Introduction
Cancer is a major public health problem worldwide.
Epidemiological and animal studies have indicated that the
consumption of fruit and vegetables containing chemopreventive
natural products, alone or in combination with others, is
associated with a reduced risk of cancer development (1,2). For
>15 years, many studies have screened natural phytochemical
products in vegetables and fruits for inhibitors of DNA metabolic
enzymes, primarily mammalian DNA polymerases (pols) and human DNA
topoisomerases (topos).
Pols (DNA-dependent DNA polymerases, E.C. 2.7.7.7)
catalyze deoxyribonucleotide addition to the 3′-hydroxyl terminus
of primed double-stranded DNA (dsDNA) molecules (3). The human genome encodes at least 15
pols that have functions in cellular DNA synthesis (4,5).
Eukaryotic cells contain three replicative pols (α, δ and ɛ), one
mitochondrial pol (γ), and at least 11 non-replicative pols [β, ζ,
η, δ, ι, κ, λ, μ, ν, terminal deoxynucleotidyl transferase (TdT)
and REV1] (6,7). Pols have a highly conserved structure
and their overall catalytic subunits show little variance among
species. Conserved enzyme structures are normally preserved over
time due to the fact that they perform important cellular functions
that confer evolutionary advantages. On the basis of sequence
homology, eukaryotic pols may be divided into four main families:
A, B, X and Y (6). Family A
includes mitochondrial pol γ in addition to pols δ and ν. Family B
includes pol ζ and the three replicative pols α, δ and ɛ. Family X
comprises TdT and pols β, λ and μ. Family Y includes pols η, ι and
κ, in addition to REV1.
Topos are nuclear enzymes that alter the DNA
topology required for the replication, transcription, recombination
and segregation of daughter chromosomes (8). Eukaryotic cells have Type I and Type
II topos. Topo I catalyzes the passage of the DNA strand through a
transient single-strand break in the absence of a high-energy
cofactor. Topo II, by contrast, catalyzes the passage of DNA double
strands through a transient double-strand break in the presence of
ATP.
Due to their antiproliferative and cytotoxic
effects, selective inhibitors of pols and topos are considered to
be useful as anticancer, antiviral, antiparasitic and birth control
agents (9–11). In screening for these enzyme
inhibitors, we focused on low molecular weight (LMW) polyphenolics
isolated from Chaga, a medicinal mushroom [Inonotus obliquus
(persoon) Pilat] (12), including
caffeic acid (CA, 3,4-dihydroxycinnamic acid, compound 1),
3,4-dihydroxybenzalacetone (DBL, compound 2), and hydroxyl benzoic
acid derivatives such as gallic acid (GA, 3,4,5-trihydroxybenzoic
acid, compound 3) and their derivatives (Fig. 1). Inonotus obliquus is used
as a folk medicine in countries such as Korea, Japan and Russia; in
Russia it is also used as a source of anticancer medicine (13). The anticancer activities of
Inonotus obliquus extract and its components have been
examined in vitro(14,15).
LMW phenolic compounds such as CA, ferulic acid and GA are among
the major phenolic compounds derived from fruits, vegetables,
grains and coffee. These diet-associated phenolic compounds are
often described as potential antioxidants and, consequently, as
inhibitors of deleterious oxidative processes associated with
cardiovascular and inflammatory diseases, and cancer (16,17).
Phenolic compounds potentially act as chemopreventive and/or
chemotherapeutic agents (16–18).
Specifically, these LMW polyphenolics exhibit antioxidant
properties in addition to biological activity towards several tumor
cells, as their growth inhibitory effects are markedly dependent on
their structural characteristics (19).
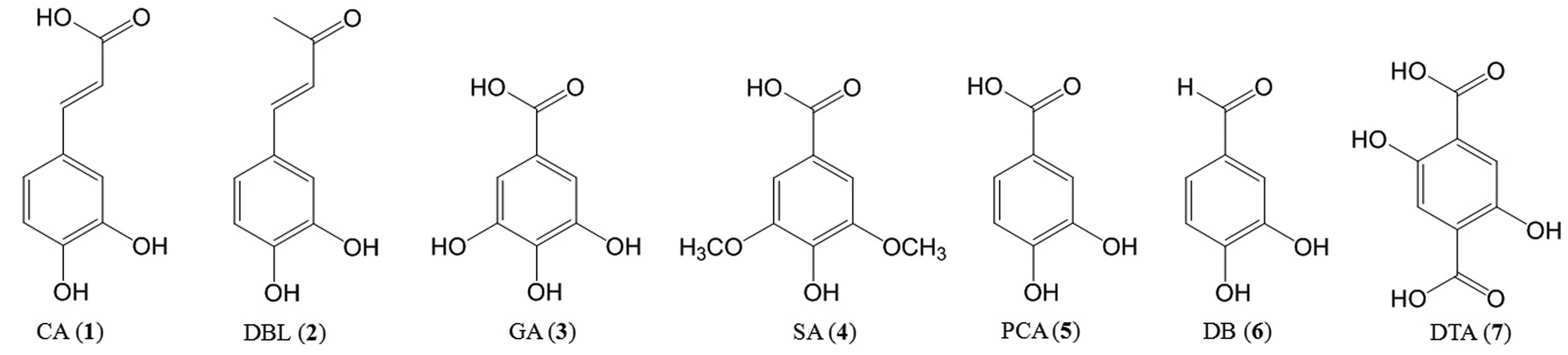 | Figure 1Structures of Inonotus
obliquus low molecular weight polyphenolic compounds. (1) Caffeic acid (CA, 3,4-dihydroxycinnamic
acid); (2)
3,4-dihydroxy-benzalacetone (DBL); (3) gallic acid (GA,
3,4,5-trihydroxy-benzoic acid); (4) syringic acid (SA,
4-hydroxy-3,5-dimethoxy-benzoic acid); (5) protocatechuic acid (PCA,
3,4-dihydroxy-benzoic acid); (6)
3,4-dihydroxy-benzaldehyde (DB); (7) 2,5-dihydroxy-terephthalic acid
(DTA). |
The purpose of this study was to discover novel
bioactivities among seven LMW polyphenolic compounds isolated from
Inonotus obliquus as shown in Fig. 1. We investigated whether these
compounds inhibit enzymes involved in DNA metabolism, such as pols
and topos, or whether they block the replication of the colorectal
cancer cell line HCT116. We identified two catechol propanoid
compounds, CA and DBL, that have possible anticancer activity.
Materials and methods
Materials
CA, GA, 3,4-dihydroxybenzaldehyde (DB, compound 6)
and 2,5-dihydroxyterephthalic acid (DTA, compound 7) were purchased
from Sigma-Aldrich (St. Louis, MO, USA). Syringic acid (SA,
4-hydroxy-3,5-dimethoxybenzoic acid, compound 4) and protocatechuic
acid (PCA, 3,4-dihydroxybenzoic acid, compound 5) were purchased
from WAKO Chemical Co. Ltd. (Tokyo, Japan). DBL was synthesized and
kindly provided by Dr Yutaka Nakamura of the Synthetic Organic
Chemistry Lab, Niigata University of Pharmacy and Applied Life
Sciences (Niigata, Japan). All compounds were primarily isolated
and purified from Inonotus obliquus(12), and their chemical structures are
shown in Fig. 1. The compounds,
purified using HPLC, were of analytical grade. A chemically
synthesized DNA template, poly(dA), was purchased from
Sigma-Aldrich and a customized oligo(dT)18 DNA primer
was purchased from Sigma-Aldrich Japan K.K. (Hokkaido, Japan).
Radioactive nucleotide [3H]-labeled
2′-deoxythymidine-5′-triphosphate (dTTP; 43 Ci/mmol) was obtained
from Moravek Biochemicals Inc. (Brea, CA, USA). Supercoiled pBR322
plasmid dsDNA was obtained from Takara Bio, Inc. (Kyoto, Japan).
All other reagents such as buffers were of analytical grade and
were obtained from Nacalai Tesque Inc. (Kyoto, Japan).
Enzymes
Pol α was purified from calf thymus by
immuno-affinity column chromatography, as described by Tamai et
al(20). Recombinant rat pol β
was purified from Escherichia coli (E. coli) JMpβ5,
as described by Date et al(21). The human pol γ catalytic gene was
cloned into pFastBac. The histidine-tagged enzyme was expressed
using the Bacto-Bac HT Baculovirus Expression system according to
the manufacturer's instructions (Life Technologies, Frederick, MD,
USA) and was purified using ProBond resin (Invitrogen Japan, Tokyo,
Japan) (22). Human pols δ and ɛ
were purified by nuclear fractionation of human peripheral blood
cancer cells (MOLT-4) using the second subunit of pol δ and
ɛ-conjugated affinity column chromatography, respectively (23). A truncated form of human pol κ
(residues 1–511) tagged with His6 at its C-terminal was
expressed in E. coli cells and purified as described by
Kusumoto et al(24). A
recombinant mouse pol ι tagged with His6 at its
C-terminal was expressed by E. coli and purified by Ni-NTA
column chromatography. A truncated form of pol κ (residues 1–560)
with six His-tags attached at the C-terminus was overproduced in
E. coli and purified as described by Ohashi et
al(25). Recombinant human
His-pol λ was overexpressed in E. coli and purified
according to a method described by Shimazaki et al(26). Recombinant human His-pol μ was
overexpressed in E. coli BL21 and purified by Glutathione
Sepharose™ 4B column chromatography (GE Healthcare Bio-Science
Corp., Piscataway Township, NJ, USA) following the same method as
for pol λ (26). Calf TdT, T7 RNA
polymerase, T4 polynucleotide kinase and bovine pancreas
deoxyribonuclease I were purchased from Takara Bio, Inc. Purified
human placenta topos I and II were purchased from TopoGen Inc.
(Port Orange, FL, USA).
Measurement of pol activity
Reaction mixtures for calf pol α and rat pol β have
been previously described by Mizushima et al(27,28);
those for pol γ and for pols δ and ɛ were as described by Umeda
et al(22) and Ogawa et
al(29), respectively.
Reaction mixtures for pols η, ι and κ were the same as those for
pol α and the mixtures for pols λ, μ and TdT were the same as those
for pol β. For the pol reactions, poly(dA)/oligo(dT)18
(A/T, 2/1) and dTTP were used as the DNA template-primer substrate
and nucleotide (dNTP; 2′-deoxynucleoside-5′-triphosphate)
substrate, respectively. For TdT reactions, oligo(dT)18
(3′-OH) and dTTP were used as the DNA primer substrate and
nucleotide substrate, respectively.
The test compounds 1–7 were dissolved in various
concentrations of distilled DMSO and sonicated for 30 sec.
Subsequently, 4-μl aliquots were mixed with 16 μl of each enzyme
(0.05 units) in 50 mM Tris-HCl at pH 7.5 that contained 1 mM
dithiothreitol, 50% glycerol (by vol) and 0.1 mM EDTA. The mixtures
were maintained at 0°C for 10 min. Next, 8 μl of each
inhibitor-enzyme mixture was added to 16 μl of the enzyme standard
reaction mixture and incubated at 37°C for 60 min. The activity of
samples without inhibitors was considered to be 100%, and the
activity was determined for each inhibitor concentration relative
to the uninhibited activity. One unit of pol activity was defined
as the amount of each enzyme that catalyzed the incorporation of 1
nmol dTTP into synthetic DNA template-primers in 60 min at 37°C
under normal reaction conditions (27,28).
Measurement of topo activity
The catalytic activity of topo I was determined by
detecting supercoiled plasmid DNA (Form I) in its nicked form (Form
II) (30). The topo I reaction was
performed in a 20-μl reaction mixture that contained 10 mM Tris-HCl
(pH 7.9), pBR322 DNA (250 ng), 1 mM EDTA, 150 mM NaCl, 0.1% BSA,
0.1 mM spermidine, 5% glycerol, 2 μl of one of the seven test
compounds dissolved in DMSO and 2 units of topo I. The catalytic
activity of topo II was analyzed in the same manner, except the
reaction mixture contained 50 mM Tris-HCl (pH 8.0), 120 mM KCl, 10
mM MgCl2, 0.5 mM ATP, 0.5 mM dithiothreitol, supercoiled
pBR322 DNA (250 ng) and 2 units of topo II (30). The reaction mixtures were incubated
at 37°C for 30 min, followed by digestion with 1% sodium dodecyl
sulfate (SDS) and 1 mg/ml proteinase K. Following digestion, 2 μl
loading buffer consisting of 5% sarkosyl, 0.0025% bromophenol blue
and 25% glycerol was added. To study the binding of enzymes to DNA
based on mobility shifts, the same procedure was followed, but the
SDS denaturation and proteinase K digestion steps were omitted. The
mixtures were subjected to 1% agarose gel electrophoresis in
Tris/borate/EDTA buffer. Agarose gel was stained with ethidium
bromide (EtBr), and the DNA band shifts from Form I to Form II by
topos I and II were detected using an enhanced chemiluminescence
detection system (Perkin Elmer Life Sciences Inc., Waltham, MA,
USA). Zero-D scan (Version 1.0, M&S Instruments Trading Inc.,
Osaka, Japan) was used for densitometric quantitation.
Other enzyme assays
Standard assays were used according to the
manufacturer's instructions to measure the activities of T7 RNA
polymerase, mouse inosine-5′-monophosphate (IMP) dehydrogenase
(type II), T4 polynucleotide kinase, and bovine deoxyribonuclease
I, as described by Nakayama and Saneyoshi (31), Mizushina et al(32), Soltis and Uhlenbeck (33), and Lu and Sakaguchi (34), respectively.
Cell culture and measurement of cancer
cell viability
Human colon carcinoma cell line HCT116 was obtained
from the American Type Culture Collection (Manassas, VA, USA).
HCT116 cells were cultured in McCoy's 5A medium supplemented with
10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100
μg/ml) at 37°C in a humid atmosphere of 5% CO2/95% air.
For the cell viability assay, cells were plated at 1×104
into each well of a 96-well microplate with 10 and 100 μM of one of
the test compounds (1–7). Cell viability was determined by WST-1
assay (35).
Cell cycle analysis
Cellular DNA content for cell cycle analysis was
determined as follows: aliquots of 3×105 HCT116 cells
were added to a 35-mm dish and incubated with a medium containing
70.0 μM CA and 49.4 μM DBL, based on the LD50 values,
for 24 h. Cells were then washed with ice-cold PBS three times by
centrifugation, fixed with 70% (v/v) ethanol and stored at −20°C.
DNA was stained with PI
{3,8-diamino-5-[3-(diethylmethylammonio)propyl]-6-phenylphenanthridinium
di iodide} staining solution for a minimum of 10 min at room
temperature in the dark. The intensity of the fluorescence was
measured using a FACSCanto flow cytometer in combination with
FACSDiVa software (Becton-Dickinson Co., Franklin Lakes, NJ,
USA).
Computational analysis
The molecular structures of compounds were
constructed using Discovery Studio (DS) 3.5 modeling software
(Accelrys Inc., San Diego, CA, USA). Energy minimization was
achieved using minimization and dynamics protocols within the DS.
Calculations were performed using the Chemistry at HARvard
Macromolecular Mechanics (CHARMm) force-field. The calculated logP
(ClogP) values and pKa values of the test compounds 1–7 were
obtained from the calculated properties in SciFinder Scholar, which
were originally calculated using Advanced Chemistry Development
(ACD/Lab) Software V8.14 for Solaris (ACD/Labs).
Results
Effect of Inonotus obliquus LMW
polyphenolic compounds 1–7 on the activity of mammalian pols
The inhibitory activity of each of the seven LMW
polyphenolics from Inonotus obliquus toward mammalian pols
was investigated using 11 mammalian pol species. These pols belong
to the A, B, X and Y families of pols (6,7).
Assessment of the relative activity of each pol at 200 μM after the
addition of the seven test compounds revealed that none of the
compounds had any effect on pol inhibition, as no compound resulted
in <90% relative pol activity (Table I). These results suggest that these
tested compounds did not affect the activity of any of the 11
mammalian pol species tested in vitro. When activated DNA
(bovine deoxyribonuclease I-treated DNA) was used as the DNA
template-primer substrate instead of synthesized DNA
[poly(dA)/oligo(dT)18 (A/T = 2/1)] and dNTP was used as
the nucleotide substrate instead of dTTP, the inhibitory effects of
these compounds did not change (data not shown).
 | Table IIC50 values of Inonotus
obliquus LMW polyphenolic compounds 1–7 for the activities of
mammalian pols, topos and other DNA metabolic enzymes. |
Table I
IC50 values of Inonotus
obliquus LMW polyphenolic compounds 1–7 for the activities of
mammalian pols, topos and other DNA metabolic enzymes.
| IC50
values (μM) |
|---|
|
|
|---|
| Enzyme | CA (1) | DBL (2) | GA (3) | SA (4) | PCA (5) | DB (6) | DTA (7) |
|---|
| Mammalian pols |
| Family A |
| Human pol γ | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| Family B |
| Calf pol α | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| Human pol δ | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| Human pol ɛ | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| Family X |
| Rat pol β | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| Human pol λ | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| Human pol μ | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| Calf TdT | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| Family Y |
| Human pol η | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| Mouse pol ι | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| Human pol κ | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| Mammalian
topos |
| Human topo I | 150±15 | 130±15 | >200 | >200 | >200 | >200 | >200 |
| Human topo II | 15±2.0 | 10±1.5 | 50±4.0 | 175±17 | 80±7.0 | 150±15 | 170±16 |
| Other DNA metabolic
enzymes |
| T7 RNA
polymerase | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| Mouse IMP
dehydrogenase (type II) | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| T4 polynucleotide
kinase | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
| Bovine
deoxyribonuclease I | >200 | >200 | >200 | >200 | >200 | >200 | >200 |
Effect of Inonotus obliquus LMW
polyphenolic compounds 1–7 on the activity of human topos I and
II
The inhibitory effect of each LMW polyphenolic was
examined against human topos I and II, which have ssDNA and dsDNA
nicking activity, respectively (8). CA and DBL inhibited topo I nicking
activity, and 50% inhibition was observed at a concentration of 150
and 130 μM, respectively (Table
I). Therefore, DBL is a more potent topo I inhibitor compared
with CA under these conditions. By contrast, the other compounds
had no affect on topo I activity, even at a concentration of 200 μM
(Table I).
These LMW polyphenolics from Inonotus
obliquus potently inhibited the nicking activity of topo II,
and the inhibition was ranked as DBL > CA >> GA > PCA
>> DB > DTA > SA (Table
I). Specifically, the compounds may be categorized into three
groups as follows: significant topo II inhibitors [CA and DBL with
50% inhibitory concentration (IC50) values of 15 and 10
μM, respectively], moderate topo II inhibitors (GA and PCA with
IC50 values of 50 and 80 μM, respectively), and weak
topo II inhibitors (SA, DB, and DTA with IC50 values of
150–175 μM).
Effect of Inonotus obliquus LMW
polyphenolic compounds 1–7 on the activity of mammalian pols, topos
and other DNA metabolic enzymes
None of the LMW polyphenolics examined affected the
activity of other DNA metabolic enzymes such as T7 RNA polymerase,
mouse IMP dehydrogenase (type II), T4 polynucleotide kinase and
bovine deoxyribonuclease I (Table
I). These results indicate that CA and DBL are potent and
selective inhibitors of human topos I and II, whereas other
compounds should be specifically classified as inhibitors of human
topo II.
Specific assays were performed in order to determine
whether the inhibitory activity of these LMW polyphenolics was due
to their ability to bind to DNA or to the enzyme. The interaction
of these compounds with dsDNA was investigated by studying changes
in the thermal transition of the DNA. To accomplish this, the
melting temperature (Tm) of dsDNA in the presence of an excess of
compound (200 μM) was measured using a spectrophotometer equipped
with a thermoelectric cell holder. When a typical intercalating
compound, EtBr (15 μM), was used as a positive control, a clear
thermal transition (Tm) was observed. However, no such thermal
transition was observed when any of the seven LMW polyphenolics
were heated with dsDNA (data not shown). We considered whether the
inhibitory effects of the seven Inonotus obliquus LMW
polyphenolics resulted from nonspecific adhesion to human topos or
from their selective binding to specific sites. This was
investigated by determining whether an excessive amount of nucleic
acid [poly(rC)] or protein (BSA) prevented the inhibitory effect of
the compounds. Poly(rC) and BSA had little or no effect on the
inhibition of topos by the isolated compounds (data not shown),
suggesting that all seven LMW polyphenolics selectively bound to
the topo enzyme molecule. These findings indicate that the
compounds do not act as DNA intercalating agents or as
template-primer substrates. Instead, the compounds directly bind to
topos and inhibit their activities.
These results suggest that compounds 1–7 are potent
and specific inhibitors of human topos. We therefore investigated
in more detail whether topo inhibition by these compounds results
in decreased human cancer cell proliferation.
Effect of Inonotus obliquus LMW
polyphenolic compounds 1–7 on cultured human cancer cells
Topos have recently emerged as important cellular
targets for chemical intervention in the development of anticancer
agents. The seven LMW polyphenolics examined in this study may be
useful in chemotherapy; therefore, we investigated the cytotoxic
effect of these compounds against the HCT116 cell line. As shown in
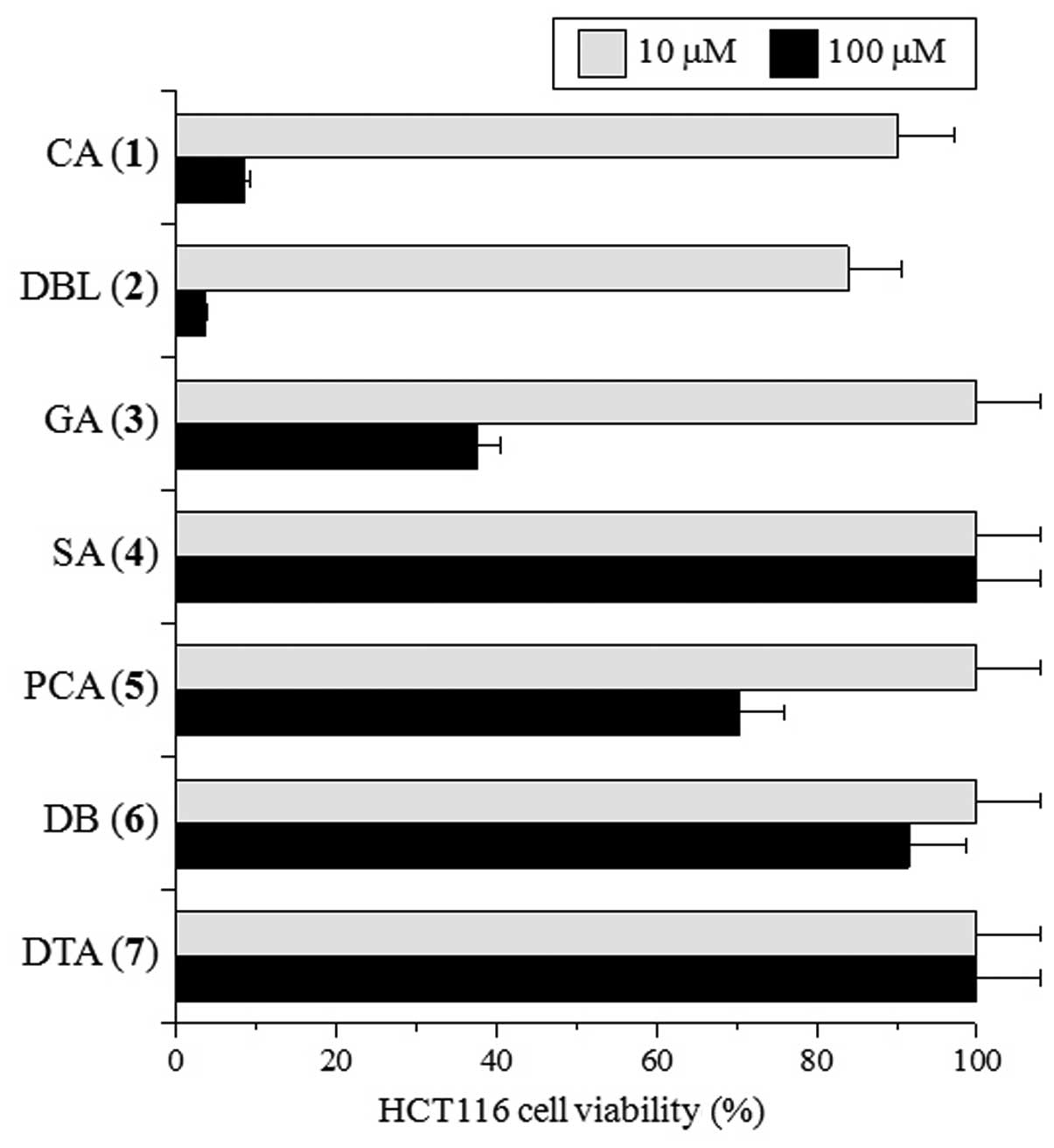
Fig. 2, 24 h of treatment with 10
μM of CA and DBL marginally suppressed HCT116 cell growth, whereas
treatment with the other compounds at 10 μM did not. On the other
hand, 100 μM of CA and DBL markedly suppressed, and GA and PCA
moderately suppressed, cell proliferation, whereas SA, DB and DTA
suppressed growth weakly or not at all (Fig. 2). The dose of these compounds
required for the suppression of cell growth was approximately the
same as that for topo II inhibition. The suppression of HCT116 cell
growth by CA and DBL was dose-dependent, with LD50 of
70.0 and 49.4 μM, respectively. These compounds suppressed the
growth of other human cancer cell lines with approximately the same
LD50 values (data not shown). These LD50
values are ~5-fold higher than the IC50 values for topo
II. This indicates that these catechol propanoid compounds may
interact with other cellular components prior to reaching the
nucleus. Subsequently, they bind and interact with topo II in order
to inhibit its activity and suppress cell growth, although they
exhibited rather specific inhibitory action to topo II.
These results suggest that, among the seven
compounds tested, CA and DBL are potent inhibitors of human topo II
rather than topo I, and were therefore selected for further
study.
Effect of CA and DBL on cell cycle
progression
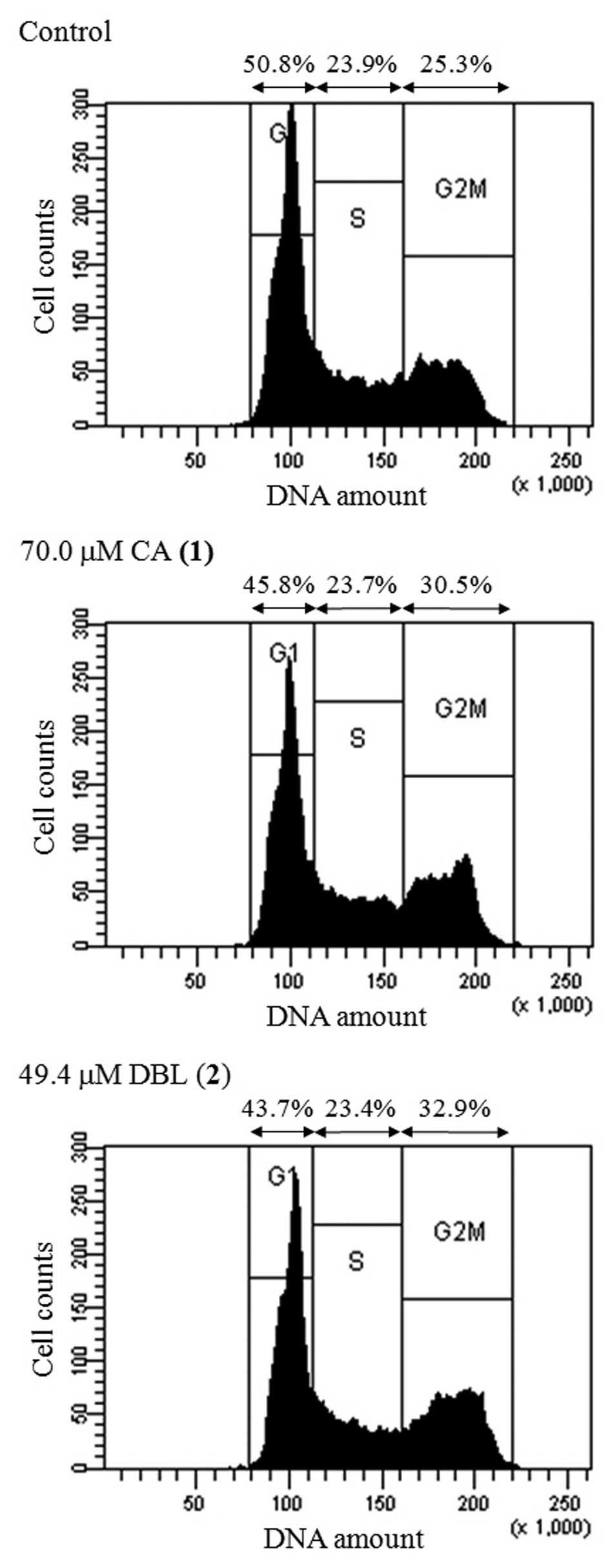
We analyzed whether CA and DBL affected the cell
cycle distribution of HCT116 cells. The cell-cycle fraction was
recorded following 24 h of treatment with the concentration of each
compound equal to its LD50. The ratio of cells in all
three phases (G1, S and G2/M) of the cell cycle is shown in
Fig. 3. Treatment with CA and DBL
increased the population of cells in the G2/M phase (1.21- and
1.30-fold, respectively), did not change the proportion of cells in
the S phase and decreased the percentage of cells in the G1 phase.
Etoposide, a classic topo II inhibitor, arrested cells in the G2/M
phase (1.40-fold increase, data not shown). These results suggest
that CA and DBL are effective inhibitors of topo II and lead to a
blockade of the cell cycle at the G2/M phase.
Three-dimensional structural simulation
of Inonotus obliquus LMW polyphenolic compounds 1–7
To obtain information regarding the molecular basis
of the inhibitory properties of compounds 1–7, we performed
computational analyses using molecular simulation. The molecular
length of the three-dimensional structure of CA and DBL were 8.0
and 8.7 Å, respectively, and other LMW polyphenolics were 5.7–6.9 Å
long (Table II); hence, CA and
DBL are longer than the others. The molecular width of the
three-dimensional structure of the compounds had the same values
(4.0–5.6 Å) (Table II). We
focused on the chemical properties of these compounds, including
the ClogP value and pKa. ClogP values, which are an indication of
hydrophobicity, revealed that CA and DBL were the first and third
highest ranking of the compounds ClogP = 1.55 and 1.01,
respectively; Table II). This
suggests that the structural difference of the propenyl side chain
in the 1-position of the polyhydroxybenzene backbone contributes to
the hydrophobicity. However, the pKa values of DBL and DB were
larger than those for other compounds (pKa = 9.41 and 7.61,
respectively; Table II). Thus,
the presence of a free carboxylic acid moiety is the major
determinant of the pKa value. These data suggest that the molecular
length and ClogP value of CA and DBL are important factors that
affect their topo II inhibitory activities, as they exhibited
marked inhibition of human topo II activity and cancer cell
proliferation.
 | Table IIMolecular length and width, ClogP and
pKa values of the three-dimensional structure of Inonotus
obliquus LMW polyphenolic compounds 1–7. |
Table II
Molecular length and width, ClogP and
pKa values of the three-dimensional structure of Inonotus
obliquus LMW polyphenolic compounds 1–7.
| Factor | CA (1) | DBL (2) | GA (3) | SA (4) | PCA (5) | DB (6) | DTA (7) |
|---|
| Length (Å) | 8.0 | 8.7 | 6.6 | 6.6 | 6.6 | 5.7 | 6.9 |
| Width (Å) | 4.0 | 4.0 | 5.2 | 5.8 | 4.0 | 4.0 | 5.2 |
| ClogP | 0.66±0.28 | 1.55±0.28 | 0.53±0.32 | 1.28±0.33 | 1.01±0.23 | 0.93±0.26 | 3.23±0.37 |
| pKa | 4.58±0.10 | 9.41±0.10 | 4.33±0.10 | 4.33±0.10 | 4.45±0.10 | 7.61±0.18 | 2.17±0.10 |
Discussion
We observed that the LMW polyphenolics 1–7 isolated
from Inonotus obliquus (Fig.
1) inhibited the activity of human topo II (Table I) and suppressed HCT116 cell growth
(Fig. 2). In particular, CA and
DBL were marked inhibitors of human topo II. The suppression of
cell growth correlated with the inhibitory effects of these
compounds on topo II, suggesting that topo II is critical for cell
survival. To analyze the role of topo II in cancer cell growth and
proliferation, we studied the effects of small interfering RNAs
(siRNA) on targeting topo II in tumor cells.
CA is found in fruit (36), wine (37,38)
and coffee (39). It exerts
diverse biological effects and has antibacterial (39), antioxidative (40) and anti-inflammatory (37) properties. The molecular length,
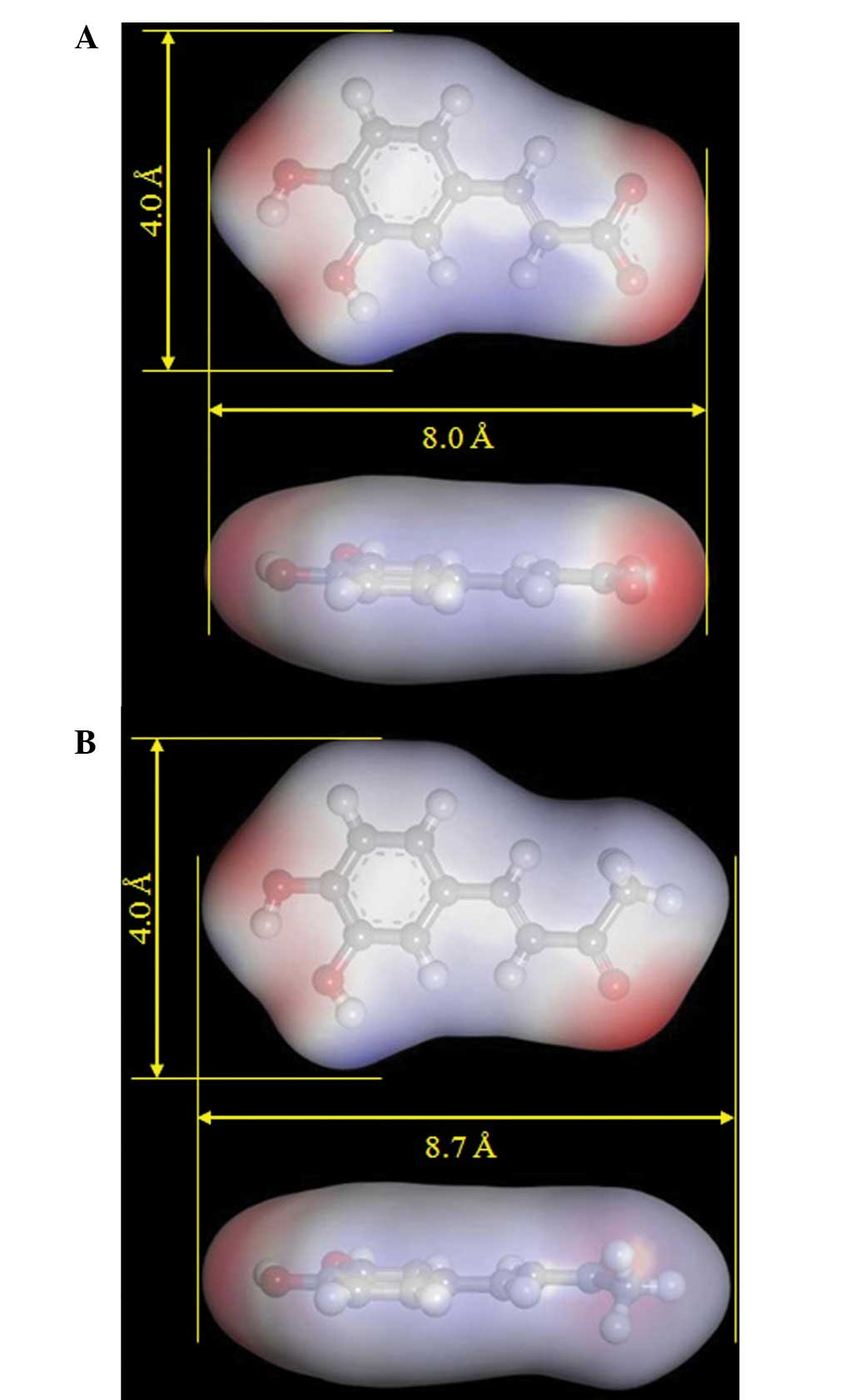
width and three-dimensional structure of CA and DBL, from which the
energy-minimized compounds were calculated, were compared in
Fig. 4. The length and width of CA
is virtually identical to that of DBL. There is a potential
inhibitor-binding pocket located on the topo II protein surface,
with a width and length of ~4.0 Å and 8.0–8.7 Å, respectively, to
accommodate the compounds. The pKa values of CA and DBL vary (pKa =
4.58 for CA and pKa = 9.41 for DBL), but these compounds have
nearly the same ClogP values (ClogP: CA = 1.01; DBL = 1.55)
(Table II). The molecular length,
width and hydrophobicity (ClogP and surface area of the functional
group negative/positive charges as shown in Fig. 4) of these compounds are important
for their bioactivity, rather than their pKa.
Topo II inhibitors such as doxorubicin, amsacrine,
ellipticine, saintopin, streptonigrin and terpentecin are DNA
intercalating agents that bind the DNA molecule directly and
subsequently indirectly inhibit topo II activity. These chemicals
inhibit the DNA chain-rejoining reactions catalyzed by topo II by
stabilizing a tight topo II protein-DNA complex called the
‘cleavable complex’. The possibility that these LMW polyphenolics,
in particular CA and DBL, also bind to DNA was examined by
measuring the Tm of dsDNA, but none of these compounds were found
to bind to dsDNA (data not shown). Therefore, we conclude that
these compounds inhibit enzyme activity via direct interaction.
Topo inhibitors are categorized into two classes, ‘suppressors’,
which are considered to interact directly with the enzyme, and
‘poisons’, which stimulate DNA cleavage and intercalation (41). CA and DBL are considered to
‘suppress’ topo function rather than act as conventional poisons,
as the compounds do not appear to stabilize topo II protein-DNA
covalent complexes. These compounds may be a new class of topo II
inhibitor.
In conclusion, several LMW polyphenolic compounds
isolated from the medicinal mushroom Inonotus obliquus
markedly suppress human cancer cell proliferation with cell cycle
arrest associated with the inhibition of cellular topo II activity.
CA and DBL should therefore be considered lead compounds in the
search for novel cancer chemotherapy agents.
Acknowledgements
This study was supported in part by the Ministry of
Education, Culture, Sports, Science and Technology, Japan
(MEXT)-Supported Program for the Strategic Research Foundation at
Private Universities, 2012–2016. I. K. acknowledges a Grant-in-Aid
for Young Scientists (B) (No. 23710262) from MEXT. Y.M.
acknowledges Grants-in-Aid for Scientific Research (C) (no.
24580205) from MEXT, Takeda Science Foundation (Japan) and the
Hyogo Science and Technology Association (Japan).
Abbreviations:
|
LMW
|
low molecular weight
|
|
CA
|
caffeic acid
|
|
DBL
|
3,4-dihydroxy-benzalacetone
|
|
GA
|
gallic acid
|
|
SA
|
syringic acid
|
|
PCA
|
protocatechuic acid
|
|
DB
|
3,4-dihydroxy-benzaldehyde
|
|
DTA
|
2,5-dihydroxy-terephthalic acid
|
|
pol
|
DNA polymerase (EC 2.7.7.7)
|
|
topo
|
DNA topoisomerase
|
|
dsDNA
|
double-stranded DNA
|
|
dTTP
|
2′-deoxythymidine 5′-triphosphate
|
|
dNTP
|
2′-deoxyribonucleotide
5′-triphosphate
|
|
DMSO
|
dimethyl sulfoxide
|
|
BSA
|
bovine serum albumin
|
|
SDS
|
sodium dodecyl sulfate
|
|
EtBr
|
ethidium bromide
|
|
IMP
|
inosine-5′-monophosphate
|
|
ssDNA
|
single-stranded DNA
|
|
DS
|
Discovery Studio
|
|
IC50
|
50% inhibitory concentration
|
|
LD50
|
50% lethal dose
|
|
Tm
|
melting temperature
|
|
ClogP
|
calculated log P
|
References
|
1
|
Liu RH: Potential synergy of
phytochemicals in cancer prevention: mechanism of action. J Nutr.
134:3479S–3485S. 2004.PubMed/NCBI
|
|
2
|
Surh YJ: Cancer chemoprevention with
dietary phytochemicals. Nat Rev Cancer. 3:768–780. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kornberg A and Baker TA: DNA Replication.
2nd edition. WD Freeman and Co; New York, NY: pp. 197–225. 1992
|
|
4
|
Bebenek K and Kunkel TA: DNA repair and
replication. Advances in Protein Chemistry and Structural Biology.
Yang W: 69. 1st Edition. Elsevier; San Diego, CA: pp. 137–165.
2004
|
|
5
|
Hubscher U, Maga G and Spadari S:
Eukaryotic DNA polymerases. Annu Rev Biochem. 71:133–163. 2002.
View Article : Google Scholar
|
|
6
|
Lange SS, Takata K and Wood RD: DNA
polymerases and cancer. Nat Rev Cancer. 11:96–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Loeb LA and Monnat RJ Jr: DNA polymerases
and human disease. Nat Rev Genet. 9:594–604. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang JC: DNA topoisomerases. Annu Rev
Biochem. 65:635–692. 1996. View Article : Google Scholar
|
|
9
|
Berdis AJ: DNA polymerases as therapeutic
targets. Biochemistry. 47:8253–8260. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu LF: DNA topoisomerase poisons as
antitumor drugs. Annu Rev Biochem. 58:351–375. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sakaguchi K, Sugawara F and Mizushina Y:
Inhibitors of eukaryotic DNA polymerases. Seikagaku. 74:244–251.
2002.(In Japanese).
|
|
12
|
Nakajima Y, Saito Y and Konishi T:
Antioxidant small phenolic ingredients in Inonotus obliquus
(persoon) Pilat (Chaga). Chem Pharm Bull (Tokyo). 55:1222–1226.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shashkina M, Shashkin P and Sergeev A:
Chemical and medicobiological properties of chaga (review).
Pharmaceut Chem J. 40:560–568. 2006. View Article : Google Scholar
|
|
14
|
Nakajima Y, Nishida H, Matsugo S and
Konishi T: Cancer cell toxicity of small phenolic compounds from
Chaga [Inonotus obliquus (persoon) Pilat]. J Med Food.
12:501–507. 2009.PubMed/NCBI
|
|
15
|
Nakajima Y, Nishida H, Nakamura Y and
Konishi T: Prevention of hydrogen peroxide-induced oxidative stress
in PC12 cells by 3,4-dihydroxybenzalacetone isolated from Chaga
[Inonotus obliquus (persoon) Pilat]. Free Radic Biol Med.
47:1154–1161. 2009.PubMed/NCBI
|
|
16
|
Fresco P, Borges F, Marques MP and Diniz
C: The anticancer properties of dietary polyphenols and its
relation with apoptosis. Curr Pharm Des. 16:114–134. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang WY, Cai YZ and Zhang Y: Natural
phenolic compounds from medicinal herbs and dietary plants:
potential use for cancer prevention. Nutr Cancer. 62:1–20. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kelloff GJ: Perspectives on cancer
chemoprevention research and drug development. Adv Cancer Res.
78:199–334. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Esteves M, Siquet C, Gaspar A, et al:
Antioxidant versus cytotoxic properties of hydroxycinnamic acid
derivatives - a new paradigm in phenolic research. Arch Pharm
(Weinheim). 341:164–173. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tamai K, Kojima K, Hanaichi T, Masaki S,
Suzuki M, Umekawa H and Yoshida S: Structural study of
immuno-affinitypurified DNA polymerase α-DNA primase complex from
calf thymus. Biochim Biophys Acta. 950:263–273. 1988.
|
|
21
|
Date T, Yamaguchi M, Hirose F, et al:
Expression of active rat DNA polymerase β in Escherichia
coli. Biochemistry. 27:2983–2990. 1988.
|
|
22
|
Umeda S, Muta T, Ohsato T, Takamatsu C,
Hamasaki N and Kang D: The D-loop structure of human mtDNA is
destabilized directly by 1-methyl-4-phenylpyridinium ion
(MPP+), a parkinsonism-causing toxin. Eur J Biochem.
267:200–206. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Oshige M, Takeuchi R, Ruike R, Kuroda K
and Sakaguchi K: Subunit protein-affinity isolation of Drosophila
DNA polymerase catalytic subunit. Protein Expr Purif. 35:248–256.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kusumoto R, Masutani C, Shimmyo S, Iwai S
and Hanaoka F: DNA binding properties of human DNA polymerase η:
implications for fidelity and polymerase switching of translesion
synthesis. Genes Cells. 9:1139–1150. 2004.
|
|
25
|
Ohashi E, Murakumo Y, Kanjo N, et al:
Interaction of hREV1 with three human Y-family DNA polymerases.
Genes Cells. 9:523–531. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shimazaki N, Yoshida K, Kobayashi T, Toji
S, Tamai T and Koiwai O: Over-expression of human DNA polymerase λ
in E. coli and characterization of the recombinant enzyme.
Genes Cells. 7:639–651. 2002.
|
|
27
|
Mizushina Y, Tanaka N, Yagi H, et al:
Fatty acids selectively inhibit eukaryotic DNA polymerase
activities in vitro. Biochim Biophys Acta. 1308:256–262. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mizushina Y, Yoshida S, Matsukage A and
Sakaguchi K: The inhibitory action of fatty acids on DNA polymerase
β. Biochim Biophys Acta. 1336:509–521. 1997.
|
|
29
|
Ogawa A, Murate T, Suzuki M, Nimura Y and
Yoshida S: Lithocholic acid, a putative tumor promoter, inhibits
mammalian DNA polymerase β. Jpn J Cancer Res. 89:1154–1159.
1998.PubMed/NCBI
|
|
30
|
Yonezawa Y, Tsuzuki T, Eitsuka T, et al:
Inhibitory effect of conjugated eicosapentaenoic acid on human DNA
topoisomerases I and II. Arch Biochem Biophys. 435:197–206. 2005.
View Article : Google Scholar
|
|
31
|
Nakayama C and Saneyoshi M: Inhibitory
effects of 9-β-D-xylofuranosyladenine 5′-triphosphate on
DNA-dependent RNA polymerase I and II from cherry salmon
(Oncorhynchus masou). J Biochem. 97:1385–1389. 1985.
|
|
32
|
Mizushina Y, Dairaku I, Yanaka N, et al:
Inhibitory action of polyunsaturated fatty acids on IMP
dehydrogenase. Biochimie. 89:581–590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Soltis DA and Uhlenbeck OC: Isolation and
characterization of two mutant forms of T4 polynucleotide kinase. J
Biol Chem. 257:11332–11339. 1982.PubMed/NCBI
|
|
34
|
Lu BC and Sakaguchi K: An endo-exonuclease
from meiotic tissues of the basidiomycete Coprinus cinereus:
its purification and characterization. J Biol Chem.
266:21060–21066. 1991.PubMed/NCBI
|
|
35
|
Ishiyama M, Tominaga H, Shiga M, Sasamoto
K, Ohkura Y and Ueno K: A combined assay of cell viability and in
vitro cytotoxicity with a highly water-soluble tetrazolium salt,
neutral red and crystal violet. Biol Pharm Bull. 19:1518–1520.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hsu FL, Chen YC and Cheng JT: Caffeic acid
as active principle from the fruit of Xanthium strumarium to
lower plasma glucose in diabetic rats. Planta Med. 66:228–230.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Giovannini L, Migliori M, Filippi C, et
al: Inhibitory activity of the white wine compounds, tyrosol and
caffeic acid, on lipopolysaccharide-induced tumor necrosis factor-α
release in human peripheral blood mononuclear cells. Int J Tissue
React. 24:53–56. 2002.PubMed/NCBI
|
|
38
|
Simonetti P, Gardana C and Pietta P:
Caffeic acid as biomarker of red wine intake. Methods Enzymol.
335:122–130. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Almeida AA, Farah A, Silva DA, Nunan EA
and Glória MB: Antibacterial activity of coffee extracts and
selected coffee chemical compounds against enterobacteria. J Agric
Food Chem. 54:8738–8743. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chung MJ, Walker PA and Hogstrand C:
Dietary phenolic antioxidants, caffeic acid and Trolox, protect
rainbow trout gill cells from nitric oxide-induced apoptosis. Aquat
Toxicol. 80:321–328. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bailly C: Topoisomerase I poisons and
suppressors as anticancer drugs. Curr Med Chem. 7:39–58. 2000.
View Article : Google Scholar : PubMed/NCBI
|