Introduction
Pancreatic cancer (PC) is a highly metastatic
malignancy. Despite advanced developments in surgery, radiation
therapy and chemotherapy for the treatment of PC, the 5-year
survival rate following surgery in PC patients remains extremely
poor (15–20%) (1). Therefore, it
is urgent to identify new therapeutic targets or early diagnosis
markers for PC.
Survivin (BIRC5) is a member of the inhibitors of
apoptosis protein (IAP) family, which includes seven other members,
as follows: X-linked inhibitor of apoptosis, cIAP1, cIAP2, NAIP,
livin, IAP-like protein 2 and BRUCE (2). Survivin functions to regulate cell
division, apoptosis, cellular stress responses and surveillance
checkpoints, and its expression is abnormally high in a number of
human malignancies, including esophageal, stomach, liver, brain,
lung, breast, ovary and hematological cancer (3–5). The
overexpression of Survivin is associated with advanced disease,
resistance to therapy, reduced survival and induced recurrence
(6). By contrast, the
downregulation of Survivin may reduce cell proliferation and
increase sensitivity to radiotherapy and cytotoxic drugs in various
cancer cell lines, including head, neck, thyroid, lung, bladder,
cervical and renal cancer (7–11).
These observations indicate that Survivin may represent a molecular
target for human cancer therapy.
MicroRNAs are a novel class of endogenous
single-stranded and non-coding RNA molecules. They are 19–24
nucleotides long and function as gene expression regulators by
targeting the 3′-untranslated region (UTR) of mRNA for degradation
or translational repression (12).
microRNAs have been identified to regulate cell proliferation,
apoptosis, migration, invasion and the cell cycle in various cancer
cell lines (13). In addition, it
was previously reported that the elevated expression of miR-203 is
associated with poor survival and may be used as a new prognostic
marker for PC (14,15). Notably, while miR-203 is
downregulated in hematopoietic malignancies and prostate cancer, it
is upregulated in ovarian, bladder and colon cancer (16–20).
These studies indicate that the role of miR-203 in tumorigenesis is
complex. More recent studies have reported that miR-203 inhibits
the proliferation of hepatocellular carcinoma (HCC) and laryngeal
cancer cells by targeting Survivin (21,22).
In the present study, PC cell lines were used as an
experimental model to investigate the expression and role of
miR-203 in PC. In addition, we explored the relationship between
miR-203 and Survivin expression and function, and aimed to
determine whether miR-203 directly targets Survivin in PC cells to
inhibit cancer progression.
Materials and methods
Cell culture
The human PC cell lines, SW1990, CFPAC-1, Panc-1 and
BxPc-3, were obtained from Shanghai Cell Bank (Shanghai, China) and
cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented
with 10% fetal bovine serum (FBS; both Wisent Inc., St-Bruno, QC,
Canada), 100 μg/ml streptomycin, 100 μg/ml penicillin and 2 mM
glutamine in a humidified chamber at 37°C with 5%
CO2.
miRNA and siRNA transfection
miRNA and Survivin shRNA virus (shSurvivin) were
designed and synthesized by Genepharma (Shanghai, China). CFPAC-1
cells were seeded in 6-cm tissue culture plates at a density of
50%. After 24 h, the cells were transfected with miRNAs or
shSurvivin virus using reduced serum medium (OPTI-MEM-I) according
to the manufacturer’s instructions. At 48 h post-transfection, the
fluorescent index of the cells reached 90%.
qPCR
Total RNA was extracted from the cells using TRIzol
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA), and
Primescript RT reagent (Takara Bio, Inc., Shiga, Japan) was used to
synthesize cDNA. qPCR was performed with a 7500 Real-Time-PCR
System (Applied Biosystems, Foster City, CA, USA) using the
following primers: miR-203 forward, 5′-GTCGTTACCAGTGCAGGGTCCGAGG
TATTCGCACTGGATACGACCTAGT-3′ and reverse,
5′-GCCCGTGAAATGTTTAGGACCAC-3′; U6 forward,
5′-ATTGGAACGATACAGAGAAGATT-3′ and reverse,
5′-GGAACGCTTCACGAATTTG-3′; Survivin forward,
5′-AGGACCACCGCATCTCTACATTC-3′ and reverse,
5′-CCTTGAAGCAGAAGAAACACTGGG-3′; and GAPDH forward,
5′-TCACCCACACTGTGCCCATCTACGA-3′ and reverse,
5′-CAGCGGAACCGCTCATTGCCAATGG-3′. GAPDH mRNA and U6 were used as
internal controls for determining the relative expression levels of
Survivin mRNA and miR-203, respectively. The comparative ΔΔCt
method was used to calculate the relative expression levels of mRNA
and miRNA, and the fold-changes were analyzed by
2−ΔΔCt.
Western blot analysis
Total protein was extracted from the cells using
RIPA buffer containing 1% phenylmethylsulfonyl fluoride (PMSF) and
the protein concentration was estimated with a BCA kit (Nanjing
KeyGen Biotech. Co., Ltd., Nanjing, China). Protein was resolved by
12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and
transferred to polyvinylidene difluoride membranes. The membranes
were blocked in Tris-buffered saline with 5% non-fat dry milk at
4°C for 12 h and subsequently incubated with rabbit polyclonal
anti-Survivin antibody (Abcam, Cambridge, MA, USA) or mouse
monoclonal anti-GAPDH antibody (Beyotime, Jiangsu, China) at 4°C
for 12 h, followed by incubation with horseradish
peroxidase-conjugated goat anti-rabbit or goat anti-mouse secondary
antibody (Beyotime, Jiangsu, China) for 2 h at room temperature.
Membranes were developed using an ECL kit (Pierce Biotechnology,
Inc., Rockford, IL, USA) and exposed onto X-ray films to visualize
the images. GAPDH served as a loading control.
Dual luciferase reporter assay
Four oligos corresponding to the 3′UTR of Survivin
were synthesized as follows: wild type,
5′-CTAGATAAAAAGCCTGTCATTTCAAACACTGC TGTGGACGGCCGG-3′ and
5′-CCGTCCACAGCA GTGTTTGAAATGACAGGCTTTTTAT-3′; and mutant,
5′-CTAGATAAAAAGCCTGTCGCACCA AACACTGCTGTGGACGGCCGG-3′ and 5′-CCGTCCA
CAGCAGTGTTTGGTGCGACAGGCTTTTTAT-3. The oligos were cloned into the
XbaI site of the pGL3 luciferase reporter gene (Promega
Corporation, Madison, WI, USA) to generate pGL3-Survivin-3′UTR and
pGL3-Survivin-3′UTR-mut vectors. CFPAC-1 cells were cultured in
24-well tissue culture plates and co-transfected with 200 ng
pGL3-Survivin or pGL3-Survivin-mut and 20 ng pRL-SV40 (Promega
Corporation) containing Renilla luciferase and 20 pmol 203M
or 203NC. At 48 h post-transfection, cells were collected and a
Dual-Luciferase Reporter assay kit (Promega Corporation) was used
to detect luciferase activity according to the manufacturer’s
instructions. All experiments were performed in triplicate.
Cell proliferation, apoptosis and cell
cycle analysis
An MTT kit (Nanjing KeyGen Biotech. Co., Ltd.) was
used to determine cell proliferation. The cells were seeded in
96-well tissue culture plates (Costar; Corning Inc., Acton, MA,
USA) at a density of 2×103 cells/well 24 h prior to
transfection with miRNA or siRNA, and then replaced with 10%
FBS-DMEM 6 h post-transfection and incubated for 48 h. MTT assays
were performed daily for 6 days as described previously (23). Cell cycle and apoptosis were
detected by flow cytometry (BD Biosciences, Franklin Lakes, NJ,
USA) as described previously (23). All experiments were performed in
triplicate.
Xenografts
A total of 16 four-week-old female nude BALB/cA-nu
(nu/nu) mice were obtained from the Shanghai Experimental Animal
Center (Chinese Academy of Sciences, Shanghai, China) and randomly
divided into four groups. CFPAC-1, CFPAC-1-203NC, CFPAC-1-203M and
CFPAC-1-shSurvivin cells were administered via a unilateral
subcutaneous injection into the flanks of the mice (106
cells/100 μl/flank). Tumors were measured using vernier calipers
every 5 days and the tumor volume was calculated using the
following formula: (width2 × length)/2. The mice were
then sacrificed after 30 days.
Statistical analysis
All data are presented as the mean ± SD. A Student’s
t-test was used to analyze the differences between groups.
Statistical analysis was performed with SPSS software (version
16.0; SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
hsa-miR-203 inhibits Survivin protein
expression in PC cells
Firstly, hsa-miR-203 and Survivin levels were
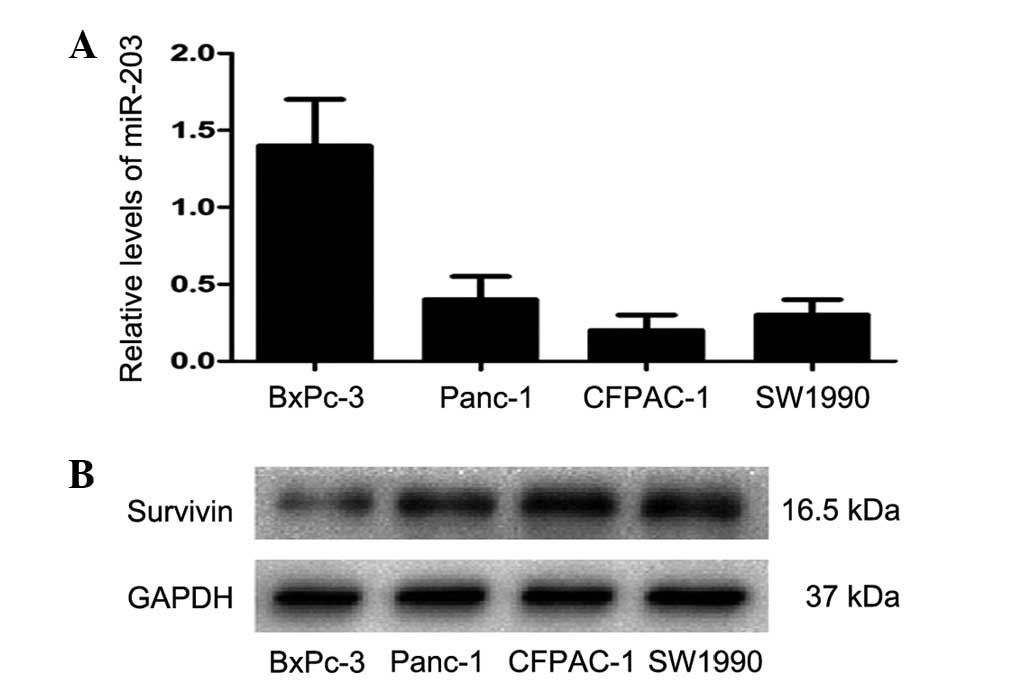
examined in four PC cell lines. qPCR revealed that miR-203
expression levels were highest in BxPc-3 cells and lowest in
CFPAC-1 cells (Fig. 1A). Western
blot analysis revealed that Survivin protein levels were highest in
CFPAC-1 cells and lowest in BxPc-3 cells (Fig. 1B). These results indicate that
Survivin protein and hsa-miR-203 levels are negatively correlated
in PC cells.
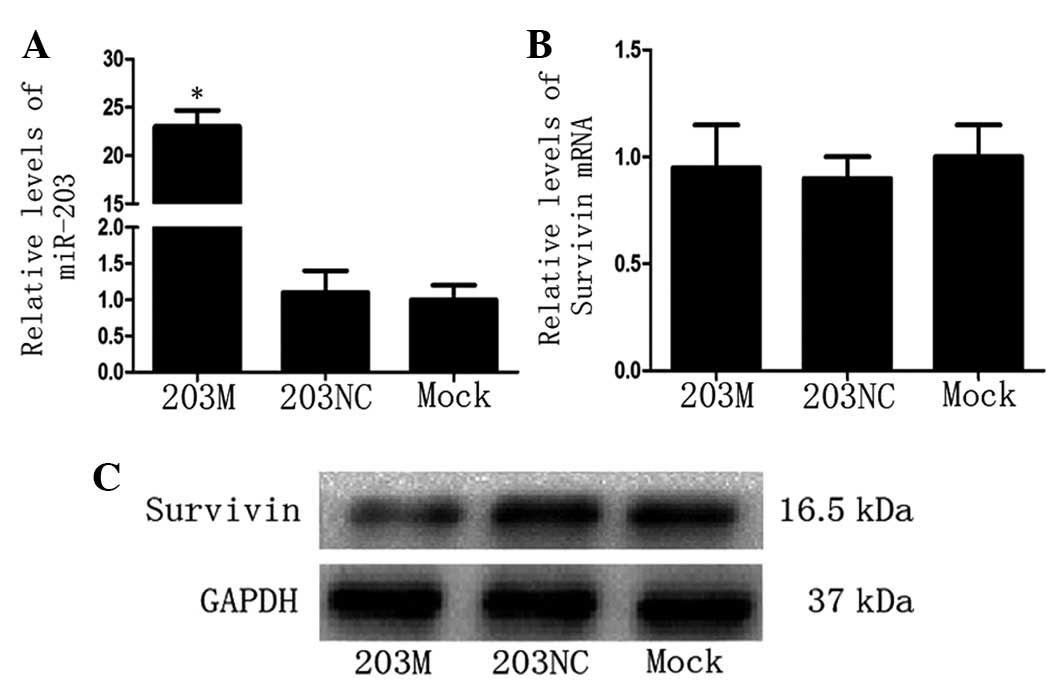
Since Survivin protein levels were high in CFPAC-1
cells, the 203M miRNA virus (miR-203 mimic) was transfected into
CFPAC-1 cells and Survivin mRNA and protein levels were detected.
As predicted, miR-203 levels increased significantly in the 203M
group (P<0.05 vs. the 203NC group; Fig. 2A). Notably, compared with the
control groups, there was no significant change in Survivin mRNA
levels in the 203M group (Fig.
2B); however, Survivin protein levels decreased significantly
in the 203M group (Fig. 2C). These
results indicate that miR-203 inhibits Survivin expression
post-transcriptionally.
Survivin is a direct target gene of
miR-203 in PC cells
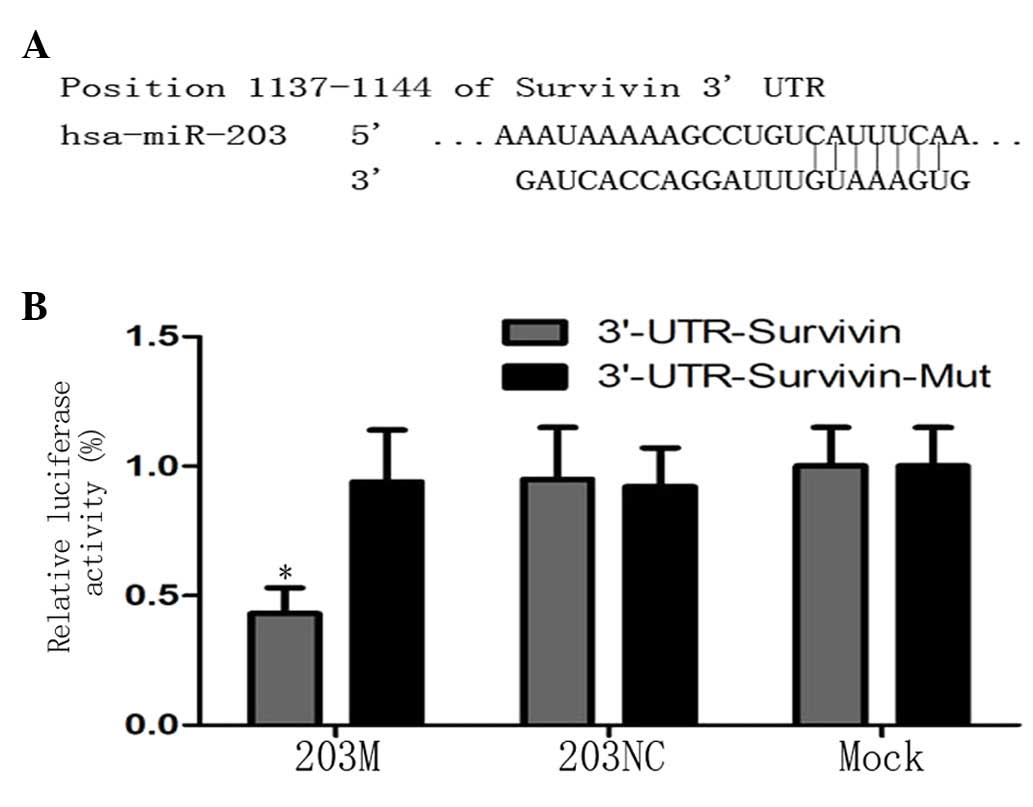
To confirm that Survivin is a direct target gene of
miR-203 in PC cells, TargetScan (http://www.targetscan.org) was used to predict the
3′UTR of Survivin and the binding site of miR-203 (Fig. 3A). Based on this prediction,
pGL3-Survivin-3′UTR and pGL3-Survivin-3′UTR-mut vectors were
constructed as a luciferase reporter and control, respectively, and
transfected into CFPAC-1 cells. The luciferase assay revealed that
luciferase activity was decreased by ~51% in the 203M group
compared with the controls (P<0.05; Fig. 3B). These results indicate that
miR-203 directly targets Survivin via the binding site in its 3′UTR
region.
hsa-miR-203 inhibits the proliferation
and promotes the apoptosis of CFPAC-1 cells
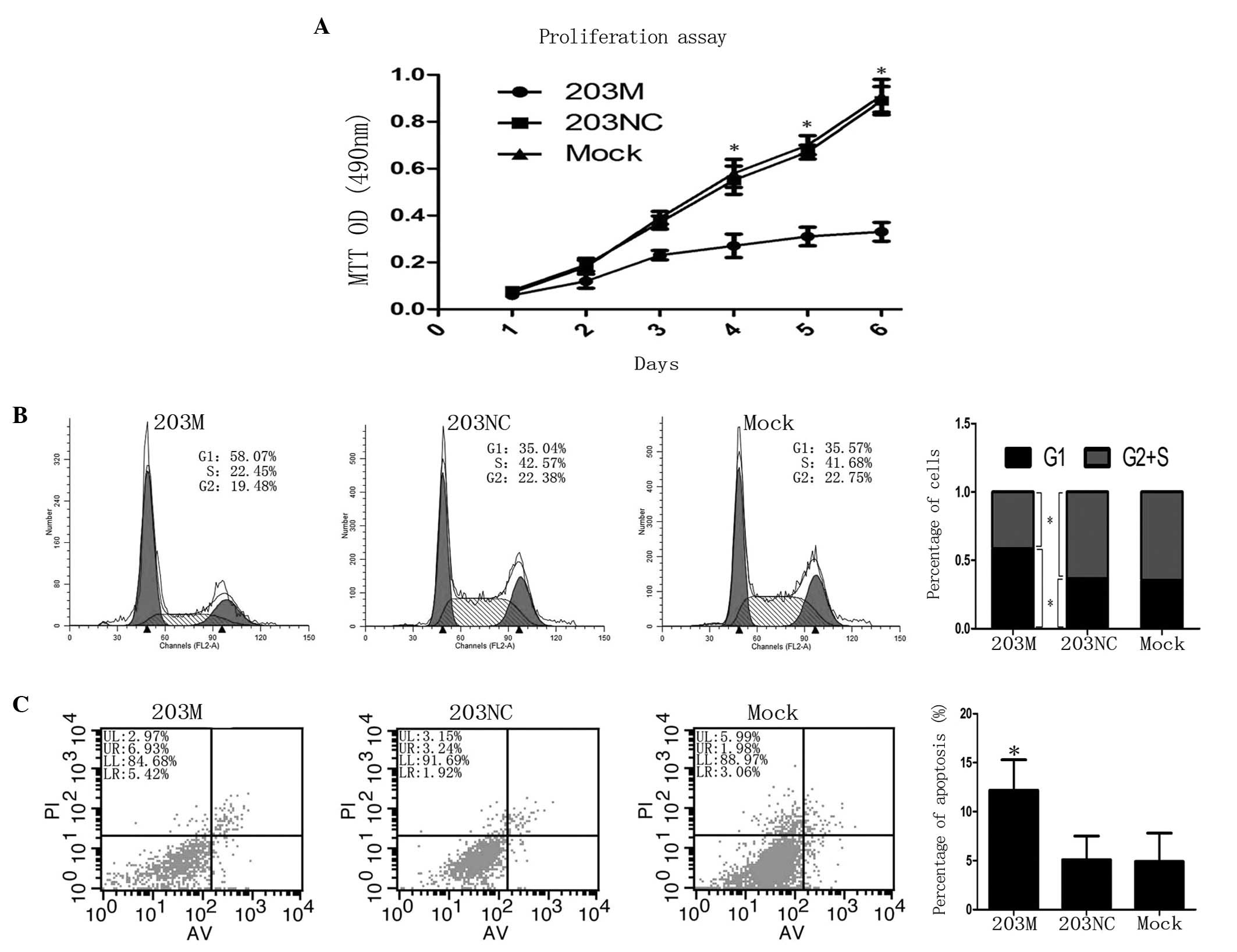
To characterize the role of hsa-miR-203 in PC cells,
hsa-miR-203 mimic 203M was transfected into CFPAC-1 cells. Cell
proliferation was observed to be significantly inhibited after day
4 when compared with the control (P<0.05; Fig. 4A). Next, flow cytometry was
employed to detect cell cycle progression and apoptosis. CFPAC-1
cells transfected with 203M revealed a reduced G2 + S
phase compared with the control (41.6±5.7 vs. 64.7±5.9%,
respectively; P<0.05), but exhibited increased G1
phase cell cycle arrest compared with the control (58.4±5.3 vs.
35.3±4.2%, respectively; P<0.05; Fig. 4B). In addition, the rate of
apoptosis was higher in CFPAC-1 cells transfected with 203M than in
the controls (12.2±2.1 vs. 5.1±1.3%, respectively; P<0.05;
Fig. 4C). Taken together, these
results indicate that miR-203 inhibits the proliferation of CFPAC-1
cells via the induction of G1 phase arrest and
apoptosis.
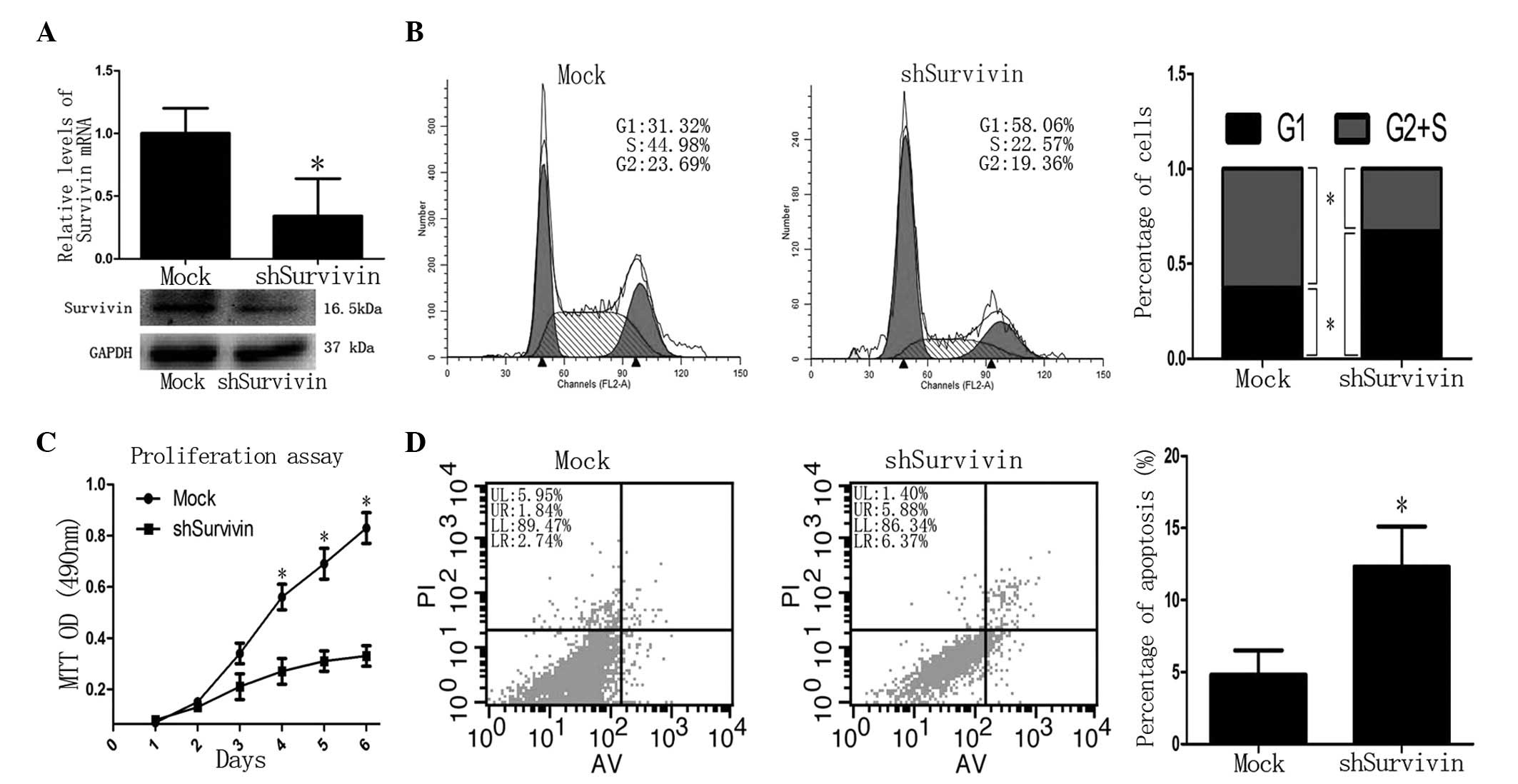
Knockdown of Survivin inhibits the
proliferation and promotes the apoptosis of CFPAC-1 cells
Next, the functional implication between miR-203 and
Survivin was investigated in PC cells. Following knockdown of
Survivin in CFPAC-1 cells by shRNA virus, Survivin mRNA and protein
levels were decreased significantly compared with the controls
(P<0.05; Fig. 5A). The MTT
assay revealed that the proliferation of shSurvivin-transfected
cells was decreased after 4 days (P<0.05 vs. control; Fig. 5C). Flow cytometric analysis
revealed that the G2 + S phase was decreased in
shSurvivin-transfected cells compared with the controls (37.3±4.6
vs. 67.2±4.8%, respectively; P<0.05) and the G1 phase
was increased in shSurvivin-transfected cells compared with the
control (62.7±5.3 vs. 32.8±3.9%, respectively; P<0.05; Fig. 5B). In addition, the rate of
apoptosis in shSurvivin-transfected cells was higher than in the
control (13.4±4.1 vs. 4.8±1.0%, respectively; P<0.05; Fig. 5D). These observations demonstrated
that the loss of Survivin produces a similar phenotype as the gain
of miR-203 in PC cells, indicating antagonism between Survivin and
miR-203.
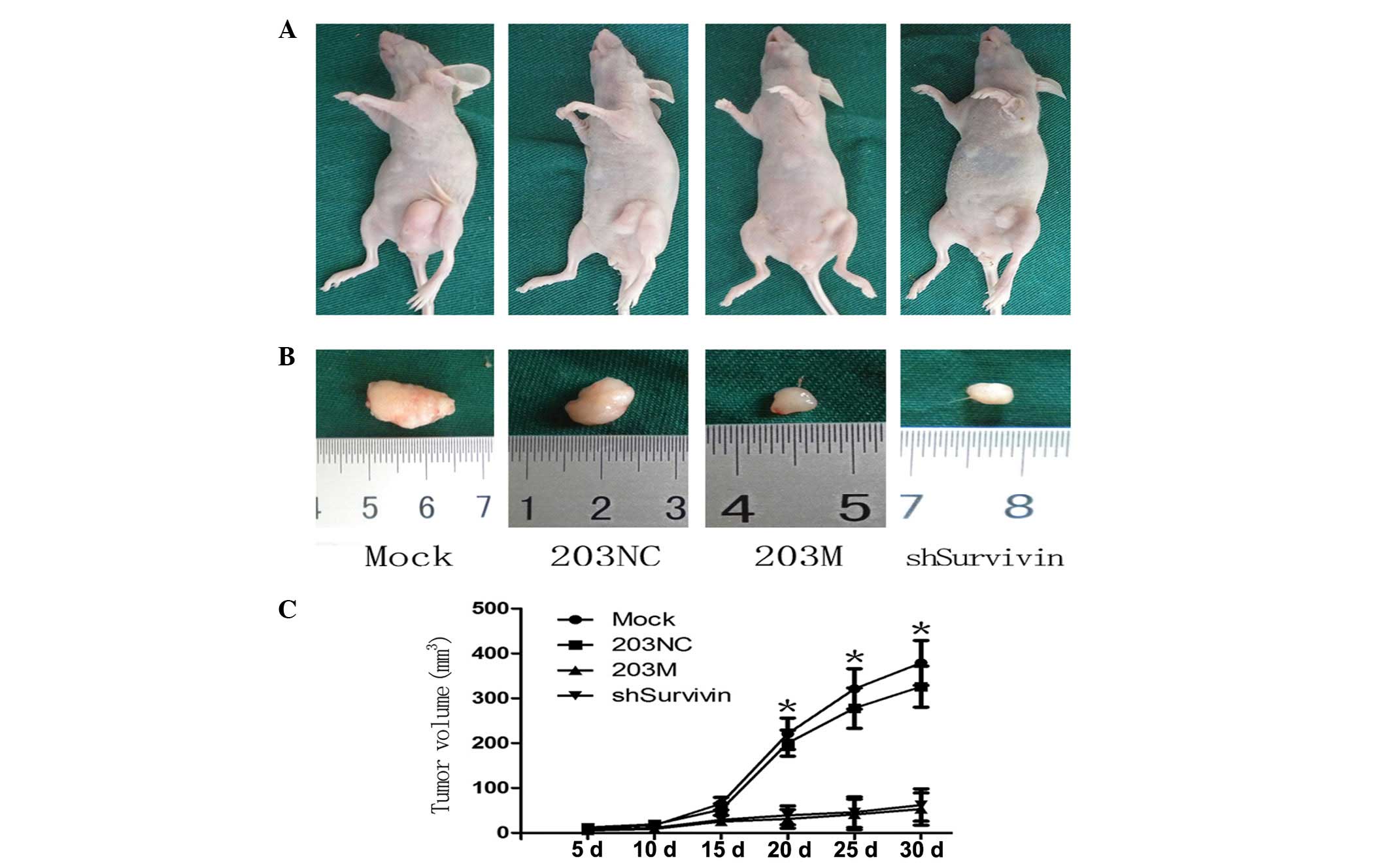
Downregulation of Survivin inhibits tumor
growth in vivo
Finally, the functional antagonism between miR-203
and Survivin in PC was investigated in vivo. Following
subcutaneous injection of transfected cells into the flanks of
BALB/cA-nu nude mice, the resulting tumors were measured every 5
days for 30 days, followed by euthanasia of the mice. Tumor growth
curves revealed that tumors of the shSurvivin group were
significantly smaller than those of the control groups after 20
days (Fig. 6A). The tumor sizes in
the 203M and shSurvivin groups were smaller than those in the
control groups (Fig. 6B). The
tumor growth curve demonstrated that tumor growth in the 203M and
shSurvivin groups was slower than that in the control groups
(Fig. 6C). These in vivo
results are consistent with the in vitro results of this
study and further confirm the antagonism between Survivin and
miR-203 in PC growth.
Discussion
In recent years, the aberrant expression of miRNAs
has been reported to be implicated in human malignancies (12,13).
While miR-203 is downregulated in hematopoietic malignancies and
prostate cancer, it is upregulated in ovarian, bladder and colon
cancer (16–20). In the present study, miR-203 levels
were found to negatively correlate with Survivin levels in PC
cells. In addition, in vitro cell proliferation and
apoptosis assays, as well as an in vivo xenograft model,
demonstrated that miR-203 mimic inhibited the malignant phenotypes
of CFPAC-1 cells. Notably, the knockdown of Survivin similarly
inhibited the malignant phenotypes of CFPAC-1 cells. A luciferase
assay further confirmed that miR-203 inhibited the expression of
Survivin by directly targeting its 3′UTR. Survivin is known to
promote cancer cell survival and drug resistance (6). It is reasonable to hypothesize that
miR-203 suppresses the expression of Survivin, leading to its loss
of oncogenic function. This prediction is consistent with the
downreguation of miR-203 and overexpression of Survivin in PC and
indicates that miR-203 is an anti-oncomir, at least in PC. These
observations are also consistent with previous results in HCC and
laryngeal cancer cells (21,22).
miR-203 was demonstrated to be overexpressed in
pancreatic adenocarcinoma samples with advanced disease, and
indicated a shorter survival time or poorer prognosis for patients
who underwent pancreatectomy (14,15).
The same outcome was demonstrated in colon cancer (20). These results indicate that miR-203
functions as an oncomir. By contrast, miR-203 has also been
reported to directly target oncogenes, including ABL1, Bcl-w,
Runx2, Scr, AKT2 and DNp63, functioning as a tumor-suppressor in
esophageal, gastric, hepatocellular, bladder, prostate and
colorectal cancer and hematological malignancies (16,23–27).
Therefore, the function of miR-203 in different tissues is complex
and further studies are required to understand its underlying
mechanisms.
In conclusion, in the present study, miR-203 was
demonstrated to inhibit the proliferation and induce the apoptosis
of PC cells. In addition, Survivin was identified as a direct
target of miR-203. These results indicate that miR-203 functions as
an anti-oncomir in PC and represents a potential molecular target
for PC therapy.
Acknowledgements
The present study was approved by the ethics
committee of Nanjing Medical University, Nanjing, Jiangsu, PR
China. (No. NJMU-ERLAUA-20120901). The authors thank Medjaden
Bioscience Limited for assistance in the preparation of this
manuscript.
References
|
1
|
Liu BB and Wang WH: Survivin and
pancreatic cancer. World J Clin Oncol. 2:164–168. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kelly RJ, Lopez-Chavez A, Citrin D, Janik
JE and Morris JC: Impacting tumor cell-fate by targeting the
inhibitor of apoptosis protein survivin. Mol Cancer. 10:352011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yamamoto H, Ngan CY and Monden M: Cancer
cells survive with survivin. Cancer Sci. 99:1709–1714. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pennati M, Folini M and Zaffaroni N:
Targeting survivin in cancer therapy. Expert Opin Ther Targets.
12:463–476. 2008. View Article : Google Scholar
|
|
5
|
Fukuda S and Pelus LM: Survivin, a cancer
target with an emerging role in normal adult tissues. Mol Cancer
Ther. 5:1087–1098. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cao C, Mu Y, Hallahan DE and Lu B: XIAP
and survivin as therapeutic targets for radiation sensitization in
preclinical models of lung cancer. Oncogene. 23:7047–7052. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sharma H, Sen S, Lo Muzio L, Mariggiò A
and Singh N: Antisense-mediated downregulation of anti-apoptotic
proteins induces apoptosis and sensitizes head and neck squamous
cell carcinoma cells to chemotherapy. Cancer Biol Ther. 4:720–727.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Du ZX, Zhang HY, Gao da X, Wang HQ, Li YJ
and Liu GL: Antisurvivin oligonucleotides inhibit growth and induce
apoptosis in human medullary thyroid carcinoma cells. Exp Mol Med.
38:230–240. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fuessel S, Kueppers B, Ning S, Kotzsch M,
Kraemer K, Schmidt U, Meye A and Wirth MP: Systematic in vitro
evaluation of survivin directed antisense oligodeoxynucleotides in
bladder cancer cells. J Urol. 171:2471–2476. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Chen ZD, Du CJ, Xu G and Luo W:
siRNA targeting survivin inhibits growth and induces apoptosis in
human renal clear cell carcinoma 786-O cells. Pathol Res Pract.
205:823–827. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li QX, Zhao J, Liu JY, Jia LT, Huang HY,
Xu YM, Zhang Y, Zhang R, Wang CJ, Yao LB, Chen SY and Yang AG:
Survivin stable knockdown by siRNA inhibits tumor cell growth and
angiogenesis in breast and cervical cancers. Cancer Biol Ther.
5:860–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hobert O: Gene regulation by transcription
factors and microRNAs. Science. 319:1785–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
14
|
Greither T, Grochola LF, Udelnow A,
Lautenschläger C, Würl P and Taubert H: Elevated expression of
microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated
with poorer survival. Int J Cancer. 126:73–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ikenaga N, Ohuchida K, Mizumoto K, Yu J,
Kayashima T, Sakai H, Fujita H, Nakata K and Tanaka M: MicroRNA-203
expression as a new prognostic marker of pancreatic adenocarcinoma.
Ann Surg Oncol. 17:3120–3128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bueno MJ, Pérez de Castro I, Gómez de
Cedrón M, Santos J, Calin GA, Cigudosa JC, Croce CM,
Fernández-Piqueras J and Malumbres M: Genetic and epigenetic
silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene
expression. Cancer Cell. 13:496–506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Viticchiè G, Lena AM, Latina A, Formosa A,
Gregersen LH, Lund AH, Bernardini S, Mauriello A, Miano R, Spagnoli
LG, Knight RA, Candi E and Melino G: MiR-203 controls
proliferation, migration and invasive potential of prostate cancer
cell lines. Cell Cycle. 10:1121–1131. 2011.PubMed/NCBI
|
|
18
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H,
Calin GA, Menard S and Croce CM: MicroRNA signatures in human
ovarian cancer. Cancer Res. 67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gottardo F, Liu CG, Ferracin M, Calin GA,
Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F,
Gomella LG, Croce CM and Baffa R: Micro-RNA profiling in kidney and
bladder cancers. Urol Oncol. 25:387–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu
CG, Calin GA, Croce CM and Harris CC: MicroRNA expression profiles
associated with prognosis and therapeutic outcome in colon
adenocarcinoma. JAMA. 299:425–436. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei W, Wanjun L, Hui S, Dongyue C, Xinjun
Y and Jisheng Z: miR-203 inhibits proliferation of HCC cells by
targeting survivin. Cell Biochem Funct. 31:82–85. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bian K, Fan J, Zhang X, Yang XW, Zhu HY,
Wang L, Sun JY, Meng YL, Cui PC, Cheng SY, Zhang J, Zhao J, Yang AG
and Zhang R: MicroRNA-203 leads to G1 phase cell cycle arrest in
laryngeal carcinoma cells by directly targeting survivin. FEBS
Lett. 586:804–809. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang F, Xue X, Wei J, An Y, Yao J, Cai H,
Wu J, Dai C, Qian Z, Xu Z and Miao Y: hsa-miR-520h downregulates
ABCG2 in pancreatic cancer cells to inhibit migration, invasion,
and side populations. Br J Cancer. 103:567–574. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bo J, Yang G, Huo K, Jiang H, Zhang L, Liu
D and Huang Y: microRNA-203 suppresses bladder cancer development
by repressing bcl-w expression. FEBS J. 278:786–792. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li J, Chen Y, Zhao J, Kong F and Zhang Y:
miR-203 reverses chemoresistance in p53-mutated colon cancer cells
through downregulation of Akt2 expression. Cancer Lett. 304:52–59.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Saini S, Arora S, Majid S, Shahryari V,
Chen Y, Deng G, Yamamura S, Ueno K and Dahiya R: Curcumin modulates
microRNA-203-mediated regulation of the Src-Akt axis in bladder
cancer. Cancer Prev Res (Phila). 4:1698–1709. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yuan Y, Zeng ZY, Liu XH, Gong DJ, Tao J,
Cheng HZ and Huang SD: MicroRNA-203 inhibits cell proliferation by
repressing ΔNp63 expression in human esophageal squamous cell
carcinoma. BMC Cancer. 11:572011.
|