Introduction
With the development of modern biotechnology,
transgene technology has been widely used in genetic engineering
and genetic therapy fields. However, low or silenced transgene
expression has become a common phenomenon (1,2) that
limits the use of transgene technology. Certain DNA elements have
been used to overcome transgene silencing and to increase transgene
expression (3,4), such as matrix attachment region (MAR)
(5–7).
MAR is a unique DNA sequence capable of attaching to
the nuclear matrix in eukaryotic chromatin. MAR is 300–1,000 bp
long, and contains the topoisomerase consensus sequence and other
AT-rich sequence motifs (8).
Previous studies demonstrated that MAR is able to overcome
transgene silencing, increase transgene expression and decrease the
expression difference in different transformants, as a boundary
regulatory element (9–11). In previous studies we demonstrated
that human β-globin MAR is capable of efficiently enhancing
transgene expression (12), and
the transgene expression level may be highly increased when two
β-globins flank the CAT expression cassette (13). Moreover, different MARs that flank
the transgene are able to significantly increase the transgene
expression level (14). As a
cis-acting DNA element, MAR is capable of enhancing transgene
expression, as it contains several characteristic motifs, such as
the AATATATTT motif, the topoisomerase-II-binding site and an Alu
element. However, the function of these motifs in enhancing gene
expression, whether these motifs are able to enhance gene
expression and identification of the MAR elements that are capable
of increasing gene expression remain to be elucidated. In this
study, we combined the characteristic elements of β-globin MAR, the
AATATATTT motif, the topoisomerase-II-binding site and an Alu
element, and removed the intermediate DNA connection for the
synthesis of an artificial splicing of the MAR fragment to
investigate the function and mechanism of the MAR characteristic
elements in regulating transgene expression. Moreover, we examined
the effect of the characteristic elements of β-globin MAR on
transgene expression.
Materials and methods
MAR cloning and artificial synthesis
The fragment of human β-globin MAR (Genebank No.
L22754) was amplified by PCR using the primers: P1: 5′-ATCGGTACC
GTAAGACATCACCTTGCATTT-3′ and P2: 5′-GCAACGCG
TCATAGTTTGAGTTACCCTTT-3′. Human β-globin MAR, ~2.15 kb in length
(site 840–2,998), was amplified using human peripheral blood as a
template. The KpnI/BglII enzyme sites (GTCAGACTC and
AGCGGTACC) with three additional nucleotides were inserted at the
5′ site of the above primers, respectively. The PCR conditions
were: 95°C for 3 min, 94°C for 40 sec, 56–60°C for 30 sec, 72°C for
2 min at two cycles every annealing temperature, 20 cycles at 55°C
and a final extension at 72°C for 5 min.
The characteristic elements of the human β-globin
MAR, including the AATATATTT motif (sites 840–848, 1149–1157 and
1636–1644), the topoisomerase-II-binding sites (1252–1266,
1456–1470) and an Alu element (site 2687–2998), were sequentially
synthesized, forming an artificial splicing of the MAR fragment
~369 bp long [Takara Biotechnology (Dailan) Co., Ltd, China]. The
KpnI/BglII enzyme sites (GTCAGACTC and AGCGGTACC)
were inserted at the two ends of the fragment.
Vector construction
The 2.15 kb MAR and 369 bp MAR splice were inserted
into the pMD18-T-vector (Takara Bio, Inc., Shiga, Japan). Following
sequence identification, two MAR fragments and a pCATG plasmid were
cut with KpnI and BglII, and then ligated using the
T4 DNA ligase, to generate the pCATGM1and pCATGM2 vectors
containing the 2.15 kb MAR and 369 bp MAR splice, respectively. The
constructs are shown schematically in Fig. 1.
Cell culture and transfection
Chinese hamster ovary cells (CHO; Institute of
Laboratory Animal Sciences, China) were inoculated in six-well
plates at 2×106 cells/well. After 24 h, the cells were
transfected with pCATG, pCATGM1 and pCATGM2 vectors according to
the manufacturer’s instructions (Xiamen Sunma Biotechnology Co.,
Ltd. Xiamen, China). Following transfection for 48 h, the cells
were screened in DMEM supplemented with 800 mg/ml G418 (Calbiochem,
La Jolla, CA, USA) for 10 days. The cells were then screened with a
maintenance concentration of 400 mg/ml G418 for 10 days. When the
colonies of stably transfected cells had formed, these were
digested with 0.25% trypsin. Each group of cells was diluted and
single colonies were selected. The colonies were then replanted in
a culture bottle to continue the culturing process. The cells were
collected when their density reached 80–90%.
CAT expression assays
Cells were collected, adjusted to the same
concentration of 1×106 cells/ml by cell counting, and
then washed three times with pre-cooled PBS. Lysis buffer (1 M;
5-fold diluted) was then added to the tubes. The tubes were gently
agitated, left to stand at room temperature for 30 min, and then
centrifuged at 13,000 × g for 10 min at 4°C. Approximately 200 μl
supernatant was extracted and placed into EP tubes. Negative and
positive controls were set. CAT content was assayed using a CAT
ELISA kit (Roche Applied Science, Indianapolis, IN, USA) according
to the manufacturer’s instructions.
Analysis of gene copy number
Real-time fluorescent quantitative PCR was used to
analyze the gene copy number using internal control primers as the
controls. Stably transfected cells were collected to extract the
genomic DNA and served as PCR templates. The primers were designed
according to the CAT gene sequence. The primers sequences used
were: P1: 5′-CATCGCTCTGGAGTGAATACC-3′ and P2: 5′-GGCATC
AGCACCTTGTCG-3′. The internal control β-actin primer sequences
were: P3: 5′-GTCTTTCTTCTGCCGTTCTC-3′ and P4:
5′-ACCAGCCTCATTAGGTTTGT-3′. The PCR amplification system was 20 μl,
and the PCR conditions were as follows: 95°C pre-degeneration for
10 min, 95°C for 15 sec for 40 cycles, and 60°C for 1 min. The
SYBR-Green I fluorescent dye method was used for fluorescent
quantitative real-time PCR. Experimental data were treated with
2−ΔΔCt (double ΔCt) method. Each experiment was
conducted in triplicate, and the mean CT and ΔCt values [ΔCt = Ct
(target gene) - Ct (internal gene)] were calculated. The
2−ΔΔCt was calculated, which is the relative fold of a
gene copy number to the internal control gene copy number.
Statistical analysis
Analysis of variance was performed with SPSS 17.0
(SPPS, Inc., Chicago, IL, USA). The Q-test was used for comparison
between two indices. P<0.05 was considered to indicate
statistical significance.
Results
Vector construction
The constructed vectors were digested with
KpnI and KpnI/BglII. The agarose gel
electrophoresis results are shown in Fig. 1. Following digestion of the
constructed pCATGM1 vector with KpnI/BglII, an
~2.15-kb fragment appeared. Following digestion of the constructed
pCATGM1 vector with KpnI, linear DNA was evident (Fig. 2). The results of the enzyme
digestion conformed to the expected fragments. The results
demonstrated that the vector containing MAR (2.15 kb) had been
constructed. The same method was used to analyze the constructed
pCATGM2 vector; a 369-bp long fragment and linear DNA were
observed. The vector containing MAR (369 bp) had also been
constructed.
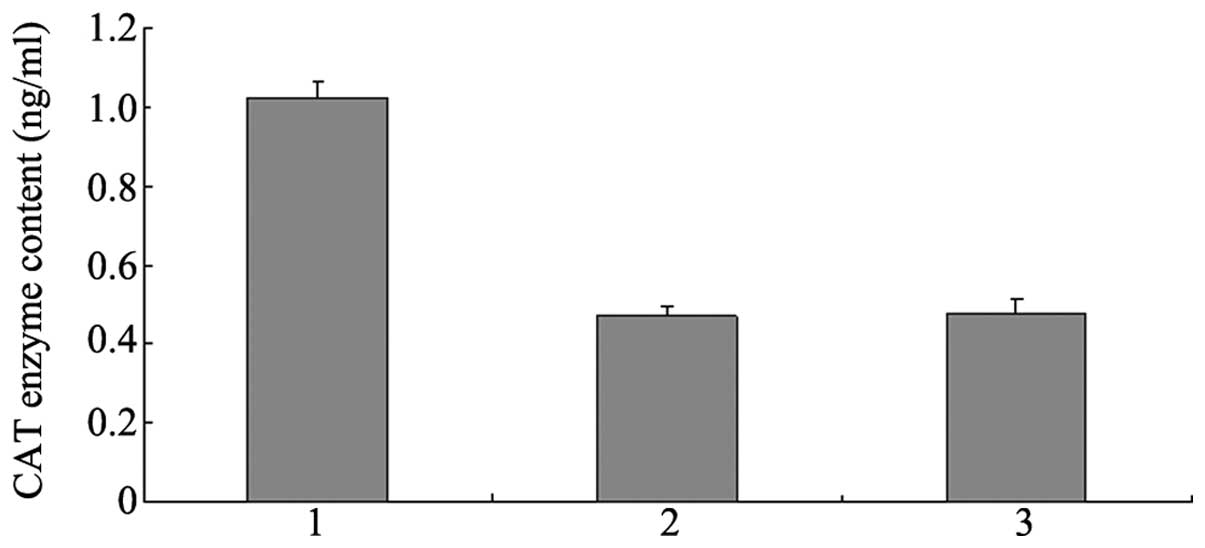
CAT assays
CHO cells were transfected with the constructed
vectors using the non-MAR pCATG plasmid as control. The cells were
screened by the use of G418. When the cell concentration reached
80–90%, the cells were adjusted to the same concentration of
1×106 cells/ml via cell counting. The CAT content was
analyzed following collection of the cells. The statistical
analysis results from 10 monoclonal cells revealed that the CAT
content of pCATGM1-transfected cells was 2.1489-fold higher than
that of non-MAR pCATG-transfected cells. However,
pCATGM2-transfected cells had the same CAT content as the non-MAR
pCATG-transfected cells (Fig. 3).
The difference in CAT expression between the different
vector-transfected cells was statistically significant
(P<0.01).
Gene copy number
Genomic DNA was extracted from six strains of cells
from each group. Following real-time fluorescent quantitative PCR,
2−ΔΔCt was calculated. Combined with ELISA analysis, the
data from Table I demonstrate that
the CAT gene copy numbers of the pCATGM1 expression vector were
1.2-fold higher compared with those of the pCATG expression vector
(P<0.01). In addition, the gene expression of the pCATGM1
expression vector was 2.1489-fold higher compared with that of the
pCATG expression vector. Therefore, the CAT gene expression of the
different strains of the pCATGM1 expression vector was not linear
with its gene copy number. However, exogenetic gene expression
enhancing may be correlated with the increase in gene copy
number.
 | Table ICAT gene relative copy number analysis
in stably transfected cell strains. |
Table I
CAT gene relative copy number analysis
in stably transfected cell strains.
| Ct mean (vector) | Ct mean
(β-actin) | ΔCt mean | ΔΔCt | Fold |
|---|
| pCATG | 0.29277 | 20.94518 | 9.34759 | 0 | 1.0 |
| pCATGM1 | 32.24645 | 23.20043 | 9.04602 | 0.30157 | 1.2 |
Discussion
Studies with regard to MAR increasing transgene
expression have been reported (6,15,16).
Kim et al(9) inserted
different MAR sources into the β-galactosidase gene expression
vector and found that human β-globin MAR enhances the frequency of
the positive colonies by 2.5-fold, and the β-galactosidase gene
expression by 7-fold. However, other MAR sources have demonstrated
little or no effect on β-galactosidase gene expression. Wang et
al found that CAT gene expression was enhanced by 5.49-fold
when human β-globin MAR was inserted in the 5′ end of the CAT
expression cassette (12).
However, CAT gene expression decreased by 0.62-fold when the
β-globin MAR was inserted at the 3′ end of the CAT expression
cassette. When the two ends of the CAT expression cassette were
also inserted into the β-globin MARs, the CAT gene expression was
10.4-fold higher than that of the non-MAR expression vector
(13). Wang et al found
that if the two ends were inserted in the β-globin MAR and
β-interferon MAR, the CAT gene expression was 4.5-fold higher than
that inserted only with β-globin MARs at the two ends of the CAT
expression cassette, and 46.4-fold higher than that of the non-MAR
expression vector (14). Due to
the different sources of MAR, different effects of MAR were
observed. Certain MAR sources have a strong effect on gene
expression, whereas other sources have little effect. Another
reason for the different effects may be the site of the MAR
insertion. The 5′ end insertion possibly has more function than the
3′ end insertion.
In this study, we inserted longer β-globin MAR,
containing connection DNA and all the characteristic elements in
the 5′ end of the CAT expression cassette, thereby constructing the
pCATGM1 expression vector. Following the transfection of CHO cells,
we found that the pCATGM1 vector was capable of increasing CAT gene
expression; the expression was 2.15-fold higher than that of the
non-MAR expression vector. Therefore, human β-globin MAR is capable
of enhancing the transgene expression and decreasing transgene
silencing. Our results are consistent with those of Kim et
al(9). The pCATGM2 vector,
which only contained characteristic elements, was not able to
increase the CAT gene expression, similar to that of the non-MAR
expression vector. These results confirmed that although the
characteristic elements of MAR may have an important effect on gene
expression enhancing in theory, the characteristic elements of MAR
alone do not have the same effect as that of the MAR sequence.
Thus, more studies are required to identify which characteristic
elements of MAR are involved in gene expression enhancing and which
have no effect, and to determine whether the connection DNA between
MARs is necessary for gene expression enhancement. Although the
characteristic elements of MAR may have an important effect on gene
expression, the connection DNA between the characteristic elements
is necessary for the shape-formation and function of MAR. If the
connection DNA between the characteristic elements is missing, MAR
is not able to form the correct shape, and thus is not capable of
enhancing gene expression.
Real-time fluorescent quantitative PCR was used to
analyze the gene copy number. The results revealed that the pCATGM1
vector was capable of increasing the gene copy number by 1.2-fold.
In addition, β-globin MAR was able to enhance the transgene copy
number. However, the CAT gene expression of different strains of
the pCATGM1 expression vector was not linear with the gene copy
number of the pCATGM1 expression vector. Thus, we deduced that
exogenetic gene expression enhancement may be correlated with the
increase in gene copy number. This result is not consistent with
those of other studies (11,12).
Therefore, the mechanism requires further investigation.
An increasing number of separated MAR fragments has
led to an improvement in our understanding of the function and
mechanism of MAR. However, investigations concerning MAR are in
their infancy. Although several studies have shown that MAR is able
to increase gene expression, decrease transgene silencing and
decrease the expression difference in different transformants,
knowledge of MAR expression regulation at the chromatin level and
the mechanism of its effect on transgene expression remains limited
and should be investigated.
References
|
1
|
Spiker S and Thompson WF: Nuclear matrix
attachment region and transgene expression in plants. Plant
Physiol. 110:15–21. 1996.PubMed/NCBI
|
|
2
|
Jenuwein T, Forrester WC,
Fernández-Herrero LA, Laible G, Dull M and Grosschedl R: Extension
of chromatin accessibility by nuclear matrix attachement regions.
Nature. 385:269–272. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zehnpfennig D, Deissler H, Polack A, Herr
D, Bornkamm GW and Kurzeder C: B-cell-specific elements enhance
sustained gene expression mediated by self-replicating
extrachromosomal vectors. Mol Med Rep. 3:689–692. 2010.PubMed/NCBI
|
|
4
|
Argyros O, Wong SP, Fedonidis C, et al:
Development of S/MAR minicircles for enhanced and persistent
transgene expression in the mouse liver. J Mol Med (Berl).
89:515–529. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Girod PA, Nguyen DQ, Calabrese D, et al:
Genome-wide prediction of matrix attachment regions that increase
gene expression in mammalian cells. Nat Methods. 4:747–753. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sjeklocha L, Chen Y, Daly MC, Steer CJ and
Kren BT: β-globin matrix attachment region improves stable genomic
expression of the Sleeping Beauty transposon. J Cell Biochem.
112:2361–2375. 2011.
|
|
7
|
Zhang J, Lu L, Ji L, Yang G and Zheng C:
Functional characterization of a tobacco matrix attachment
region-mediated enhancement of transgene expression. Transgenic
Res. 18:377–385. 2009. View Article : Google Scholar
|
|
8
|
van der Geest AHM, Hall GE, Spiker S and
Hall TC: The β-phaseolin gene is flanked by matrix attachment
regions. Plant J. 6:413–423. 1994.
|
|
9
|
Kim JM, Kim JS, Park DH, et al: Improved
recombinant gene expression in CHO cells using matrix attachment
regions. J Biotechnol. 107:95–105. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Girod PA, Zahn-Zabal M and Mermod N: Use
of the chicken lysozyme 5′ matrix attachment region to generate
high producer CHO cell lines. Biotechnol Bioeng. 91:1–11. 2005.
|
|
11
|
Buceta M, Galbete JL, Kostic C,
Arsenijevic Y and Mermod N: Use of human MAR elements to improve
retroviral vector production. Gene Ther. 18:7–13. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang TY, Yang R, Qin C, Wang L and Yang
XJ: Enhanced expression of transgene in CHO cells using matrix
attachment region. Cell Biol Int. 32:1279–1283. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang TY, Zhang JH, Jing CQ, Yang XJ and
Lin JT: Positional effects of the matrix attachment region on
transgene expression in stably transfected CHO cells. Cell Biol
Int. 34:141–145. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang F, Wang TY, Tang YY, Zhang JH and
Yang XJ: Different matrix attachment regions flanking a transgene
effectively enhance gene expression in stably transfected Chinese
hamster ovary cells. Gene. 500:59–62. 2012. View Article : Google Scholar
|
|
15
|
Harraghy N, Regamey A, Girod PA and Mermod
N: Identification of a potent MAR element from the mouse genome and
assessment of its activity in stable and transient transfections. J
Biotechnol. 154:11–20. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Van der Geest AH, Welter ME, Woosley AT,
et al: A short synthetic MAR positively affects transgene
expression in rice and Arabidopsis. Plant Biotechnol J. 2:13–26.
2004.PubMed/NCBI
|