Introduction
Lung cancer is the leading cause of cancer
mortalities worldwide. The main type of lung cancer is non-small
cell lung cancer (NSCLC), which may be divided into three
histologic subtypes: squamous-cell carcinoma (SCC), adenocarcinoma
(AC), and large-cell lung carcinoma (LCC) (1). Data from recent clinical trials
suggest a significant interaction between treatment efficacy and
tumor histology (2–7). Analysis of a phase III study with
pemetrexed demonstrated that survival was superior in non-squamous
NSCLCs compared to SCC (2,3). This finding may be explained by the
fact that thymidylate synthase, one of the main targets of
pemetrexed, was found to be differentially expressed among the
histotypes of lung cancer, with a lower expression in AC and a
higher expression in SCC and small-cell lung cancer (4,5).
Although the most important prediction factor of an epidermal
growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) has
been reported to be the EGFR mutation status rather than the
histology, the histology of AC remains an important clinical
predicting factor for EGFR-TKI, particularly gefitinib, in the case
that the mutation status is not obtained (6,7).
Bevacizumab, a targeted therapy agent that inhibits tumor
angiogenesis, has only been registered for the treatment of non-SCC
in the USA, Europe and other countries due to serious hemorrhagic
complications in SCC (3,8). Therefore, histological
characterization is the milestone for tailored therapy, which is
important in order to maximize the benefits of novel drugs and to
avoid hazardous side effects.
Immunohistochemical markers including TTF-1, p63,
cytokeratin 7 and high-molecular-weight cytokeratins have been
identified for subtyping AC and SCC. However, the sensitivity and
specificity of these diagnostic markers requires further definition
(5). Interobserver variability,
tumor heterogeneity and the degree of differentiation may also
contribute to variability in the histological subtyping of NSCLC.
microRNAs (miRNAs) are an abundant class of small non-protein
coding RNAs that function as negative gene regulators. miRNAs
regulate diverse biological processes, and evidence has shown that
miRNA mutations or the aberrant expression of miRNA are correlated
with various types of human cancer (9). Several miRNAs have been shown to
repress the expression of important cancer-related genes and have
proven to be useful in the diagnosis and treatment of cancer
(10,11).
We hypothesized that certain miRNA signatures may
provide a highly sensitive and specific diagnostic test for
distinguishing between SCC and AC. An initial screening of miRNA
was performed in 86 test samples from NSCLC patients using a
quantitative polymerase chain reaction (qPCR)-based miRNA array
analysis and three miRNAs whose expression levels varied
significantly between SCC and AC were identified. Algorithms
combined with the expression levels of these three miRNAs
accurately divided SCC and AC. The diagnostic assay developed was
validated with an additional set of 88 clinical NSCLC samples.
These newly identified miRNAs may prove to be highly attractive
molecular markers for the classification of NSCLC histological
subtypes, and may contribute to the pathogenesis of each subtype of
NSCLC.
Materials and methods
Patients and histologic diagnosis
Anonymous surgically resected NSCLC samples obtained
at the Keio University Hospital were obtained as indicated in
Table I. Institution review and
ethics approval was obtained for all the samples in accordance with
the institutional review board of Keio University (Institutional
Review Board #16-90-1). Tumor tissues were intraoperatively
dissected with the surrounding normal tissues, and paired normal
lung tissues were obtained from the same patients at an adjacent
area from their tumors. Serial cryostat sections from the specimens
were stained with HE for the histological diagnosis based on the
most recent WHO classification lung tumors (12,13).
HE-stained slides of all the cases were reviewed by Y.H. In case of
any discrepancy, the slide was also reviewed by M.S. and the
original pathologist who signed out the case, in order to achieve a
consensus.
 | Table IClinical characteristics of 86 test
samples obtained from NSCLC patients. |
Table I
Clinical characteristics of 86 test
samples obtained from NSCLC patients.
| Characteristics | AC | SCC | LCC | LCNEC | Total |
|---|
| Number of cases | 54 | 25 | 4 | 3 | 86 |
| Median age
(range) | 65 (36–83) | 72 (56–83) | 59 (51–79) | 76 (62–80) | 68 (36–83) |
| Gender |
| Male | 27 | 22 | 4 | 3 | 56 |
| Female | 27 | 3 | 0 | 0 | 30 |
| Smoking |
| Ever | 29 | 22 | 3 | 2 | 57 |
| Never | 23 | 2 | 1 | 1 | 26 |
| NA | 2 | 1 | 0 | 0 | 3 |
| Stage |
| I | 40 | 19 | 3 | 2 | 64 |
| II | 5 | 3 | 1 | 1 | 10 |
| I | 5 | 3 | 0 | 0 | 8 |
| IV | 1 | 0 | 0 | 0 | 1 |
| NA | 3 | 0 | 0 | 0 | 3 |
RNA extraction
Total RNA containing small RNA was extracted from
the tumor and adjacent normal tissues obtained during surgical
resection for a carcinoma located elsewhere in the lung. Frozen
tissue fragments were placed in TRIzol® reagent
(Invitrogen, Carlsbad, CA, USA) and homogenized with a Polytron
homogenizer (Kinematica AG, Lucerne, Switzerland), followed by RNA
extraction according to the manufacturer’s instructions.
miRNA array
The Megaplex RT (Applied Biosystems, Carlsbad, CA,
USA) was used to reverse transcribe 377 miRNAs using 50 ng total
RNA from each sample. The expression levels of 377 miRNAs that
included control U6 snRNA were measured using TaqMan microRNA array
human A v 2.0 (Applied Biosystems) according to the manufacturer’s
instructions.
qRT-PCR
The relative levels of miR-196b, miR-205 and miR-375
were determined by stem loop real-time qPCR using gene-specific
primers according to the TaqMan microRNA assay instructions. U6
snRNA was selected as the normalizer, as this miRNA demonstrated
minimal variation in expression among ACs and SCCs (data not
shown). Each miRNA was individually amplified in triplicate. The
relative levels of individual miRNAs, with reference to U6, were
calculated using the ΔCT method.
Data analysis and statistics
MATLAB software (MathWorks Inc., Natick, MA, USA)
was used for the statistical analyses of the array data. The tumor
expression level of each miRNA was calculated as the decadic
logarithm of the average of all the normal samples. The difference
in the decadic logarithm for each miRNA between SCC and AC was
calculated using the t-test with MATLAB software, followed by
extracting five miRNAs using a Pearson’s correlation of P<0.001.
As a result, miR-196b, miR-205 and miR-375 were selected as the
log10AC-log10SCC absolute values (AC/SCC
ratio) of the miRNAs, with a value of >0.6.
Unpaired t-test, receiver-operator characteristic
(ROC) curve, correlation coefficient and discriminant formula were
obtained using IBM SPSS statistics software (ver. 17.0, SPSS Inc.,
Chicago, IL, USA). The average Ct values of the triplicates
(AvgCtmiR196b, AvgCtmiR205,
AvgCtmiR375, AvgCtU6) detected by qRT-PCR
were calculated (outliers differing by a SD >1 were excluded).
The sample score was obtained using the formula: Score =
AvgCtmiR - AvgCtU6. The difference in the
sample score for each miRNA between SCC and AC was calculated using
the Mann-Whitney test as the distribution of miR-205 scores in the
SCC group and miR-375 scores in the AC group were not assumed to be
normal. ROC curves were constructed and the area under the ROC
curve (AUC) was calculated with SPSS software using the score for
each miRNA.
The sensitivity (%) for each miRNA was calculated by
dividing the sample number of each subtype, determined either from
the cut-off value or the discriminant function by the number
determined histologically. Cut-off values dividing the SCC and AC
groups were selected to reveal the maximum value of the Youden
index (sensitivity + specificity − 1), calculated from the ROC
curves. Coefficients and constants of discriminant function were
developed from discriminant analysis with SPSS software. If the
z-value was positive, the sample was judged to be AC and if the
z-value was negative, the sample was judged to be SCC. The
discriminant formulae for the miRNAs are as follows:
ZmiR196b=0.516 × CtmiR196b−5.956,
ZmiR205=0.393 × CtmiR205−2.169,
ZmiR375=−0.446 × CtmiR375+2.501,
ZmiR-196b and miR-205=0.269 × CtmiR196b+0.26
× CtmiR205−4.541, ZmiR-196b and miR-375=0.277
× CtmiR196b−0.314 × CtmiR375−1.422,
ZmiR-205 and miR-375=0.216 × CtmiR205−0.282 ×
CtmiR375+0.407, Zcombination=0.201 ×
CtmiR196b + 0.144 × CtmiR205−0.243 ×
CtmiR37−1.736.
Results
Identification of miRNAs differentially
expressed between AC and SCC
We initially performed miRNA profiling of 86
Japanese lung cancer samples that included 54 lung ACs, 25 SCCs, 4
LCCs and 3 large cell neuroendocrine carcinomas (LCNECs) using
miRNA arrays (Table I). We
compared the tumor miRNA expression levels relative to those in
adjacent normal tissues in SCC and those in AC, and identified five
differentially expressed miRNAs between the SCC and AC tumors
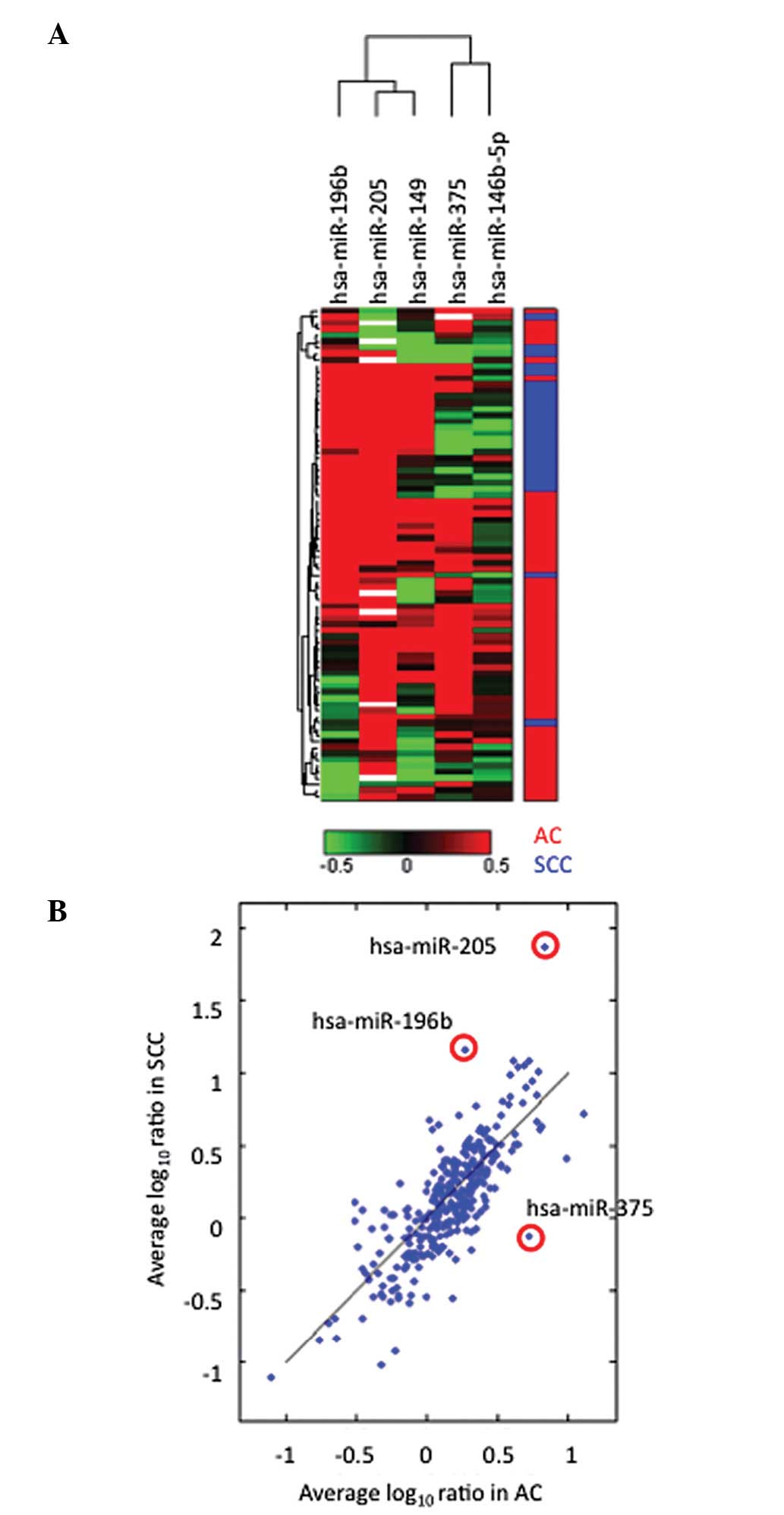
(Fig. 1A, P<0.001). Of the five
miRNAs, miR-196b, miR-205 and miR-375 exhibited significant
differential expression between SCC and AC (Fig. 1B). The difference in
log10SCC and log10AC was −0.86, −1.12 and
0.81 for miR-196b, miR-205 and miR-375, respectively. While
miR-196b and miR-205 were overexpressed in SCC, miR-375 was
overexpressed in AC.
Expression levels of miR-196b, miR-205
and miR-375
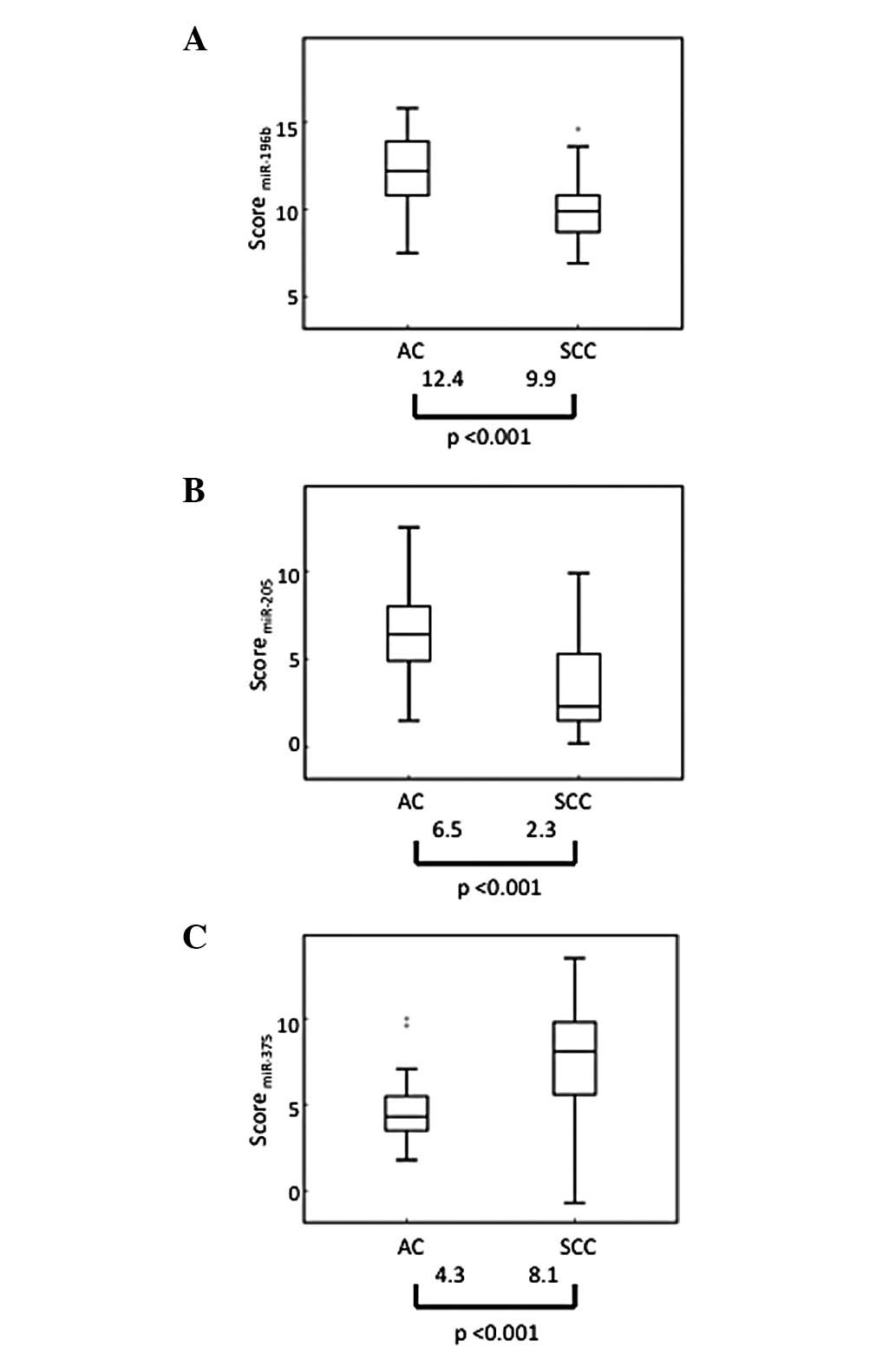
To confirm the array results, the expression levels
of the three miRNAs and the U6 snRNA of 86 samples were measured in
triplicate by qRT-PCR, independently from the array. The expression
patterns of these miRNAs in LCCs were similar to those in ACs,
while their expression patterns in LCNECs were similar to those in
SCCs (data not shown). The median values of each miRNA score varied
significantly between the SCC and AC groups (P<0.001) (Fig. 2).
Optimization of miRNA markers to
distinguish SCC from AC
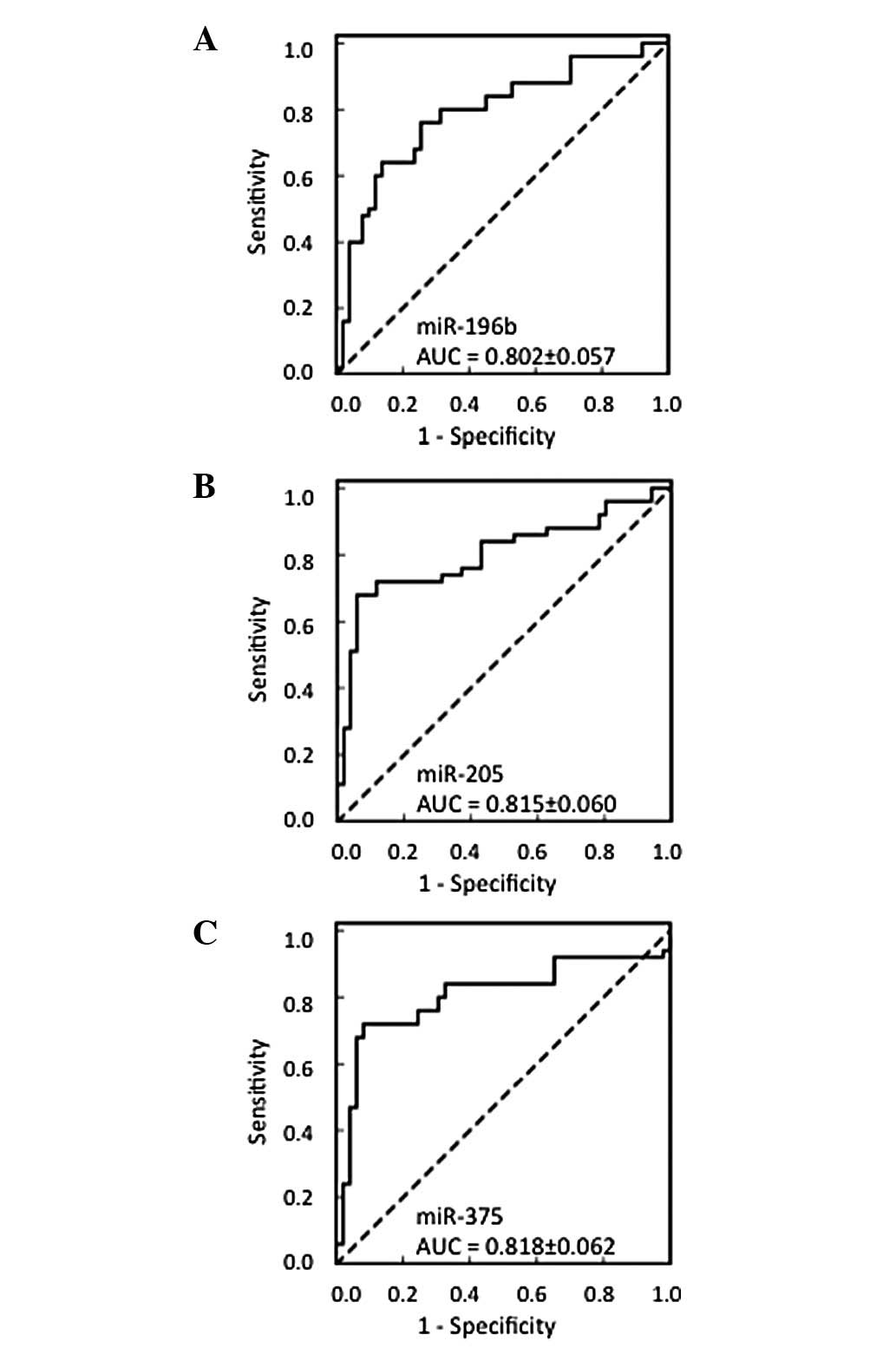
The AUC of the three miRNAs was >0.8, indicating
that a diagnosis by these markers would provide moderate accuracy
even as a single biomarker (Fig.
3). The cut-off value for each miRNA and the sensitivity from
each cut-off value used to distinguish SCC and AC are listed in
Table II. While the three miRNAs
demonstrate moderate correlation with each other, 0.558 for
miR-196b and -205, -0.440 for miR-196b and -375 and -0.568 for
miR-205 and -375, we introduced discriminant analysis by
calculating the z-value using scores of not only single miRNA but
two or three miRNAs (Table
III).
 | Table IISensitivity (%) by cut-off values
calculated from ROC curves. |
Table II
Sensitivity (%) by cut-off values
calculated from ROC curves.
| Variables | miR-196b | miR-205 | miR-375 |
|---|
| Cut-off value | 10.15 | 2.55 | 6.65 |
| AC | 88% | 96% | 92% |
| SCC | 64% | 68% | 72% |
 | Table IIISensitivity (%) by discriminant
analysis. |
Table III
Sensitivity (%) by discriminant
analysis.
| miRNAs |
|---|
|
|
|---|
| Samples | 196b | 205 | 375 | 196b and 205 | 196b and 375 | 205 and 375 | 196b, 205 and
375 |
|---|
| Test |
| AC | 63 | 63 | 76 | 69 | 71 | 80 | 80 |
| SCC | 80 | 76 | 72 | 80 | 76 | 76 | 76 |
| Validation |
| AC | 63 | 62 | 89 | 58 | 75 | 78 | 83 |
| SCC | 88 | 86 | 78 | 91 | 80 | 85 | 85 |
Validation
These classification methods with novel identified
miRNA markers were validated using an independent set of 88 NSCLC
samples that included 44 SCCs and 44 ACs. The expression level was
measured and the sample score was calculated using the established
method. The sensitivity of identifying SCC by all three miRNAs was
85% and the specificity was 83% (Table III).
Discussion
In this study, we demonstrated that three miRNAs,
hsa-miR-196b, hsa-miR-205 and hsa-miR-375, may be used for the
classification of AC and SCC of the lung. miR-196b is a novel
distinguishing marker of AC and SCC that we identified, while
miR-205 and miR-375 have already been reported to be candidates of
specific markers for lung SCC (14–16).
We observed that a combination of these three markers was more
accurate than a single marker for distinguishing SCC from AC.
Initially, we attempted to predict the histology
using cut-off values calculated from the ROC curves. The AUC of the
three miRNAs was >0.8, indicating that a diagnosis by these
markers would provide moderate accuracy even with a single
biomarker (Fig. 3). The
sensitivity for predicting AC was high (range, 88–96%), compared
with the sensitivity of predicting SCC (range, 64–72%). The
specificity of predicting AC was relatively low, (range, 64–72%)
for each of the three miRNAs using this classification method.
However, it is important to accurately diagnose SCC in order to
avoid adverse events and ineffective drug administration since in
the clinic there are more agents that are safe and effective for
non-SCC cases only. As the correlation between the three markers
was low, we believed that a higher sensitivity would be obtained by
combining the data for the three markers. We calculated the
prediction sensitivities by discriminant function analysis for each
miRNA. In contrast to the result using cut-off values, we observed
a higher sensitivity for predicting SCC and a higher specificity
for predicting AC. The results also suggested that subtyping by two
or three miRNAs is more appropriate for diagnosis, due to the
increased sensitivity for SCC and AC rather than by subtyping for a
single miRNA. The result was confirmed with validation samples and
only the combination of all three samples demonstrated >80%
sensitivity for the two histological types. It is thought that the
z-value calculated by discriminant analysis would have more
generality than the cut-off value determined by Ct score as the
z-value is normalized, while the Ct score itself is a relative
value and may differ in each instituion or equipment used.
Therefore, classification by discriminant analyses using three
miRNAs is recommended for the future classification of SCC and AC.
As the size of the coefficient is proportional to the magnitude of
the impact of each miRNA, miR-375 is thought to be the most
effective factor among the three miRNAs for discrimination in all
combinations. The significance of a combination might be increased
by opposite-type markers, such as miR-375, which is overexpressed
in AC, and miR-196b or miR-205, which are overexpressed in SCC.
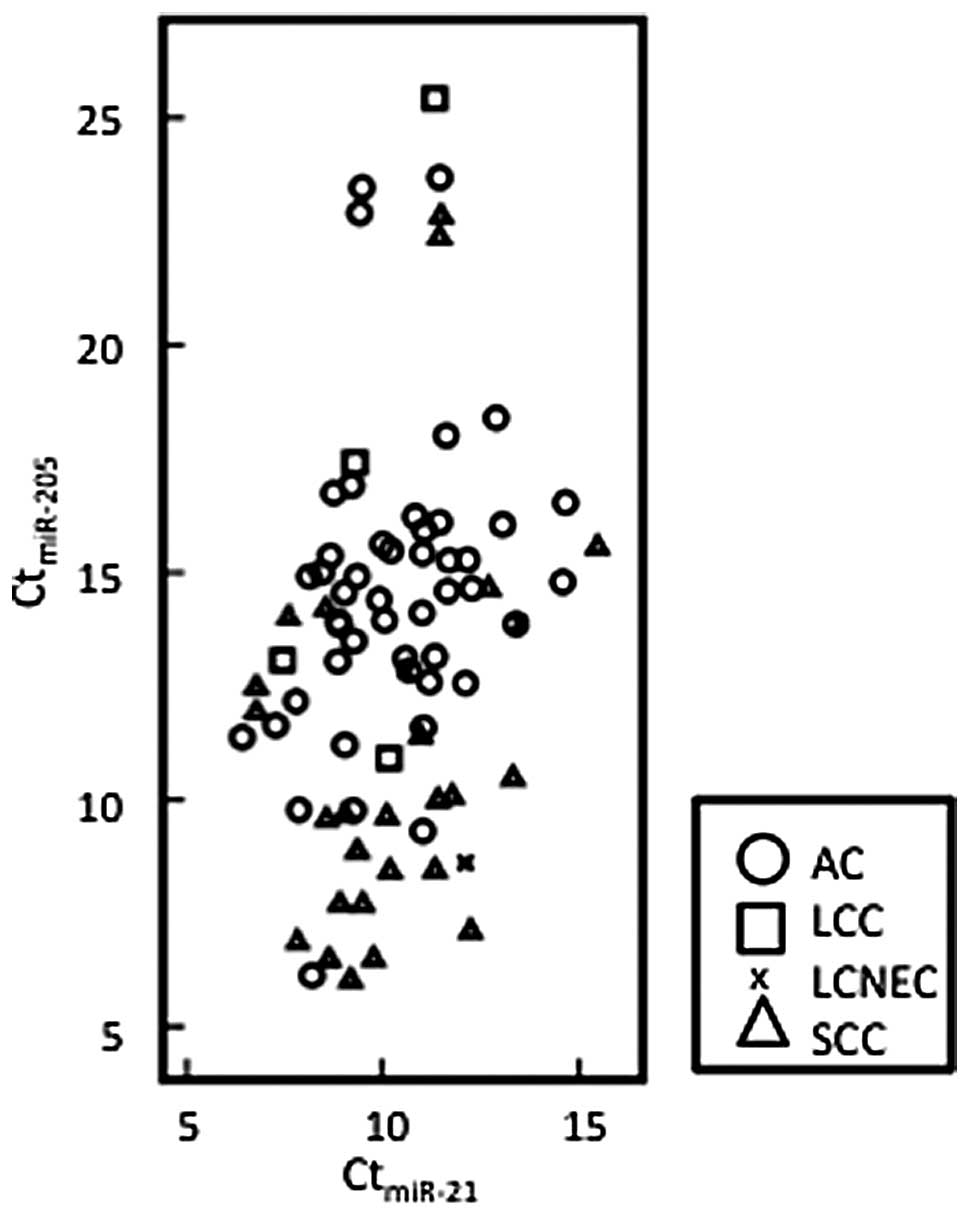
Lebanony et al and Bishop et al
reported that miR-205 alone sufficiently distinguished SCC from
non-SCC and they obtained a straight line between SCC and non-SCC
when they plotted the expression levels of normalized miR-205 and
miR-21 (14,16). We applied the same approach to our
test samples, however, we did not obtain a clear line to
distinguish histology types as shown in Fig. 4. Although we also observed that SCC
expressed higher levels of miR-205 compared with AC, the expression
pattern of miR-205 was more complex. Their formula, using U6 and
miR-21, revealed only 72% sensitivity in the identification of SCC
in our experiments, although 96 or 100% was previously reported
(14,16). Although it is difficult to explain
this discrepancy, it may be due to ethnic differences or
heterogeneity of the tissue samples.
Overexpression of miR-196b has been observed in
colon cancer or in leukemia associated with the mixed-lineage
leukemia (MLL) gene, leading to increased proliferative capacity
and survival, in addition to a partial block in the differentiation
specifically in bone marrow progenitor cells (17,18).
miR-196a, a member of the miR-196b family, was reported to increase
the development of lung metastases in mice following tail vein
injection (19). High levels of
miR-196a were found to activate the AKT signaling pathway as
indicated by the increased phosphorylation of AKT and promoted
cancer cell detachment, migration and invasion but did not have an
impact on proliferation or apoptosis (19).
It has been verified that miR-205 is not only a
tumor suppressor in certain cancers, including breast, and head and
neck cancer (20,21), but is also a candidate as a highly
specific marker for lung SCCs. A recent study determined that the
expression of PTEN is regulated by miR-205 (22).
In our study miR-375 was overexpressed in AC in
accordance with a previous report which found that miR-375, in
combination with miR-21, miR-486 and miR-200b, distinguishes lung
AC patients from normal subjects (15). miR-375 was also elevated in AC
compared with SCC in esophageal carcinoma (23). miR-375 is reported to target PDK1
leading to inactivation of the PI3K pathway (24). The PI3K pathway is more activated
in SCC than in AC (1). The
expression levels of miR-196b, miR-205 and miR-375 may explain the
difference in the PI3K activation status between AC and SCC,
although this requires further clarification.
LCC is further divided into LCNEC, basaloid
carcinoma, and lymphoepithelioma-like carcinoma. The LCNEC samples
in our study expressed higher levels of miR-196b and miR-205, but
lower levels of miR-375, demonstrating SCC-like properties.
However, other LCC samples, with the exception of LCNECs, were
accurately classified into the AC group by the three miRNA markers
identified in this study. This finding suggests that it
inappropriate to classify LCNEC into LCC according to the miRNA
marker method. LCNEC is actually known to be genetically and
immunohistochemically more similar to SCLC than NSCLC (25,26).
Conventional histological subtyping of NSCLC,
including immunohistochemical staining, has limitations with
respect to the accurate diagnosis of NSCLC (5). There are certain cases in which
samples are not capable of being evaluated by immunohistochemistry
due to their small biopsy size or loss of immunoreactivity.
Therefore, we believe that measuring miRNA expression levels may be
a complementary diagnostic technique that assists with the
immunohistochemical profiling of cancer biopsies with unknown
histology.
In conclusion, we demonstrated that hsa-miR-196b,
hsa-miR-205 and hsa-miR-375 are highly valuable molecular markers
for the classification of NSCLC histologic subtypes.
Acknowledgements
The authors would like to thank Ms. Mikiko Shibuya
for her technical assistance. This study was supported in part by
Grants-in-Aid for Scientific Research on Priority Areas from the
Ministry of Education, Culture, Sports, Science and Technology of
Japan to K.S. (grant no. 22590870) and J.H. (grant no.
23790920).
References
|
1
|
Herbst RS, Heymach JV and Lippman SM: Lung
Cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Scagliotti GV, Parikh P, von Pawel J, et
al: Phase III study comparing cisplatin plus pemetrexed in
chemotherapy-naive patients with advanced-stage non-small-cell lung
cancer. J Clin Oncol. 26:3543–3551. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ciuleanu T, Brodowicz T, Zielinski C, et
al: Maintenance pemetrexed plus best supportive care versus placebo
plus best supportive care for non-small-cell lung cancer: a
randomised, double-blind, phase 3 study. Lancet. 374:1432–1440.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gridelli C, Ardizzoni A, Douillard JY, et
al: Recent issues in first-line treatment of advanced
non-small-cell lung cancer: Results of an International Expert
Panel Meeting of the Italian Association of Thoracic Oncology. Lung
Cancer. 68:319–331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rossi G, Pelosi G, Graziano P, Barbareschi
M and Papotti M: A reevaluation of the clinical significance of
histological subtyping of non-small-cell lung carcinoma: diagnostic
algorithms in the era of personalized treatments. Int J Surg
Pathol. 17:206–218. 2009. View Article : Google Scholar
|
|
6
|
Mok TS, Wu YL and Thongprasert S:
Gefitinib or carboplatinpaclitaxel in pulmonary adenocarcinoma. N
Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kawada I, Soejima K, Watanabe H, et al: An
alternative method for screening EGFR mutation using RFLP in
non-small cell lung cancer patients. J Thorac Oncol. 3:1096–1103.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sandler A, Gray R, Perry MC, et al:
Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell
lung cancer. N Engl J Med. 355:2542–2550. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 4:259–269. 2006.
View Article : Google Scholar
|
|
10
|
Raponi D, Dossey L, Jatkoe T, et al:
MicroRNA classifiers for predicting prognosis of squamous cell lung
cancer. Cancer Res. 69:5776–5783. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 3:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Colby TV, Noguchi M, Henschke C, et al:
Adenocarcinoma. World Health Organization classification of
tumours. Pathology and Genetics. Tumours of the Lung, Pleura,
Thymus and Heart. Travis WD, Brambilla E, Muller-Hemelink HK and
Harris CC: IARC Press; Lyon: pp. 35–44. 2005
|
|
13
|
Hammar SP, Brambilla C, Pugatch B, et al:
Squamous cell carcinoma. World Health Organization classification
of tumours. Pathology and Genetics. Tumours of the Lung, Pleura,
Thymus and Heart. Travis WD, Brambilla E, Muller-Hemelink HK and
Harris CC: IARC Press; Lyon: pp. 26–30. 2005
|
|
14
|
Lebanony D, Benjamin H, Gilad S, et al:
Diagnostic assay based on hsa-miR-205 expression distinguishes
squamous from nonsquamous non-small-cell lung carcinoma. J Clin
Oncol. 27:2030–2037. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu L, Todd NW, Xing L, et al: Early
detection of lung adenocarcinoma in sputum by a panel of microRNA
markers. Int J Cancer. 127:2870–2878. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bishop JA, Benjamin H, Cholakh H, Chajut
A, Clark DP and Westra WH: Accurate classification of non-small
cell lung carcinoma using a novel microRNA-based approach. Clin
Cancer Res. 16:610–619. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen
XM and Gao HJ: Initial study of microRNA expression profiles of
colonic cancer without lymph node metastasis. J Dig Dis. 11:50–54.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Popovic R, Riesbeck LE, Velu CS, et al:
Regulation of mir-196b by MLL and its overexpression by MLL fusions
contributes to immortalization. Blood. 113:3314–3322. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schimanski CC, Berger M, Rahman F, et al:
High miR-196a levels promote the oncogenic phenotype of colorectal
cancer cells. World J Gastroenterol. 15:2089–2096. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu H and Mo YY: Targeting miR-205 in
breast cancer. Expert Opin Ther Targets. 13:1439–1448. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Childs G, Fazzari M, Kung G, et al:
Low-level expression of microRNAs let-7d and miR-205 are prognostic
markers of head and neck squamous cell carcinoma. Am J Pathol.
17:736–745. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Greene SB, Gunaratne PH, Hammond SM and
Rosen JM: A putative role for microRNA-205 in mammary epithelial
cell progenitors. J Cell Sci. 123:606–618. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mathé EA, Nguyen GH, Bowman ED, et al:
MicroRNA expression in squamous cell carcinoma and adenocarcinoma
of the esophagus: associations with survival. Clin Cancer Res.
15:6192–6200. 2009.PubMed/NCBI
|
|
24
|
El Ouaamari A, Baroukh N, Martens GA,
Lebrun P, Pipeleers D and van Obberghen E: miR-375 targets
3’-phosphoinositide-dependent protein kinase-1 and regulates
glucose-induced biological responses in pancreatic beta-cells.
Diabetes. 57:2708–2717. 2008.
|
|
25
|
Saji H, Tsuboi M, Matsubayashi J, et al:
Clinical response of large cell neuroendocrine carcinoma of the
lung to perioperative adjuvant chemotherapy. Anticancer Drugs.
21:89–83. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kozuki T, Fujimoto N, Ueoka H, et al:
Complexity in the treatment of pulmonary large cell
neuroendocrinecarcinoma. J Cancer Res Clin Oncol. 131:147–51. 2005.
View Article : Google Scholar : PubMed/NCBI
|