Introduction
Autophagy is the major cellular pathway for the
degradation of long-lived proteins and cytoplasmic organelles
(1). Autophagy is becoming an
important area in cancer research (2,3).
Abnormal autophagy has certain associations with the formation of
tumors (4,5), however, autophagy in lung cancer has
not yet been studied.
BECLIN-1 (the mammalian counterpart of the yeast
Atg6 gene), which is part of a type III phosphatidylinositol
3-kinase complex, is required for autophagic vesicle formation
(6,7). BECLIN1 suppresses tumor formation via
the lysosomal degradation pathway (8). BECLIN1 is monoallelically deleted in
~75% of ovarian cancers (9,10),
50% of breast cancers (11) and
40% of prostate cancers (12). The
expression of BECLIN1 has been observed to be significantly reduced
in non-small cell lung cancer (NSCLC) compared with that in
non-cancerous tissue (13),
however, the role of BECLIN1 in lung cancer is unclear. In the
present study, BECLIN1 was knocked down in the A549 human lung
cancer cell line and its role in cell proliferation and apoptosis
was studied.
Materials and methods
Main reagents
The lentiviral small interfering (si)RNA
pRNAT-U6.2/Lenti vectors were obtained from GenScript Biotechnology
(Piscataway, NJ, USA). The lentiviral packaging cell line 293T and
the human lung cancer cell line A549 were purchased from the China
Center for Type Culture Collection (CCTCC; Wuhan, Hubei, China).
The E. coli DH5α restriction enzymes (BamHI and
XhoI) and the T4 DNA ligase were purchased from Promega
(Madison, WI, USA). The RPMI-1640 medium and the fetal bovine serum
(FBS) were purchased from Hyclone Biotechnology (Waltham, MA, USA).
The liposomes (Lipofectamine™ 2000) and G418 were purchased from
Invitrogen Biotechnology (Carlsbad, CA, USA). The RNA Extraction,
Plasmid DNA Extraction and Gel DNA Isolation kits were obtained
from Qiagen Biotechnology (Hilden, Germany). The study was approved
by the ethics committee of Zhengzhou University.
Construction of the siRNA plasmid against
BECLIN1
Promega, Ambion and Invitrogen siRNA target sequence
analysis and design software were used to scan the human-specific
BECLIN1 cDNA coding sequences (NM_003766). The siRNA target
sequences were based on basic design principles and a NCBI Basic
Local Alignment Search Tool (BLAST) homology analysis. The target
sequences were ascertained in 356–374 bits (GAGAGGAGCCATTTATTGA).
The small hairpin (sh)DNA single-chains were synthesized and
BamHI and XhoI enzyme residues were added at the two
ends. This process was performed by Shanghai Bio-engineering
Biotechnology (Shanghai, China). The designed sequences encoding
the region of BECLIN1 were inserted between the BamHI and
XhoI sites of the pRNAT-U6.2/Lenti plasmid. Competent
E.coli were transformed with the recombinants following the
manufacturer's recommendations. The positive clones were confirmed
by PCR amplification screening and sequencing.
siRNA transfection
Three recombinant lentiviral vectors, empty
lentiviral vectors and secondary packaging plasmids were
co-transfected into the 293T cells using 36 ml Lipofectamine 2000
reagent, according to the manufacturer's instructions. The obtained
BECN1 siRNA lentiviral fake virus particle solutions were
designated as Lv-si356 and the Lv-control and were stored at −80°C
until use. The human NSCLC A549 cell lines were cultured in
RPMI-1640 medium supplemented with 10% defined FBS. The day prior
to infection, the cells were plated into a tissue culture plate to
80% confluence. The human NSCLC A549 cell lines were infected by
the Lv-si356- and Lv-Control-prepared lentiviral particles. After
24 h, the medium was replaced and the culture was continued. To
obtain a stable infection, the cell lines were screened by G418 for
3 weeks.
RNA isolation and cDNA synthesis
Total RNA was isolated using TRIzol reagent
(Invitrogen) following the manufacturer's instructions. The amount
of RNA was measured spectrophotometrically by absorbance at 260 nm
and the purity of the RNA was estimated by the ratio of the
absorbance at A260/280. Reverse transcription of 2 μg total RNA was
performed with Moloney murine leukemia virus (MMLV) reverse
transcriptase (Promega, USA) using oligo (dT)18
(Promega) as the reverse transcription primer. The reaction
conditions for the reverse transcription were as follows: 2 μg of
total RNA was incubated with 100 pmol/l oligo (dT)18
primer at 72°C for 10 min and rapidly chilled on ice, then 2 μl 10
mM dNTPs (Promega) and 4 μl 5X MMLV buffer (Promega) were added.
The mixtures were incubated at 37°C for 5 min. The mixture
containing 200 units Moloney murine leukemia virus reverse
transcriptase (Promega) was run at 42°C for 1 h. The reaction was
then heated at 72°C for 10 min and stored at −20°C until use.
Quantitative real-time reverse
transcription (RT)-PCR
To quantitatively determine the level of the mRNA
expression of BECLIN1, the primer pairs were tested empirically for
amplification from 100 ng cDNA. The optimal annealing temperature
was determined by testing the generation of a single band of the
primers on the gels and then a specific primer was successfully
identified for BECLIN1. Quantitative real-time PCR reactions were
performed on the ABI Prism 7000 Sequence Detection System (Applied
Biosystems, Foster City, CA, USA). For the SYBR-Green II-based
quantitative real-time PCR reactions (Takara, Shiga, Japan), each
30 μl reaction contained 0.4 μM primer pairs, 100 ng cDNA, 15 μl
SYBR-Green, 0.6 μl ROX as a fluorescence internal control and
ddH2O. The primers were as follows: BECLIN1 forward,
5′-GGCTGAGAGACTGGATCAGG-3′ and reverse, 5′-CTGCGTCTGGGCATAACG-3′;
GAPDH forward, 5′-GGACTGACCTGCCGTCTAG-3′ and reverse,
5′-TAGCCCAGGATGCCCTTGAG-3′. The amplification program comprised two
stages, with an initial 95°C Taq activation stage for 10 min
followed by 45 cycles of 95°C denaturation for 5 sec and 60°C
annealing for 35 sec. Subsequent to the amplification, a melting
curve analysis was performed by collecting fluorescence data. GAPDH
served as an internal control. The comparative threshold cycle
(2−ΔΔCT) method was used to enable quantification of the
mRNA of these genes. All sample analyses were performed in
duplicate. The relative amount of the target gene was calculated
using the 2−ΔΔCT method. The relative amplification
efficiencies of the primers were tested and shown to be
similar.
Western blotting
The uninfected A549 cells and the four groups of
infected cells were added into the RLT buffer. Each group of cells
was harvested and the protein concentrations were determined with a
Bicinchoninic Acid (BCA) Protein Assay kit. The samples were
dissolved in loading buffer, boiled for 5 min and then loaded onto
10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE). The protein bands were transferred onto a
polyvinylidene difluoride membrane, which was blocked overnight at
4°C by Tris-buffered saline with Tween 20 (TBST) containing 5%
skimmed milk. The blocked membranes were then incubated with rat
anti-human BECLIN1 antibodies (1:150) for 2 h. The blocked
membranes were then washed three times and incubated with
horseradish peroxidase-conjugated rabbit anti-rat antibodies
(1:2000) for 1 h. The bands were visualized using
3,3′-diaminobenzidine (DAB).
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
method
The cells were seeded onto 96-well plates at a
density of 5×104 cells per well in 100 μl medium. All
the cells were maintained in a humidified incubator at 37°C with 5%
CO2. A total of 20 μl MTT (5 g/l) was added to each well
and the absorbance at 570 nm was measured by a microplate reader.
Subsequent to 4 h incubation, the number of surviving cells was
measured. There were 5 parallel wells for each group.
Detection of apoptosis by flow
cytometry
Subsequent to 48-h incubation, each group of cells
was digested by trypsin. The cells were further incubated at a
density of 5×105–1×106/ml nutrient solution
containing 75% cold ethanol overnight. Subsequent to
centrifugation, 1 ml of the cells were fixed with 100 μl RNA
enzymes (1 mg/ml) at 37°C for 30 min. Then 100 μl 10 μg/ml
propidium iodide (PI) dye liquor was added in the dark. Following
fixation, the cells were incubated in the dark for 30 min. The
samples were filtered through a 300 mesh nylon net, then the cell
apoptosis rates were detected by flow cytometry.
Detection of active caspase-3 and
caspase-9
Each group of ~5×105 cells was
centrifuged at 500 × g for 5 min. The collected cells were fixed
into ice cold cell lysate at a density of 50 μl/2×105
cells for 10 min. The microtubes were centrifuged at 20,000 × g for
5 min, maintaining a consistent temperature of 4°C. Thereafter, the
supernatants were transferred to another set of ice-cold
microtubes. The 50 μl samples were pipetted into a 96-well plate,
mixed and sealed with paraffin wax. Subsequent to a 2-h incubation
in the dark, the fluorescence was determined by a plate reader
using an excitation wavelength of 380 nm and an emission wavelength
of 460 nm.
Statistical analysis
Statistical analyses were performed using SPSS 13.0
(SPSS, Chicago, IL, USA). Quantitative data are presented as the
mean ± standard deviation. Comparisons among all the groups were
performed with a one-way analysis of variance (ANOVA) test.
Comparisons between two groups were evaluated with a t-test.
Results
Lentivirus-mediated siRNA vector
transfection in the A549 human lung cancer cell line
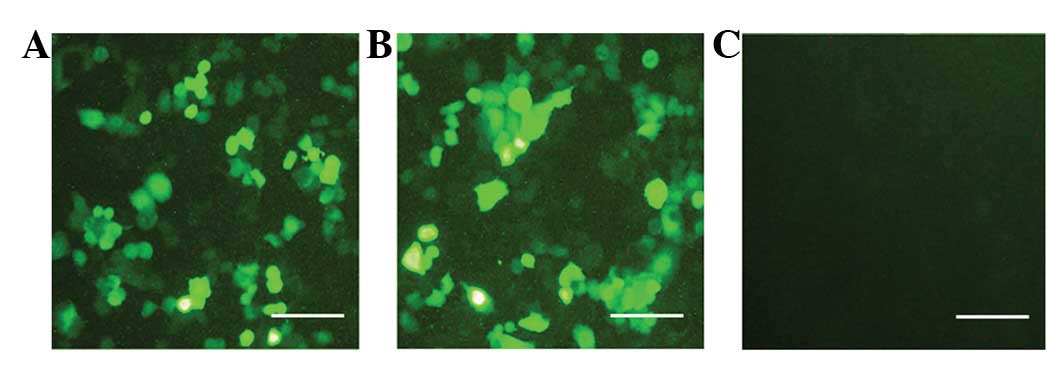
Following transfection with the siRNA virus against
BECLIN1 for 48 h, the A549 cells expressed enhanced green
fluorescent protein (EGFP). In total, >90% of the cells were
EGFP-positive in the cell culture (Fig. 1). Subsequent to 2 weeks of G418
selection, the BECLIN1 knockdown A549 cell line was
established.
Decrease of BECLIN1 expression following
siRNA virus transfection in the A549 cells
Subsequent to transfection with the siRNA and
scramble viruses for 48 h, BECLIN1 expression was detected using
RT-PCR and western blotting (Fig.
2). The mRNA and protein expression of BECLIN1 decreased in the
siRNA virus-transfected A549 cells.
Knockdown of BECLIN1 increases A549 cell
proliferation
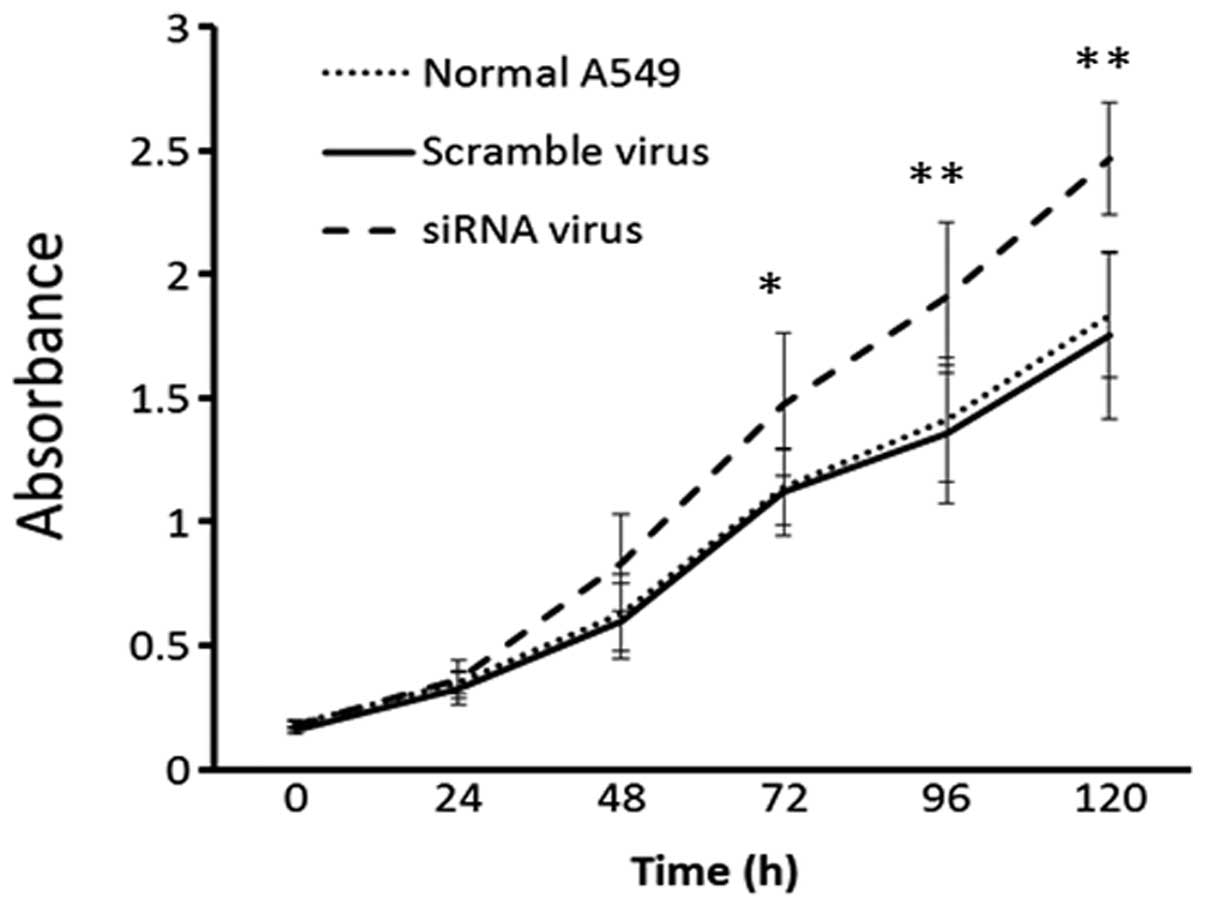
Cell proliferation was detected by MTT assay in the
A549 cells transfected with the siRNA or scramble viruses. Normal
A549 cells were assayed as the blank control. The knockdown of
BECLIN1 increased cell proliferation significantly (Fig. 3).
Knockdown of BECLIN1 inhibits apoptosis
in the A549 cells
Apoptosis was detected by PI staining and flow
cytometry in the A549 cells transfected with the siRNA or scramble
viruses. Normal A549 cells were assayed as the blank control. The
apoptosis index (AI) was less in the BECLIN1-knockdown A549 cells
than that in the scramble virus-transfected A549 cells and blank
controls (P<0.05; Fig. 4).
Knockdown of BECLIN1 inhibits the
activity of caspase-3 and caspase-9
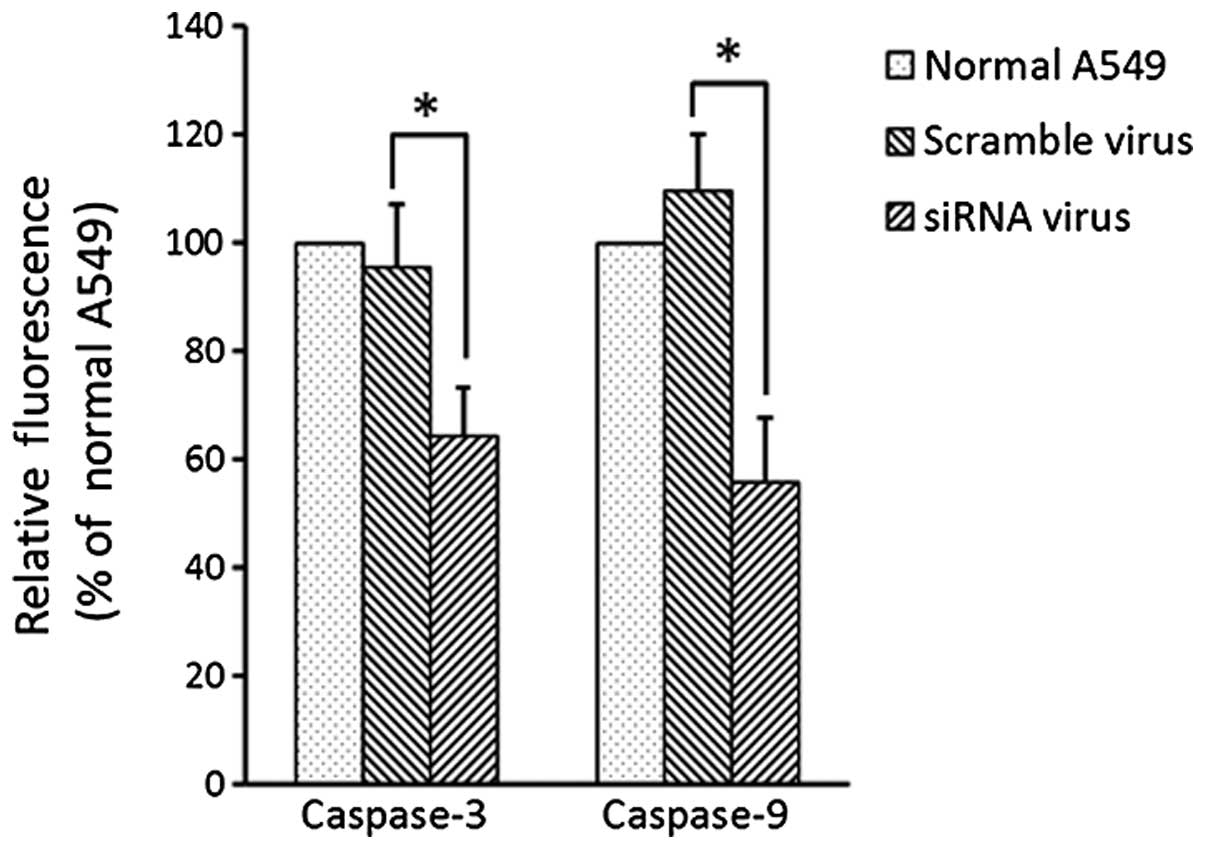
The activity of caspase-3 and caspase-9 decreased in
the A549 cells following siRNA transfection in contrast with the
activity in the normal A549 cells and the A549 cells following
scramble transfection (Fig. 5).
There was no difference in the activity of caspase-3 and caspase-9
between the normal A549 and scramble virus-transfected A549
cells.
Discussion
The role of autophagy in tumorigenesis is
controversial. Autophagy inhibitors (chloroquine) and autophagy
promoters (rapamycin) block tumorigenesis by an unknown
mechanism(s) (14,15). This is known as the ‘Autophagy
Paradox’. Little is known about the role of autophagy in the
tumorigenesis of NSCLC.
BECLIN1 is a key regulator of autophagy (16) and is involved in cell proliferation
and tumor formation. BECLIN1 triggers autophagy in human breast
carcinoma cells: this autophagy-promoting activity was observed to
be associated with the inhibition of cell proliferation in
vitro and with tumorigenesis in nude mice (17). Exogenous BECLIN1 expression in
MCF-7 breast cancer cells inhibits the cell proliferation and
tumorigenesis of these cells (17). In BECLIN1-knockout transgenic mice,
the incidence of lung adenocarcinoma, liver cancer and lymphoma
increases significantly (18,19).
The expression of BECLIN1 is significantly decreased in lung cancer
tissues, suggesting that autophagy may be involved in the
pathogenesis of lung cancer (20).
In the present study, BECLIN1 knockdown increased cell
proliferation in the A549 cells. This suggested that BECLIN1
expression may be a significant factor in maintaining the normal
level of cell proliferation and that the downregulation or
depletion of BECLIN1 in lung cancer may be an etiology responsible
for the increased cell proliferation in cancer.
Certain studies have demonstrated the paradoxical
correlation between BECLIN1 and apoptosis. Liang et
al(17) showed that the
BECLIN1 gene reduced apoptosis of the central nervous system. Wang
(7) demonstrated that the
depletion of BECLIN1 activated apoptosis. However, BECLIN1 was also
suggested to bind the Bcl-2 family members, which may have a direct
role in initiating apoptotic signaling (21,22).
In the present study, the inhibition of apoptosis and the decrease
in caspase-3 and caspase-9 activity by BECLIN1 knockdown was
validated in the A549 cells. It appears that BECLIN1 may play a
different role in the regulation of apoptosis, which therefore
requires further elucidation. When considering the promotive effect
of BECLIN1 in the apoptosis of the A549 cells, we considered the
theory that BECLIN1 downregulation may aid lung cancer cells to
escape the body's normal clearance mechanism by apoptosis
inhibition. This would contribute to the development of malignant
tumors.
Taking these results together, BECLIN1 knockdown
increases cell proliferation and decreases apoptosis in lung cancer
cells. Therefore, BECLIN1 may be a biological target for gene
therapy and the upregulation of BECLIN1 may be a promising
treatment method for lung cancer, or even other tumor cells. This
may provide further insight into the treatment of cancer.
Acknowledgements
The authors would like to express their particular
appreciation to Dr Yongmin Liu who provided statistical
assistance.
References
|
1
|
Kanzawa T, Germano IM, Komata T, Ito H,
Kondo Y and Kondo S: Role of autophagy in temozolomide-induced
cytotoxicity for malignant glioma cells. Cell Death Differ.
11:448–457. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carew JS, Kelly KR and Nawrocki ST:
Autophagy as a target for cancer therapy: new developments. Cancer
Manag Res. 4:357–365. 2012.
|
|
3
|
Meschini S, Condello M, Lista P and
Arancia G: Autophagy: molecular mechanisms and their implications
for anticancer therapies. Curr Cancer Drug Targets. 11:357–379.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang ZJ, Chee CE, Huang S and Sinicrope
FA: The role of autophagy in cancer: therapeutic implications. Mol
Cancer Ther. 10:1533–1541. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lockshin RA and Zakeri Z:
Caspase-independent cell deaths. Curr Opin Cell Biol. 14:727–733.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun Q, Fan W and Zhong Q: Regulation of
Beclin 1 in autophagy. Autophagy. 5:713–716. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang J: Beclin 1 bridges autophagy,
apoptosis and differentiation. Autophagy. 4:947–948. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shintani T and Klionsky DJ: Autophagy in
health and disease: a double-edged sword. Science. 306:990–995.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ibragimova I and Cairns P: Assays for
hypermethylation of the BRCA1 gene promoter in tumor cells to
predict sensitivity to PARP-inhibitor therapy. Methods Mol Biol.
780:277–291. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tangir J, Muto MG, Berkowitz RS, Welch WR,
Bell DA and Mok SC: A 400 kb novel deletion unit centromeric to the
BRCA1 gene in sporadic epithelial ovarian cancer. Oncogene.
12:735–740. 1996.PubMed/NCBI
|
|
11
|
Saito H, Inazawa J, Saito S, et al:
Detailed deletion mapping of chromosome 17q in ovarian and breast
cancers: 2-cM region on 17q21.3 often and commonly deleted in
tumors. Cancer Res. 53:3382–3385. 1993.PubMed/NCBI
|
|
12
|
Gao X, Zacharek A, Salkowski A, et al:
Loss of heterozygosity of the BRCA1 and other loci on chromosome
17q in human prostate cancer. Cancer Res. 55:1002–1005.
1995.PubMed/NCBI
|
|
13
|
Liu Q, Wang JJ, Pan YC, Meng LF, Zhan X
and Zheng QF: Expression of autophagy-related genes Beclin1 and
MAPLC3 in non-small cell lung cancer. Ai Zheng. 27:25–29. 2008.(In
Chinese).
|
|
14
|
Martinez-Outschoorn UE, Whitaker-Menezes
D, Pavlides S, et al: The autophagic tumor stroma model of cancer
or ‘battery-operated tumor growth’: a simple solution to the
autophagy paradox. Cell Cycle. 9:4297–4306. 2010.
|
|
15
|
Maycotte P, Aryal S, Cummings CT, Thorburn
J, Morgan MJ and Thorburn A: Chloroquine sensitizes breast cancer
cells to chemotherapy independent of autophagy. Autophagy.
8:200–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ruck A, Attonito J, Garces KT, et al: The
Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde
transport in addition to autophagy in C. elegans. Autophagy.
7:386–400. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liang XH, Jackson S, Seaman M, et al:
Induction of autophagy and inhibition of tumorigenesis by beclin 1.
Nature. 402:672–676. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lozy F and Karantza V: Autophagy and
cancer cell metabolism. Semin Cell Dev Biol. 23:395–401. 2012.
View Article : Google Scholar
|
|
19
|
Yue Z, Jin S, Yang C, Levine AJ and Heintz
N: Beclin 1, an autophagy gene essential for early embryonic
development, is a haploinsufficient tumor suppressor. Proc Natl
Acad Sci USA. 100:15077–15082. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang ZF, Shao LJ, Wang WM, Yan XB and Liu
RY: Decreased expression of Beclin-1 and LC3 in human lung cancer.
Mol Biol Rep. 39:259–267. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Oberstein A, Jeffrey PD and Shi Y: Crystal
structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a
novel BH3-only protein. J Biol Chem. 282:13123–13132. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Maiuri MC, Le Toumelin G, Criollo A, et
al: Functional and physical interaction between Bcl-X(L) and a
BH3-like domain in Beclin-1. EMBO J. 26:2527–2539. 2007. View Article : Google Scholar : PubMed/NCBI
|