Introduction
Obesity results from the interaction between
genetic, environmental and psychosocial factors. Obesity poses an
important public health issue in the developed world and a growing
health issue in the developing world. The current worldwide
epidemic of obesity, along with its implications for public health,
emphasizes the importance of understanding the complex mechanisms
implicated in its development. Although >600 genes, markers and
chromosomal regions have been identified to be associated with or
linked to human obesity phenotypes (1), the responsible genes remain unknown
in >95% of severe obesity cases (2). Thus, identification of novel genes
and proteins associated with the development of obesity remains an
important issue.
Data from our previous study (3) demonstrated that C10orf116 is
highly expressed in adipose tissue and is localized primarily
within the nucleus. Overexpression studies in 3T3-L1 cells
indicated that C10orf116 upregulated the transcription
levels of CCAAT-enhancer-binding protein (C/EBP)α and peroxisome
proliferator-activated receptor (PPAR)γ, and promoted adipogenic
differentiation during the early stages of adipogenesis. However,
more precise functional properties of this gene need to be
clarified and require further investigation.
In the present study, we report on the effects of
C10orf116 on cell proliferation and apoptosis in
vitro. Our results indicate that the overexpression of
C10orf116 in preadipocytes stimulates proliferation and
inhibits apoptosis. Furthermore, we investigated the effects of
C10orf116 on glucose uptake and demonstrated that the
ectopic expression of C10orf116 significantly increases
insulin-stimulated glucose uptake in adipocytes by increasing
glucose transporter type 4 (GLUT4) expression levels.
Materials and methods
Cell culture and differentiation
3T3-L1 preadipocytes [American Type Culture
Collection (ATCC), Manassas, VA, USA] were maintained in Dulbecco’s
medified Eagle’s medium (DMEM) supplemented with 10% fetal calf
serum (FCS) (Biomedia, Boussens, France), 100 U/ml penicillin and
50 μg/ml streptomycin (Invitrogen, Life technologies, Carlsbad, CA,
USA) at 37°C in 5% CO2. The 3T3-L1 cells that were
stably integrated into either the pcDNA3.1Myc/HisB-C10orf116
plasmid or the empty vector were established as previously
described (3). To induce
differentiation, 2-day post-confluent 3T3-L1 preadipocytes (day 0)
were exposed to differentiation cocktail (100 μM
methylisobutylxanthine, 0.25 μM dexamethasone and 1 μg/ml insulin).
Two days later (day 2), cells were switched to medium containing 1
μg/ml insulin for 2 days (day 4). The cells were then switched back
to DMEM containing only 10% FCS until day 8. Cultures were
replenished every 2 days.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
Adipocytes (2×102 cells/well) were seeded
in 96-well culture plates and maintained in serum-free DMEM for 24
h until they were adherent. The cells were then cultured in DMEM
supplemented with 10% FCS. Cell growth was monitored for 7 days
successively using the Cell Proliferation MTT kit (Roche
Diagnostics GmbH, Mannheim, Germany) according to the
manufacturer’s instructions. Following this, 10 μl of MTT labeling
reagent were added to each well and incubated for 4 h at 37°C.
Solubilization solution (100 μl) was then added to each well and
cells were incubated overnight. The absorbance values at 560 and
660 nm were recorded by an ELISA reader and the difference between
these values was recorded as the optical density (OD).
Cell cycle assay
Cells were incubated at a density of
2×106 cells/750 mm2 in DMEM supplemented with
10% FCS, then washed with PBS and starved in serum-free DMEM for 24
h for synchronization. The cell cycle was initiated by replacement
of the starvation medium with full medium (DMEM with 10% FCS) at
various time points (0, 6, 12, 18, 24 h) following serum
deprivation. Cultured cells were harvested using trypsin/EDTA and
washed twice with PBS. Aliquots of 2×106 cells were
centrifuged, fixed in 70% ethanol and stained with 500 μl propidium
iodide (PI) solution (100 μg/ml RNase and 50 μg/ml PI in 1X PBS).
Labeled cells were analyzed using a BD FACScan (BD Biosciences,
Franklin Lakes, NJ, USA) and data were analyzed using CellQuest
software (BD Biosciences).
Evaluation of apoptotic index
Cells were cultured in FCS-free DMEM for 24 h to
induce apoptosis. Cells were then harvested using trypsin/EDTA,
washed with PBS, resuspended in 1 ml of binding buffer and stained
with 10 μl fluorescein isothiocyanate (FITC)-labeled Annexin V and
10 μl PI at room temperature for 5 min (BioVision, Mountain View,
CA, USA). The fluorescence of FITC and PI was analyzed by flow
cytometry.
Hoechst 33342 staining
Following the induction of apoptosis, cells were
treated with the apoptosis Hoechst 33324 staining kit (Sigma, St.
Louis, MO, USA) for 5–10 min according to the manufacturer’s
instructions. The cells were incubated with phenol red-free Hanks’
balanced salt solution containing 3 μM of Hoechst 33342 and washed
with PBS twice. Images were observed under a fluorescence
microscope.
Caspase-3 and -8 activity
Following the induction of apoptosis, cells were
collected and washed with PBS. The activity of caspase-3 and -8 was
assayed using commercially available kits (Sigma) according to the
manufacturer’s instructions. The measurements were based on the
hydrolysis of acetyl-Asp-Glu-Val-Asp p-nitroanilide (Ac-DEVD-pNA).
The reaction results in the release of the p-nitroaniline (pNA)
moiety. The pNA moiety was detected spectrophotometrically at 405
nm. The concentration of pNA released from the substrate was
calculated from a calibration curve prepared with pNA standards. In
order to confirm the measurements, additional experiments using
adequate inhibitors included in the kit were performed.
Glucose uptake
2-Deoxy-D-[3H]glucose (CIC,
Beijing, China) uptake was assayed as previously described
(4). The cells were cultured in
6-well plates and were serum starved in DMEM containing 0.5% FCS
for 3 h prior to the experiments. The cells were then washed twice
with PBS and incubated in KRP-HEPES buffer [30 mmol/l HEPES (pH
7.4), 10 mmol/l NaHCO3, 120 mmol/l NaCl, 4 mmol/l
KH2PO4, 1 mmol/l MgSO4 and 1
mmol/l CaCl2] in the presence or absence of 100 nmol/l
insulin for 30 min at 37°C. Labeled
2-deoxy-D-[3H] glucose was added to a final
concentration of 2 μCi/ml. After 10 min at 37°C, the reaction was
terminated by washing with ice-cold PBS 3 times, supplemented with
10 mmol/l D-glucose. The cells were solubilized by adding
200 μl of 1 mol/l NaOH to each well and aliquots of the cell lysate
were transferred to scintillation vials for radioactivity counting;
the remainder was used for the protein assay.
GLUT4 expression
Total RNA was extracted from 3T3-L1 adipocytes using
TRIzol reagent (Invitrogen Life Technologies) and the extracted RNA
was quantified by spectrophotometry at 260 nm. cDNA was synthesized
from 1 μg of total RNA by using an AMV Reverse Transcriptase kit
(Promega A3500; Promega Corp., Madison, WI, USA) according to the
manufacturer’s instructions. Real-time RT-PCR was performed using
the TaqMan Sequence Detection System and the DNA
double-strand-specific SYBR-Green I dye (Roche Diagnostics GmbH)
according to the manufacturer’s instructions. The samples were
briefly incubated at 95°C for 10 min for an initial denaturation,
followed by 40 PCR cycles. Each cycle consisted of incubation at
95°C for 15 sec and 60°C for 1 min. We used β-actin as the
reference in a comparative CT method and obtained the relative
changes in the target samples. We used the following primers for
the PCR analyses: GLUT4 homologous genes in mouse forward, 5′-ATT
GGA CGC TCT CTC TCC AA-3′ and reverse, 5′-GAT TCT GCT GCC CTT
CTGTC-3′; and β-actin forward, 5′-ATC TGG CAC CAC ACC TTC-3′ and
reverse, 5′-AGC CAG GTC CAG ACG CA-3′.
Statistical analysis
All data are expressed as the means ± SEM.
Statistical analysis was performed using the paired Student’s
t-test using the SPSS 10.0 statistical software package (SPSS,
Inc., Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Effects of C10orf116 on cell
proliferation
As indicated by MTT assay (Fig. 1), the overexpression of
C10orf116 in 3T3-L1 preadipocytes resulted in a
significantly higher rate of proliferation compared with the cells
transfected with the empty vector at each time point.
C10orf116 also had an effect on the cell cycle. The
percentage of 3T3-L1 cells overexpressing C10orf116 in the S
phase was significantly higher compared to the control cells
following the replacement of the starvation medium with full medium
at 12, 18 and 24 h and following serum deprivation, as shown by
flow cytometry (Fig. 2).
Effects of C10orf116 on cell
apoptosis
Annexin V was shown to interact strongly and
specifically with phosphatidylserine and may be used to detect the
early stages of apoptosis by targeting the loss of plasma membrane
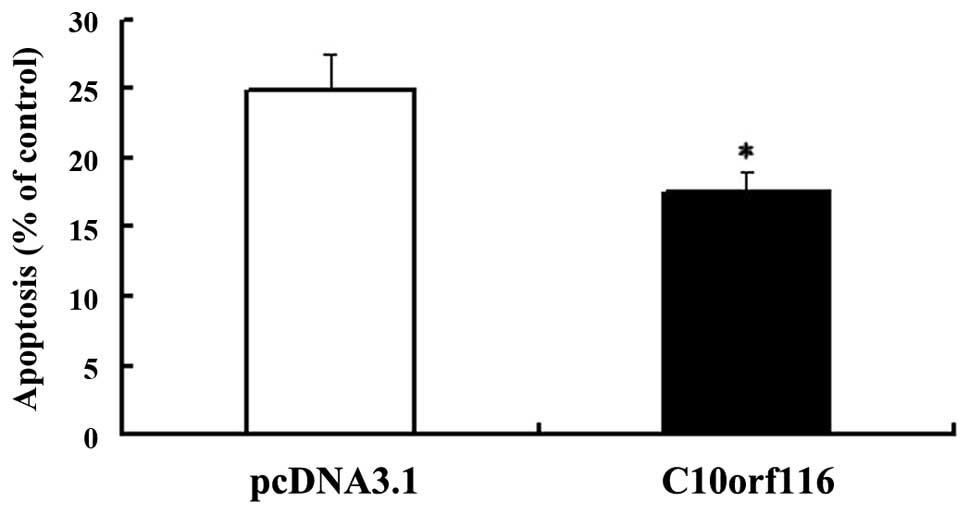
asymmetry (5). To examine the
effects of C10orf116 on cell apoptosis, cells were cultured
in FCS-free DMEM for 24 h to induce apoptosis and the apoptotic
index was analyzed. As shown in Fig.
3, C10orf116 protects 3T3-L1 preadipocytes from serum
deprivation-induced apoptosis. In addition, the results of the
Hoechst 33342 staining demonstrated that the 3T3-L1 cells
transfected with the pcDNA.3.1 vector exhibited more significant
apoptotic morphological changes, including chromatin condensation
and the formation of apoptotic bodies (Fig. 4C) compared with the cells
transfected with the C10orf116-pcDNA3.1 vector (Fig. 4D). However, no significant
differences were observed in the control cells (Fig. 4A) and the cells overexpressing
C10orf116 without the induction of apoptosis (Fig. 4B). Caspase-3 and -8 activity was
also determined. The data (Fig. 5)
also showed that 3T3-L1 preadipocytes transfected with the
C10orf116-pcDNA3.1 vector had a lower caspase-3 and -8
activity compared with the cells transfected with the pcDNA.3.1
vector. These results indicate that C10orf116 inhibits
apoptosis in preadipocytes induced by serum deprivation.
Effects of C10orf116 on basal and
insulin-stimulated glucose uptake and GLUT4 expression
The transfected 3T3-L1 preadipocytes were induced to
differentiate as described in Materials and methods. Glucose uptake
was then assayed in the 3T3-L1 adipocytes with or without
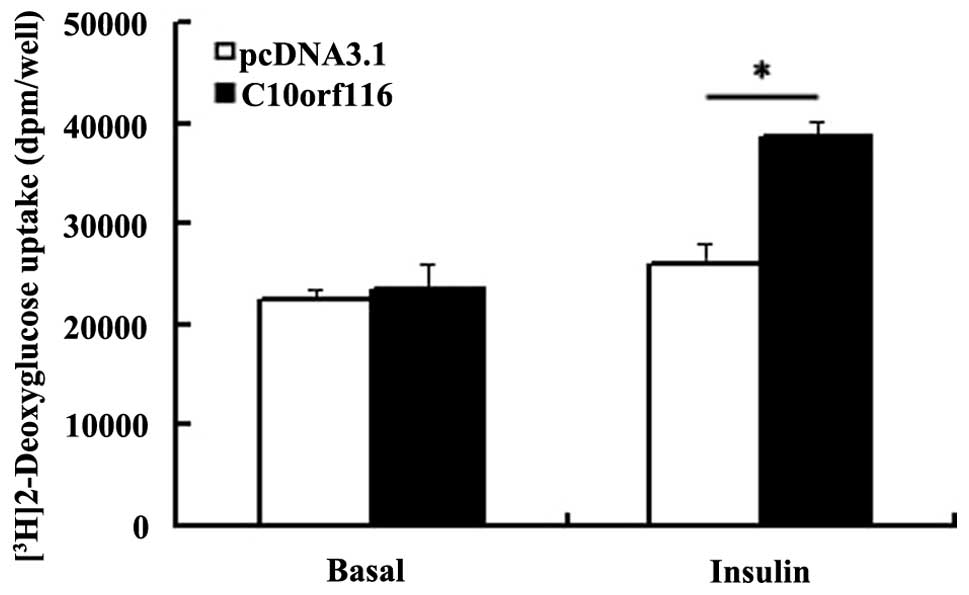
C10orf116 overexpression. As shown in Fig. 6, the insulin-stimulated glucose
uptake was significantly enhanced in the adipocytes overexpressing
C10orf116; however, the basal glucose uptake was similar
compared with the control cells. In the adipocytes,
insulin-stimulated glucose uptake is dependent on the expression,
the activity or the translocation of the insulin-responsive glucose
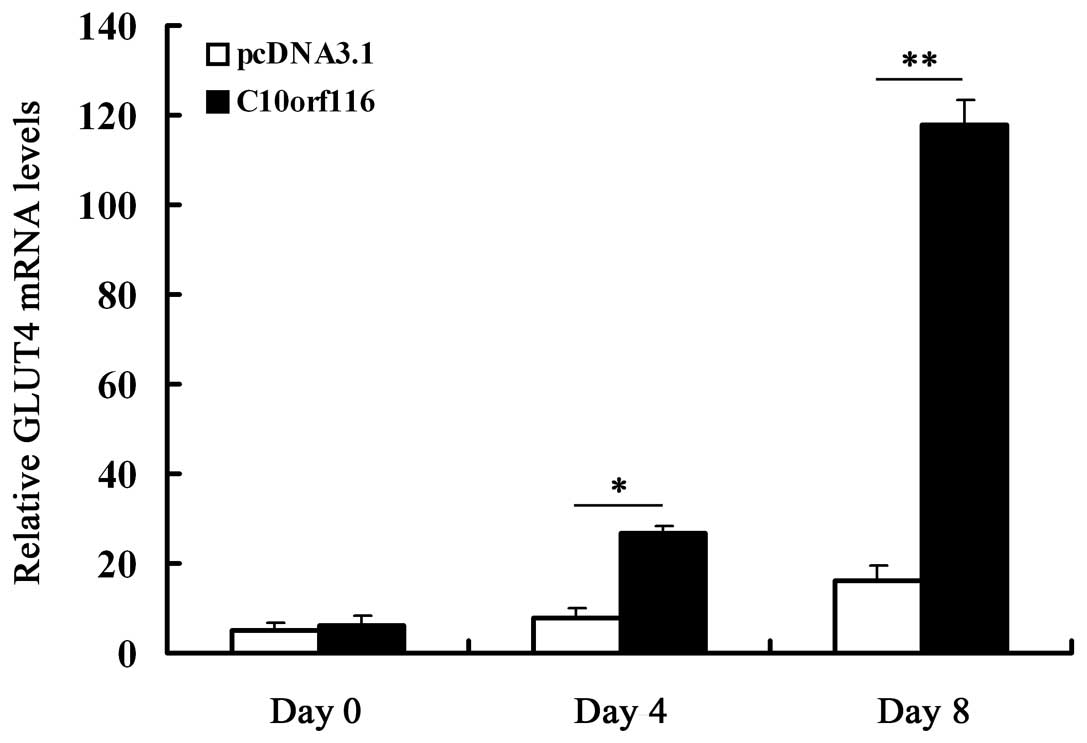
transporter GLUT4 (6–10). Therefore, we further examined the
effects of C10orf116 on GLUT4 expression during the
differentiation of 3T3-L1 preadipocytes. The results demonstrated
that C10orf116 overexpression significantly increased GLUT4
expression on days 4 and 8 of differentiation (Fig. 7).
Discussion
Obesity is an increasing global health issue that is
usually accompanied by a number of serious health impairments,
including type 2 diabetes and cardiovascular disease (11). Considerable evidence suggests that
obesity is caused by the interaction between multiple genes and the
environment (12,13). A better understanding of the
candidate genes required for the development of obesity may form
the basis for novel therapies that directly target the molecular
mechanisms underlying obesity. In our previous study (3), we identified C10orf116 as a
novel gene that may be important in the development of obesity.
Overexpression studies in 3T3-L1 cells indicated that
C10orf116 upregulated the transcription levels of C/EBPα and
PPARγ, and promoted adipogenic differentiation beginning from the
early stage of adipogenesis. In the present study, we further
investigated the association between C10orf116 and
obesity.
As a metabolic and endocrine organ, adipose tissue
is important in the regulation of energy balance (14). Accordingly, adipocytes are emerging
as a potential therapeutic target for obesity, type 2 diabetes and
cardiovascular disease (15).
Adipose tissue mass reflects the number and average volume of
adipocytes, in particular the balance between cell acquisition and
cell loss (16–18). The proliferation of adipocyte
precursors and their differentiation into mature adipocytes
contribute to the development of obesity in mammals. Apoptosis is
another important mechanism regulating adipose tissue mass
(19–22). Therefore, we further investigated
the effects of C10orf116 on 3T3-L1 preadipocyte
proliferation and apoptosis by establishing a stably transfected
3T3-L1 cell line overexpressing C10orf116 and demonstrated
that: i) C10orf116 causes the promotion of cell population
growth in 3T3-L1 preadipocytes indicated by results of MTT assay,
and cell cycle analysis by flow cytometry showed a significantly
increased percentage of cells in the S phase in
C10orf116-overexpressing preadipocytes; ii) cell apoptosis
analysis by Annexin V-FITC, Hoechst 33342, caspase-3 and -8
activity demonstrated that C10orf116 may prevent apoptosis
induced by serum deprivation. In conclusion, our data demonstrate
that by increasing cell proliferation and lowering the apoptotic
rate, C10orf116 may affect the size of the preadipocyte pool
and influence adipose tissue homeostasis. The function and
mechanism of C10orf116 in adipocytes requires further
investigation.
In adipocytes, insulin plays a role in multiple
stages of glucose metabolism. One of its most important effects is
the ability to increase the rate of cellular glucose transport
(23). In this study, we observed
that C10orf116 overexpression significantly increases
insulin-stimulated glucose transport in mature adipocytes and
exerted no effect on basal glucose uptake. We examined GLUT4
expression to determine the mechanism by which C10orf116
increases insulin-stimulated glucose uptake. The results indicated
that C10orf116 affected insulin-stimulated glucose uptake by
increasing GLUT4 expression levels.
Collectively, these data further support the
hypothesis that C10orf116 is important in regulating the
number of preadipocytes and glucose transport in adipocytes and
aids in the understanding of the complex mechanisms responsible for
obesity. However, in vivo research is required to verify the
physiological functions of this gene. Future studies addressing the
biochemical and functional properties of C10orf116 may
provide further insight into its role in obesity.
Acknowledgements
This study was supported by grants from the National
Key Basic Research Program of China (2013CB530604), the National
Natural Science Foundation of China (no. 81000349) and the Science
and Technology Development Fund of Nanjing Medical University
(09njmum052).
References
|
1
|
Pérusse L, Rankinen T, Zuberi A, et al:
The human obesity gene map: the 2004 update. Obes Res. 13:381–490.
2005.
|
|
2
|
Flier JS: Obesity wars: molecular progress
confronts an expanding epidemic. Cell. 116:337–350. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ni Y, Ji C, Wang B, Qiu J, Wang J and Guo
X: A Novel pro-adipogenesis factor abundant in adipose tissues and
over-expressed in obesity acts upstream of PPARγ and C/EBPα. J
Bioenerg Biomembr. Dec 13–2012.(Epub ahead of print).
|
|
4
|
Ceddia RB, Somwar R, Maida A, Fang X,
Bikopoulos G and Sweeney G: Globular adiponectin increases GLUT4
translocation and glucose uptake but reduces glycogen synthesis in
rat skeletal muscle cells. Diabetologia. 48:132–139. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vermes I, Haanen C, Steffens-Nakken H and
Reutelingsperger C: A novel assay for apoptosis. Flow cytometric
detection of phosphatidylserine expression on early apoptotic cells
using fluorescein labelled Annexin V. J Immunol Methods. 184:39–51.
1995. View Article : Google Scholar
|
|
6
|
Bryant NJ, Govers R and James DE:
Regulated transport of the glucose transporter GLUT4. Nat Rev Mol
Cell Biol. 3:267–277. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kanzaki M: Insulin receptor signals
regulating GLUT4 translocation and actin dynamics. Endocr J.
53:267–293. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tozzo E, Gnudi L and Kahn BB: Amelioration
of insulin resistance in streptozotocin diabetic mice by transgenic
overexpression of GLUT4 driven by an adipose-specific promoter.
Endocrinology. 138:1604–1611. 1997.PubMed/NCBI
|
|
9
|
Garvey WT, Hardin D, Juhaszova M and
Dominguez JH: Effects of diabetes on myocardial glucose transport
system in rats: implications for diabetic cardiomyopathy. Am J
Physiol. 264:H837–H844. 1993.PubMed/NCBI
|
|
10
|
Park SY and Lee W: The depletion of
cellular mitochondrial DNA causes insulin resistance through the
alteration of insulin receptor substrate-1 in rat myocytes.
Diabetes Res Clin Pract. 77(Suppl 1): S165–S171. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mokdad AH, Ford ES, Bowman BA, et al:
Prevalence of obesity, diabetes, and obesity-related health risk
factors, 2001. JAMA. 289:76–79. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee YS: The role of genes in the current
obesity epidemic. Ann Acad Med Singapore. 38:45–43. 2009.PubMed/NCBI
|
|
13
|
Walley AJ, Asher JE and Froguel P: The
genetic contribution to non-syndromic human obesity. Nat Rev Genet.
10:431–442. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rosen ED and Spiegelman BM: Adipocytes as
regulators of energy balance and glucose homeostasis. Nature.
444:847–853. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nawrocki AR and Scherer PE: Keynote
review: the adipocyte as a drug discovery target. Drug Discov
Today. 10:1219–1230. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Furuyashiki T, Nagayasu H, Aoki Y, et al:
Tea catechin suppresses adipocyte differentiation accompanied by
down-regulation of PPARgamma2 and C/EBPalpha in 3T3-L1 cells.
Biosci Biotechnol Biochem. 68:2353–2359. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Brook CG, Lloyd JK and Wolf OH: Relation
between age of onset of obesity and size and number of adipose
cells. Br Med J. 2:25–27. 1972. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Prins JB and O’Rahilly S: Regulation of
adipose cell number in man. Clin Sci (Lond). 92:3–11.
1997.PubMed/NCBI
|
|
19
|
Sorisky A, Magun R and Gagnon AM: Adipose
cell apoptosis: death in the energy depot. Int J Obes Relat Metab
Disord. 24(Suppl 4): S3–S7. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Della-Fera MA, Qian H and Baile CA:
Adipocyte apoptosis in the regulation of body fat mass by leptin.
Diabetes Obes Metab. 3:299–310. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Margareto J, Aguado M, Osés-Prieto JA, et
al: A new NPY-antagonist strongly stimulates apoptosis and
lipolysis on white adipocytes in an obesity model. Life Sci.
68:99–107. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Della-Fera MA, Li C and Baile CA:
Resistance to IP leptin-induced adipose apoptosis caused by
high-fat diet in mice. Biochem Biophys Res Commun. 303:1053–1057.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ducluzeau PH, Fletcher LM, Vidal H,
Laville M and Tavaré JM: Molecular mechanisms of insulin-stimulated
glucose uptake in adipocytes. Diabetes Metab. 28:85–92.
2002.PubMed/NCBI
|