 2.7993±1.45805 | 0.3743±0.31283 |
Effects of drug 5-aza-dc on the
regulation of gastric cancer cell viability and apoptosis
We evaluated a DNA demethylation agent, 5-aza-dc, on
the regulation of gastric cancer cell viability using the MTT
assay. Our data showed that different concentrations of 5-aza-dc
reduced AGS cell viability (Table
VII), and the optimal dose was 2.0 μm. The apoptosis rate was
detected following treatment with 5-aza-dc and it was found that
2.0 μm of 5-aza-dc significantly induced AGS cells to undergo
apoptosis (Table VIII, Fig. 8).
 | Table VIIReduction of cell viability by
5-aza-dc treatment of gastric cancer cells. |
Table VII
Reduction of cell viability by
5-aza-dc treatment of gastric cancer cells.
| 5-aza-dc (μM) | N | OD | P-value | t-test |
|---|
| 0.5 | 3 |
0.8305±0.08756a | 0.017 | 3.514 |
| 1.0 | 3 |
1.0295±0.04447a | 0.042 | 2.709 |
| 2.0 | 3 |
0.8150±0.04272a | 0.001 | 6.957 |
| 5.0 | 3 |
0.9933±0.03007a | 0.004 | 5.025 |
| 10.0 | 3 |
1.2333±0.07932a | 0.403 | −0.913 |
| Control | 3 | 1.1652±0.03686 | | |
 | Table VIIIInduction of apoptosis by 5-aza-dc
treatment of gastric cancer cells (%). |
Table VIII
Induction of apoptosis by 5-aza-dc
treatment of gastric cancer cells (%).
| 5-aza-dc dose
(μM) | N | Apoptosis (%) | P-value |
|---|
| 0.5 | 3 | 15.52±0.28 | 0.06 |
| 1.0 | 3 | 15.89±0.09 | 0.011 |
| 2.0 | 3 | 33.39±0.4 | 0.000 |
| 5.0 | 3 | 23.44±0.38 | 0.001 |
| 10.0 | 3 | 16.11±0.08 | 0.122 |
| Control | 3 | 14.51±0.25 | |
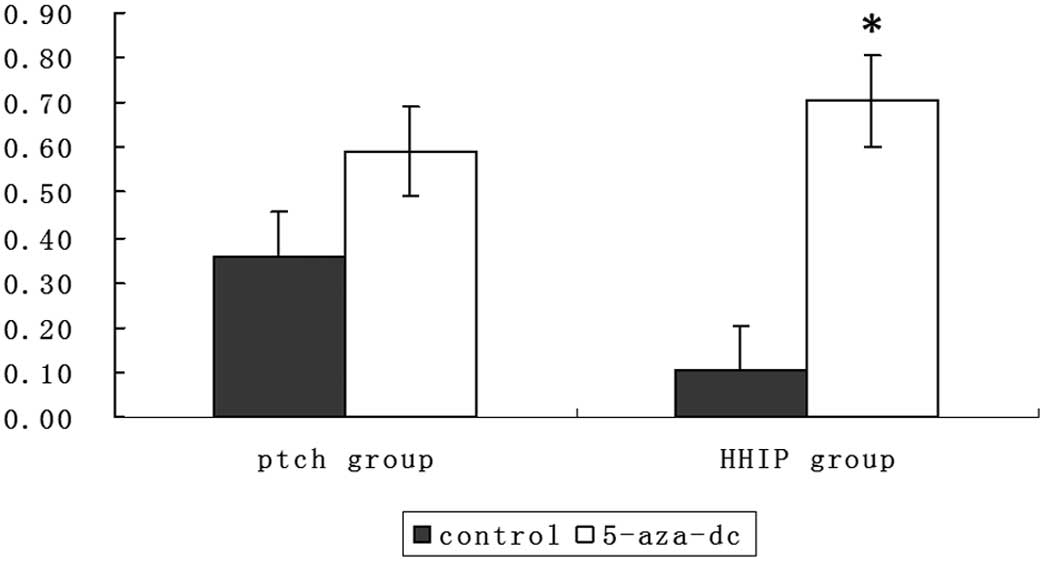
To determine whether the effects of 5-aza-dc
treatment occurred through upregulation of PTCH and HHIP
expression, we assessed PTCH and HHIP expression. As shown in
Fig. 9, PTCH and HHIP expression
following treatment with 5-aza-dc was upregulated, and HHIP mRNA
expression was much higher in the presence of 5-aza-dc treatment
compared to no treatment (P<0.05). The expression of PTCH mRNA
had no statistically significant changes. As shown in Fig. 10, methylation of PTCH and HHIP
gene promoters in AGS cell was reversed by 5-aza-dc treatment. The
5-aza-dc treatment lead to a marked decrease in the methylation of
PTCH and HHIP gene promoters.
Discussion
In this study, we first analyzed the expression of
PTCH and HHIP mRNA and protein in gastric cancer tissues and found
that expression of the two genes was reduced in gastric cancer
tissues compared to adjacent normal tissues. We also associated the
expression of PTCH and HHIP protein with their mRNA levels and
detected a positive association between them. Methylation of PTCH
and HHIP gene promoters was identified and our data showed that a
number of gastric cancer tissues have methylated PTCH and HHIP gene
promoters compared to the adjacent normal tissues, which was
inversely associated with expression of these two genes. Gastric
cancer AGS cells were then treated with 5-aza-dc and it was found
that 5-aza-dc reduced AGS cell viability and induce the cells to
undergo apoptosis, which was associated with upregulation of the
HHIP mRNA expression in AGS cells. Data from the present study
indicate that PTCH and HHIP genes may be novel targets for the
future control of gastric cancer.
The findings of this study on the reduced expression
of PTCH and HHIP mRNA and protein are consistent with those of a
previous study (22).
Specifically, the Hedgehog family of ligand proteins appears to be
important in the initiation of tissue repair, while the gastric
epithelial cells are constantly replenished from progenitor
populations, and these proteins are also essential for the
regeneration process. However, aberrant expression and regulation
of these proteins could alter homeostasis and neoplastic
transformation of the stomach epithelium (22,23).
Nevertheless, our current data showed that expression of both PTCH
and HHIP mRNA and protein are not associated with
clinicopathological characteristics, such as gender, age, TNM
stages, and lymph node metastasis, with the exception of the fact
that expression of PTCH mRNA is associated with tumor
differentiation in gastric cancer tissues. However, these data need
to be confirmed using a large sample size.
We also determined the potential cause of the loss
of PTCH and HHIP expression in gastric cancer tissues by examining
their gene promoter methylation status. The findings of the present
study show that PTCH and HHIP gene promoters were aberrantly
methylated in gastric cancer, which was reversely associated with
expression of their mRNA and proteins in gastric cancer. Moreover,
we found that the degree of methylation of the two gene promoters
in the gastric cancer AGS cell line was 99.7±0.67 and 99.2±0.45%,
respectively. Specifically, methylation of gene promoters is a
common epigenetic modification to silence gene expression, which is
occasionally reversed using DNA de-methylation agents, such as
5-aza-dc. Thus, the reverse of HHIP and Ptched gene promoter
methylation may provide a novel therapy for gastric cancer.
In vitro experiments were conducted to verify
whether 5-aza-dc was able to restore expression of the two genes
via de-methylation of Ptched and HHIP gene promoters. The results
showed that 5-aza-dc induced PTCH and HHIP expression, which was
associated with a reduction in tumor cell viability and induction
of apoptosis in AGS cells. Of note, the proportion of apoptosis was
not associated with drug concentration. When the concentration of
5-aza-dc was 2.0 μm, the apoptosis rate was significantly higher
than at other concentrations. Furthermore, we analyzed the
methylation status of 46 CpG dinucleotides in the promoter region
of the HHIP gene and the methylation status of 19 CpG dinucleotides
in the promoter region of the PTCH gene. We found that the
demethylation reagent 5-aza-dc reversed the methylation of PTCH and
HHIP gene promoters. Thus, we can conclude that methylation is the
cause of downregulated PTCH and HHIP gene expression in gastric
cancer tissue samples. DNA methylation is one of the most important
mechanisms in the regulation of gene expression, and may also play
a role in carcinogenesis. Our data suggest that the enhanced
expression of PTCH and HHIP genes could strengthen the negative
feedback function of PTCH and HHIP, which may provide novel targets
for the future treatment of gastric cancer.
In summary, we have demonstrated that the silencing
of PTCH and HHIP expression in gastric cancer tissue specimens is
due to methylation of their gene promoters. Epigenetic reactivation
of PTCH and HHIP expression likely modulates hedgehog pathway
activity in gastric cancer. Thus, 5-aza-dc treatment was able to
upregulate the expression of HHIP and PTCH genes and lead to a
reduction in tumor cell viability and induction of apoptosis.
Limitations of this study include its small sample size and the
fact that it is not a molecular mechanistic study of PTCH and HHIP
downstream genes.
Acknowledgements
We thank Medjaden Bioscience Limited for assisting
in the preparation of this manuscript.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Lum L and Beachy PA: The Hedgehog response
network: sensors, switches, and routers. Science. 304:1755–1759.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hooper JE and Scott MP: Communicating with
Hedgehogs. Nat Rev Mol Cell Biol. 6:306–317. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chaudary N, Pintilie M, Hedley D, Fyles
AW, Milosevic M, Clarke B, Hill RP and Mackay H: Hedgehog pathway
signaling in cervical carcinoma and outcome after chemoradiation.
Cancer. 118:3105–3115. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zardawi SJ, O’Toole SA, Sutherland RL and
Musgrove EA: Dysregulation of Hedgehog, Wnt and Notch signalling
pathways in breast cancer. Histol Histopathol. 24:385–398.
2009.PubMed/NCBI
|
|
6
|
Pasca di Magliano M and Hebrok M: Hedgehog
signalling in cancer formation and maintenance. Nat Rev Cancer.
3:903–911. 2003.PubMed/NCBI
|
|
7
|
Katoh M: WNT and FGF gene clusters
(Review). Int J Oncol. 21:1269–1273. 2002.PubMed/NCBI
|
|
8
|
Scherz PJ, Harfe BD, McMahon AP and Tabin
CJ: The limb bud Shh-Fgf feedback loop is terminated by expansion
of former ZPA cells. Science. 305:396–399. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schnorrer F and Dickson BJ: Axon guidance:
morphogens show the way. Curr Biol. 14:R19–R21. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han ME, Lee YS, Baek SY, Kim BS, Kim JB
and Oh SO: Hedgehog signaling regulates the survival of gastric
cancer cells by regulating the expression of Bcl-2. Int J Mol Sci.
10:3033–3043. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tada M, Kanai F, Tanaka Y, Tateishi K,
Ohta M, Asaoka Y, Seto M, Muroyama R, Fukai K, Imazeki F, Kawabe T,
Yokosuka O and Omata M: Down-regulation of hedgehog-interacting
protein through genetic and epigenetic alterations in human
hepatocellular carcinoma. Clin Cancer Res. 14:3768–3776. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chuang PT and McMahon AP: Vertebrate
Hedgehog signalling modulated by induction of a Hedgehog-binding
protein. Nature. 397:617–621. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yoon JW, Kita Y, Frank DJ, Majewski RR,
Konicek BA, Nobrega MA, Jacob H, Walterhouse D and Iannaccone P:
Gene expression profiling leads to identification of GLI1-binding
elements in target genes and a role for multiple downstream
pathways in GLI1-induced cell transformation. J Biol Chem.
277:5548–5555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Saqui-Salces M and Merchant JL: Hedgehog
signaling and gastrointestinal cancer. Biochim Biophys Acta.
1803:786–795. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cul’bová M, Lasabová Z, Stanclová A,
Tilandyová P, Zúbor P, Fiolka R, Danko J and Visnovský J:
Methylation of selected tumor-supressor genes in benign and
malignant ovarian tumors. Ceska Gynekol. 76:274–279. 2011.(In
Slovak).
|
|
16
|
Caffarelli E and Filetici P: Epigenetic
regulation in cancer development. Front Biosci. 16:2682–2694. 2011.
View Article : Google Scholar
|
|
17
|
Wolf I, Bose S, Desmond JC, Lin BT,
Williamson EA, Karlan BY and Koeffler HP: Unmasking of
epigenetically silenced genes reveals DNA promoter methylation and
reduced expression of PTCH in breast cancer. Breast Cancer Res
Treat. 105:139–155. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pritchard JI and Olson JM: Methylation of
PTCH1, the Patched-1 gene, in a panel of primary medulloblastomas.
Cancer Genet Cytogenet. 180:47–50. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fang JY and Xiao SD: Alteration of DNA
methylation in gastrointestinal carcinogenesis. J Gastroenterol
Hepatol. 16:960–968. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cretnik M, Musani V, Oreskovic S, Leovic D
and Levanat S: The Patched gene is epigenetically regulated in
ovarian dermoids and fibromas, but not in basocellular carcinomas.
Int J Mol Med. 19:875–883. 2007.PubMed/NCBI
|
|
21
|
Sasaki M, Anast J, Bassett W, Kawakami T,
Sakuragi N and Dahiya R: Bisulfite conversion-specific and
methylation-specific PCR: a sensitive technique for accurate
evaluation of CpG methylation. Biochem Biophys Res Commun.
309:305–309. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Merchant JL, Saqui-Salces M and El-Zaatari
M: Hedgehog signaling in gastric physiology and cancer. Prog Mol
Biol Transl Sci. 96:133–156. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Martin J, Donnelly JM, Houghton J and
Zavros Y: The role of sonic hedgehog reemergence during gastric
cancer. Dig Dis Sci. 55:1516–1524. 2010. View Article : Google Scholar : PubMed/NCBI
|