Introduction
Non-syndromic cleft lip with or without cleft palate
(NSCL/P) is a complex disorder with a multifactorial etiology
involving genetic and environmental factors. Through the use of a
wide range of genetic approaches, various candidate genes and
chromosomal regions associated with NSCL/P have been identified
(1). However, these findings
remain controversial, due in part to phenomena such as genetic
heterogeneity and incomplete dominance.
Interferon regulatory factor 6 (IRF6) is a key
factor involved in the development of the maxillofacial region and
teeth. Several studies have suggested that variations in IRF6 may
be important in the etiology of NSCL/P (2). The SNP rs2235371 (V274I) was the
first marker in IRF6 shown to be associated with NSCL/P, notably in
Asian and South American populations (3). This association was subsequently
identified in additional populations (4–7).
Several genetic markers in IRF6 have also demonstrated linkage and
linkage disequilibrium (LD) in studies of non-syndromic oral
clefts. However, there are inconsistencies across these studies
(8,9).
The MSX1 gene, located on human chromosome 4p16, is
a member of the MSX gene family, and is crucial in the development
of teeth and the craniofacial skeleton. Knock-out mouse models have
shown that cleft palate results from the complete loss of MSX1
(10), with 39% of these mutants
exhibiting unilateral or bilateral cleft lip. Linkage studies have
also demonstrated that the region of chromosome 4 containing MSX1,
may have a causal mutation (11),
and several studies have indicated an association between the MSX1
gene and oral clefts in a variety of populations (12–14).
The transcription factor PAX9 has also been shown to
have a direct relationship with craniofacial development,
particularly the formation of the palate and teeth. Notably, PAX9
and MSX1 are coexpressed during craniofacial development (15), and mice that are homozygous mutant
for either one of these genes exhibit cleft palate and an early
arrest of tooth formation. The combination of PAX9 and MSX1
nullizygosity generates a cleft lip phenotype that results from
interactions between these loci (11). In addition, in a Japanese family a
heterozygous missense mutation was identified in exon 3 of PAX9 in
two siblings with NSCL/P and their phenotypically normal mother
(16). Moreover, Lee et al
recently demonstrated that PAX9 contributes to the risk of NSCL/P
in a Korean population (17).
However, there are some inconsistencies across studies in
Singaporean, Taiwanese and Korean populations (18).
The abovementioned studies suggest that IRF6, MSX1
and PAX9 are crucial for the development of dentition and the
maxillofacial region (19).
Notably, NSCL/P is often associated with missing teeth, and recent
analyses have indicated that the incidence of missing teeth is
significantly higher outside the cleft region and in the mandible
(20). However, whether any
interactions among the three genes are associated with NSCL/P
remains to be clarified. The aim of the present study was to
determine the genetic variations in IRF6, MSX1 and PAX9, and to use
the data from case-control studies to clarify the association of
single nucleotide polymorphisms (SNPs), haplotypes and gene-gene
interactions with the risk of NSCL/P.
Materials and methods
Study population
This case-control study included 204 patients with
NSCL/P and 226 normal controls; the control group had no congenital
malformations of the body, no family history of genetic disease,
were born in the same region as the patients with NSCL/P and had a
male:female ratio as close to the NSCL/P group as possible. The
study participants were recruited between January, 2010 and
January, 2012 from the Department of Cleft Lip and Palate, Plastic
Surgery Hospital, Chinese Academy of Medical Sciences (Beijing,
China). Informed consent was obtained from each participant prior
to enrollment in the study, which was approved by the local ethics
committee of Department of Cleft Lip and Palate, Plastic Surgery
Hospital, Chinese Academy of Medical Sciences (Beijing, China).
General characteristics including age, gender, ethnicity, health
status and birth defects, were recorded.
SNP identification and selection
Using the HapMap genome browser (http://www.hapmap.org/cgi-perl/gbrowse/hapmap3r2_B36)
based on Han Chinese individuals from Beijing, China (CHB), 9 tag
SNPs (r2 coefficient cut-off of 0.80 with a minor allele
frequency of 0.05), were selected to capture the IRF6 region of
chromosome 1. Two tag SNPs were selected for the MSX1 region of
chromosome 4 and 8 tag SNPs for the PAX9 region of chromosome
4.
Genotype
Peripheral blood samples (2 ml) were collected from
each participant and frozen. Genomic DNA was extracted from a 200
μl aliquot of each sample using a Tiangen™ Genomic DNA Kit (Tiangen
Biotech Co., Ltd., Haidian, China) according to the manufacturer’s
instructions, and stored at −70ºC. The SNPs were genotyped using
the Sequenom MassARRAY matrix-assisted laser desorption/ionization
time-of-flight mass spectrometry analyzer (Sequenom Inc., San
Diego, CA, USA). Primers were designed using a semi-automated
method (Assay Design 3.1, Sequenom Inc.); primer sequences are
available on request. The call rate for each assay was set at
>90%.
Statistical analysis
Data from the control and NSCL/P groups were
compared and analyzed with SPSS software, version 17.0 (SPSS Inc.,
Chicago, IL, USA) and Hardy-Weinberg equilibrium (HWE) software.
Based on the multivariate logistic regression method, the
case-control association of genotypes in three inheritance models
(dominant, recessive and additive) were tested, and these models
were coded as follows for genotypes AA, AB and BB (assuming B is
the minor allele): Dominant 0, 1 and 1 (AA vs. AB+BB), recessive 0,
0 and 1 (AA+AB vs. BB) and additive 0, 1 and 2 (trend test on B
allele count). The odds ratio (OR) and 95% confidence intervals
(CI) were calculated. The association between each SNP and
haplotype with NSCL/P was estimated using Haploview software
(http://www.broad.mit.edu/mpg/haploview/).
For haplotype construction, genotypic data from the
case and control groups was used to estimate intermarker linkage
disequilibrium (LD) by measuring pairwise D′ and r2 and
defining LD blocks. The CI method was employed in the Haploview
software to define an LD block with an extended spine if
r2=0.8. P-values were corrected for multiple tests with
10,000 permutations. To identify gene-gene interactions in our
samples, logistic regression analysis was used to calculate ORs and
95% CIs to search for gene-gene interactions in the NSCL/P and
control groups. Interactions between SNPs were classified in four
groups using a dominant model. In each case, the group carrying the
least risk served as a reference, i.e., if the variant (B) was
considered a risk factor, wild type (AA) served as a reference,
while if the variant (B) was considered a protective factor, AB+BB
served as a reference. Bonferroni corrections for multiple SNPs
were performed, and P<0.05 and 0.019 were regarded as
statistically significant.
Results
Characteristics of study subjects
A total of 19 SNPs were genotyped from the 204
patients with NSCL/P and 226 normal controls of Han Chinese origin.
The mean age was 3.23±0.91 years and the male:female ratio was
1:0.76 in the NSCL/P group. The mean age was 3.96±1.03 years and
the male:female ratio was 1:0.70 in the control group. The genomic
position, nucleic acid composition and minor allele frequencies of
the SNPs are summarized in Table
I. We calculated the HWE for all the SNPs; none of the SNPs
deviated from the HWE among these groups.
 | Table ICharacteristics of the polymorphisms
of IRF6, MSX1 and PAX9 genes. |
Table I
Characteristics of the polymorphisms
of IRF6, MSX1 and PAX9 genes.
| Gene symbol | rs no. | SNP function | Location | Allelesa | MAF | P-HWE |
|---|
| IRF6 | rs17317411 | UTR-3 | Chr1 | 209961314 | c/T | 0.042 | 0.629242 |
| rs2073485 | Intron | Chr1 | 209962794 | a/G | 0.376 | 0.597578 |
| rs2235371 | Missense | Chr1 | 209964080 | C/t | 0.310 | 0.701308 |
| rs2013196 | Intron | Chr1 | 209968411 | C/t | 0.123 | 0.744881 |
| rs7552506 | Intron | Chr1 | 209969902 | c/G | 0.188 | 0.609989 |
| rs2236909 | Intron | Chr1 | 209971655 | a/G | 0.497 | 0.133192 |
| rs861019 | UTR-5 | Chr1 | 209975386 | A/g | 0.257 | 0.122725 |
| rs861020 | Intron | Chr1 | 209977111 | a/G | 0.240 | 0.052702 |
| rs3753518 | Intron | Chr1 | 209977964 | C/g | 0.066 | 0.329238 |
| MSX1 | rs1042484 | Intron | Chr4 | 4864381 | c/T | 0.057 | 0.879223 |
| rs12532 | UTR-3 | Chr4 | 4865146 | a/G | 0.376 | 0.441807 |
| PAX9 | rs2236007 | Intron | Chr14 | 37132769 | a/G | 0.270 | 0.479949 |
| rs2295218 | Intron | Chr14 | 37134491 | a/T | 0.468 | 0.856159 |
| rs8004560 | Intron | Chr14 | 37140781 | a/G | 0.255 | 0.167655 |
| rs17104928 | Intron | Chr14 | 37142554 | a/G | 0.415 | 0.220053 |
| rs17176643 | Intron | Chr14 | 37142969 | a/C | 0.498 | 0.311689 |
| rs11156926 | Intron | Chr14 | 37143237 | a/T | 0.271 | 0.957203 |
| rs10131337 | Intron | Chr14 | 37144516 | C/t | 0.297 | 0.646023 |
| rs7144276 | Intron | Chr14 | 37144607 | A/t | 0.198 | 0.543028 |
Single SNP analysis
To evaluate the association between genetic variants
and the risk of NSCL/P, we compared IRF6, MSX1 and PAX9 genotype
frequency distributions in the NSCL/P and control groups (Table II). For IRF6, there was a
significant difference in the allele frequency of rs2073485,
rs2235371, rs2236909 and rs861020 between the NSCL/P and control
groups after correction with 10,000 permutations (P=0.0263, 0.0232,
0.0239 and 0.0104, respectively). The association between the
dominant model of rs2073485, rs2235371 and rs861020 with NSCL/P
remained significant (P=0.002, 0.002 and 0.007, respectively). The
distribution of the recessive model of rs2236909 and rs861020
differed significantly between the NSCL/P and control groups
(P=0.000 and 0.009, respectively). Cochran-Armitage trend tests of
rs2073485, rs2235371, rs2236909 and rs861020 showed a significant
difference between the NSCL/P and control groups (P=0.007, 0.008,
0.001 and 0.003, respectively). For PAX9, there was a significant
difference in the allele frequency of rs17176643 between the NSCL/P
and control groups (P=0.0199), but not a significant difference
after correction with 10,000 permutations (P=0.1349). The dominant
model and Cochran-Armitage trend test of rs17176643 remained
significant between the NSCL/P and control groups (P=0.030 and
0.049, respectively). Our results indicate that the MSX1 gene does
not influence susceptibility to NSCL/P.
 | Table IIAssociation of the polymorphisms of
IRF6, MSX1 and PAX9 genes with NSCL/P. |
Table II
Association of the polymorphisms of
IRF6, MSX1 and PAX9 genes with NSCL/P.
| Gene symbol | rs no. |
Alleleb | Genotype | Dominant model | Recessive
model | Cochran-Armitage
trend test |
|---|
|
|
|
|
|---|
| Alleles | P-value | Permuted P | Patients | Control | OR | 95% CI | P-value | OR | 95% CI | P-value |
|---|
| IRF6 | rs17317411 | c/T | 0.7535 | 0.9996 | 186/18/0 | 210/14/0 | 0.787 | 0.390–1.588 | 0.504 | - | - | - | - |
| rs2073485 | a/G | 0.0043 | 0.0263 | 97/81/26 | 74/114/38 | 0.733 |
0.603–0.891 |
0.002a | 0.850 | 0.649–1.113 | 0.238 | 0.007 |
| rs2235371 | C/t | 0.0033 | 0.0232 | 114/72/17 | 93/106/27 | 0.739 |
0.610–0.895 |
0.002a | 0.821 | 0.596–1.130 | 0.226 | 0.008 |
| rs2013196 | C/t | 0.9522 | 1 | 156/46/2 | 174/48/4 | 1.015 | 0.811–1.296 | 0.898 | 0.741 | 0.316–1.741 | 0.492 | 0.753 |
| rs7552506 | c/G | 0.8812 | 1 | 134/64/6 | 147/72/7 | 0.986 | 0.808–1.203 | 0.889 | 0.974 | 0.560–1.694 | 0.925 | 0.988 |
| rs2236909 | a/G | 0.0034 | 0.0239 | 48/88/68 | 63/123/40 | 1.121 | 0.902–1.393 | 0.304 | 1.525 |
1.218–1.908 |
0.000a |
0.001a |
| rs861019 | A/g | 0.7365 | 1 | 109/83/12 | 122/94/10 | 1.101 | 0.836–1.222 | 0.909 | 1.612 | 0.755–1.787 | 0.495 | 0.790 |
| rs861020 | a/G |
0.0016a | 0.0104 | 103/86/15 | 142/80/4 | 1.300 |
1.072–1.569 | 0.007 | 2.099 |
1.199–3.674 | 0.009 | 0.003 |
| rs3753518 | C/g | 0.7749 | 0.9997 | 178/26/0 | 197/27/2 | 0.996 | 0.750–1.323 | 0.979 | 0.000 | 0.000- | 0.999 | 0.394 |
| MSX1 | rs1042484 | c/T | 0.5081 | 0.7833 | 183/21/0 | 199/26/1 | 0.920 | 0.680–1.244 | 0.587 | 0.000 | 0.000- | 1 | 0.583 |
| rs12532 | a/G | 0.7524 | 0.9479 | 80/97/30 | 84/112/30 | 0.985 | 0.788–1.163 | 0.663 | 0.998 | 0.574–1.754 | 0.990 | 0.902 |
| PAX9 | rs2236007 | a/G | 0.7659 | 1 | 106/84/14 | 124/84/18 | 1.060 | 0.877–1.282 | 0.546 | 0.923 | 0.642–1.326 | 0.664 | 0.675 |
| rs2295218 | a/T | 0.3955 | 0.9529 | 62/98/43 | 61/111/53 | 0.920 | 0.746–1.134 | 0.434 | 0.934 | 0.743–1.173 | 0.557 | 0.546 |
| rs8004560 | a/G | 0.3787 | 0.9492 | 113/71/19 | 133/75/17 | 1.073 | 0.886–1.300 | 0.472 | 1.124 | 0.799–1.582 | 0.503 | 0.698 |
| rs17104928 | a/G | 0.8497 | 1 | 73/94/37 | 81/101/44 | 1.001 | 0.822–1.220 | 0.990 | 0.957 | 0.751–1.220 | 0.724 | 0.929 |
| rs17176643 | a/C | 0.0199 | 0.1349 | 37/112/55 | 61/120/45 | 1.292 |
1.026–1.627 | 0.030 | 1.218 | 0.973–1.526 | 0.085 | 0.049 |
| rs11156926 | a/T | 0.2891 | 0.885 | 112/79/12 | 113/91/18 | 0.918 | 0.758–1.111 | 0.378 | 0.844 | 0.578–1.232 | 0.379 | 0.548 |
| rs10131337 | C/t | 0.4247 | 0.9648 | 99/82/22 | 114/95/17 | 1.034 | 0.855–1.250 | 0.729 | 1.222 | 0.877–1.703 | 0.235 | 0.491 |
| rs7144276 | A/t | 0.8171 | 1 | 133/60/11 | 148/68/10 | 1.006 | 0.825–1.228 | 0.950 | 1.110 | 0.715–1.721 | 0.643 | 0.894 |
Haplotype analysis
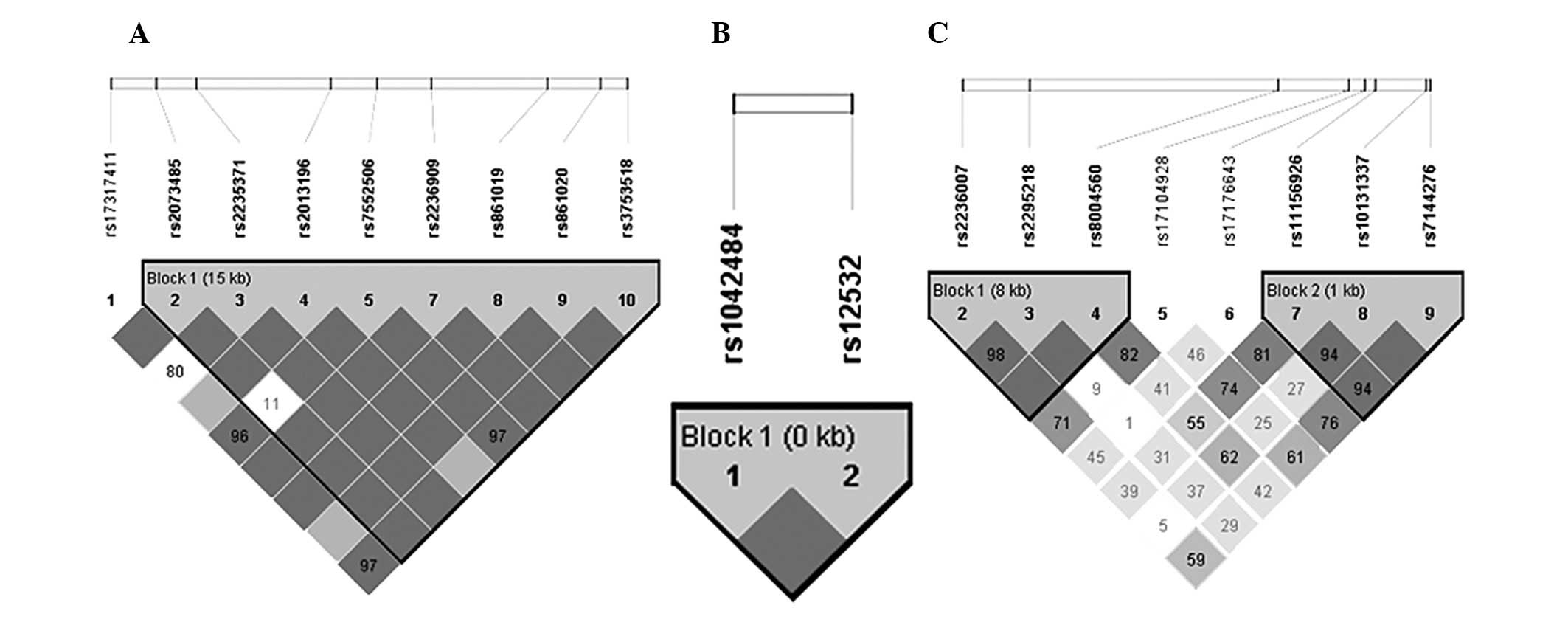
For IRF6, the Haploview program revealed rs2073485,
rs2235371, rs2013196, rs7552506, rs2236909, rs861019, rs861020 and
rs3753518, are positioned in the same LD block (Fig. 1A). Haplotype analysis of
polymorphisms in the IRF6 gene revealed SNP combinations associated
with the risk of NSCL/P (Table
III). The A-T-C-G-G-A-G-C and G-C-C-G-A-A-A-C haplotypes showed
an increased risk of NSCL/P following correction with 10,000
permutations (P=0.0109 and 0.0036, respectively). In MSX1,
rs1042484 and rs12532 were in the same LD block (Fig. 1B). The results of the haplotype
analysis showed no statistical differences between the NSCL/P and
control groups. For PAX9, rs2236007, rs2295218 and rs8004560 were
in the same LD block (block 1), while rs11156926, rs10131337 and
rs7144276 were in block 2 (Fig.
1C, Table III). No
difference at block 1 or 2 between the NSCL/P and control groups
was observed.
 | Table IIIHaplotype analysis of IRF6, MSX1 and
PAX9 gene polymorphisms. |
Table III
Haplotype analysis of IRF6, MSX1 and
PAX9 gene polymorphisms.
| | Frequencies | | | | |
|---|
| |
| | | | |
|---|
| Gene | Haplotype | Case | Control | OR | (95% CI) | Nominal P | Permuted P |
|---|
| IRF6 |
A-T-C-G-G-A-G-C | 0.26110 | 0.35400 | 1 | Reference |
0.002493a | 0.0109 |
|
G-C-C-G-A-A-A-C | 0.28820 | 0.19250 | 2.03 | 1.401–2.94 |
0.001291a | 0.0036 |
|
G-C-C-G-A-G-G-C | 0.26110 | 0.25220 | 1.404 | 0.9784–2.013 | 0.8237 | 1 |
|
A-C-C-C-G-A-G-G | 0.06404 | 0.06637 | 1.308 | 0.7327–2.336 | 0.8638 | 1 |
|
G-C-T-C-G-A-G-C | 0.12320 | 0.12390 | 1.348 | 0.8563–2.121 | 0.9358 | 1 |
| MSX1 | T-G | 0.62990 | 0.61950 | 1 | Reference | 0.7524 | 0.9442 |
| C-A | 0.05147 | 0.06195 | 0.8171 | 0.4527–1.475 | 0.5081 | 0.7935 |
| T-A | 0.31860 | 0.31860 | 0.9836 | 0.735–1.316 | 0.9989 | 1 |
| PAX9 |
| Block 1 | G-A-G | 0.4530 | 0.47990 | 1 | Reference | 0.4136 | 0.9708 |
| G-T-A | 0.2673 | 0.24110 | 1.175 | 0.8432–1.637 | 0.3894 | 0.9461 |
| A-T-G | 0.2772 | 0.26560 | 1.106 | 0.7992–1.53 | 0.7181 | 0.9996 |
| Block 2 | T-T-A | 0.3062 | 0.27950 | 1 | Reference | 0.4242 | 0.9686 |
| A-C-A | 0.2493 | 0.27690 | 0.8217 | 0.5716–1.181 | 0.3336 | 0.9066 |
| T-C-T | 0.2005 | 0.19330 | 0.9468 | 0.638–1.405 | 0.8237 | 0.9997 |
| T-C-A | 0.2408 | 0.24110 | 0.912 | 0.6281–1.324 | 0.9517 | 1 |
Gene-gene interaction analysis
A logistic regression model was constructed to
analyze the 4 SNPs in IRF6 (rs2073485, rs2235371, rs2236909 and
rs861020) and 1 SNP in PAX9 (rs17176643). The combinations of
rs2073485 (GG) and rs17176643 (aC+CC), rs2235371 (CC) and
rs17176643 (aC+CC), and rs2236909 (aG+aa) and rs17176643 (aC+CC),
were significantly associated with NSCL/P following Bonferroni
corrections (Table IV).
 | Table IVResults of gene-gene interactions
using a logistic regression method. |
Table IV
Results of gene-gene interactions
using a logistic regression method.
| | Frequencies | | | |
|---|
| |
| | | |
|---|
| IRF6a | PAX9a | Case | Control | OR | 95% CI | P-value |
|---|
| rs2073485 | rs17176643 | | | | | |
| aG+aa | CC | 0.073529 | 0.163717 | 1 | Reference | |
| GG | CC | 0.107843 | 0.106195 | 2.261 | 0.983–5.203 | 0.055 |
| aG+aa | aC+CC | 0.450980 | 0.508850 | 1.973 | 1.020–3.816 | 0.043 |
| GG | aC+CC | 0.367647 | 0.221239 | 3.700 | 1.840–7.440 |
0.000b |
| rs2235371 | rs17176643 | | | | | |
| Ct+tt | CC | 0.058824 | 0.141593 | 1 | Reference | |
| CC | CC | 0.122549 | 0.128319 | 2.299 | 0.980–5.390 | 0.056 |
| Ct+tt | aC+CC | 0.377451 | 0.446903 | 2.033 | 0.983–4.205 | 0.056 |
| CC | aC+CC | 0.441176 | 0.283186 | 3.750 | 1.795–7.835 |
0.000b |
| rs2236909 | rs17176643 | | | | | |
| GG | CC | 0.044118 | 0.079646 | 1 | Reference | |
| aG+aa | CC | 0.137255 | 0.190265 | 3.433 | 1.391–8.474 | 0.007 |
| GG | aC+CC | 0.191176 | 0.199115 | 1.387 | 0.607–3.170 | 0.438 |
| aG+aa | aC+CC | 0.627451 | 0.530973 | 7.002 | 3.154–15.543 |
0.000b |
| rs861020 | rs17176643 | | | | | |
| GG | CC | 0.093137 | 0.163717 | 1 | Reference | |
| aG+aa | CC | 0.088235 | 0.106195 | 1.045 | 0.462–2.367 | 0.915 |
| GG | aC+CC | 0.406863 | 0.464602 | 1.398 | 0.739–2.646 | 0.303 |
| aG+aa | aC+CC | 0.411765 | 0.265487 | 2.177 | 1.132–4.184 | 0.020 |
Discussion
In the present study, an association with NSCL/P was
demonstrated for the SNPs rs2073485, rs2235371, rs2236909 and
rs861020, in the IRF6 gene. Haplotype analysis supported these
findings. We also found that there was a marginally significant
association between NSCL/P and rs17176643 in the PAX9 gene.
Moreover, a logistic regression model showed that gene-gene
interactions between IRF6 and PAX9 increased the risk of
NSCL/P.
Previous studies have shown that variations in IRF6
are responsible for 12% of the genetic contribution to cleft lip or
palate and a significant association was demonstrated between
NSCL/P and the rs235371 polymorphism in IRF6, which was also
observed in certain populations (21). However, this finding remains
controversial in the Chinese population. In the present study, the
results of allelic frequencies and a dominant model analysis showed
a positive association between the rs235371 polymorphism and
NSCL/P. Jia et al reported that rs2073485 polymorphism was
associated with NSCL/P in Chinese patients (22), which was consistent with our
results. To the best of our knowledge, the relationships
demonstrated in the present study between rs2236909 and rs861020
with NSCL/P, have not been previously observed. In this study, the
allele frequency of rs2236909 in the NSCL/P group showed
significant differences from that of the control group. The
recessive model and the Cochran-Armitage trend test identified the
A variant as a risk factor for NSCL/P. Huang et al
demonstrated that there was no significant difference between the
NSCL/P and control groups at rs2013162; however, there was a
significant difference at rs2235375 (23). These SNPs are located in the same
LD block as rs2236909 in the HapMap CHB database. However, the
samples in the study by Huang et al were obtained from
subjects in West China, while the samples in our study were
obtained primarily from patients in North China, which may explain
the observed differences. Our results also showed that there was a
significant difference in the allele frequency of rs861020 between
the NSCL/P and control groups. The dominant and recessive models,
as well as the Cochran-Armitage trend test showed differences
between the NSCL/P and control groups, although these differences
were not significant following Bonferroni corrections. These
results suggest that the common A variant is a potential risk
factor for NSCL/P in the Chinese population. Other SNPs located in
the same LD block as rs861020 in the HapMap CHB database, have not
been previously studied. Pegelow et al suggested that the A
allele of rs861019 and the G allele of SNP rs7552506 showed an
association with cleft lip and palate in Swedish families (24); however, no significant association
was observed in the present study. This may be due to the use of
different geographical populations, sample sizes or study methods
(family-based versus a case-control study).
MSX1 mutations have been identified in 2% of cases
of NSCL/P and should be considered for genetic counseling. Variants
such as CA polymorphisms, A34G, P147Q, G110G, C565T, M37L and
G267A, are associated with NSCL/P in various populations (12,25,26);
however, this finding remains controversial. In the present study,
the case-control analysis using tag SNPs (rs1042484 and rs12532)
showed no statistical differences in the allelic frequency or
haplotype between the NSCL/P and control groups, which is
consistent with the data of Huang et al(13). PAX9 is important in the development
of the teeth and lips. Ichikawa et al demonstrated that a
heterozygous missense mutation in PAX9 (640A>G, S214G) was
linked to the susceptibility for NSCL/P in the Japanese population
(16). However, the remaining SNPs
in PAX9 did not have a significant association with NSCL/P
(17). To the best of our
knowledge, a study of PAX9 tag SNPs in the Chinese population has
not been previously conducted. In the present study, the allele
frequency of rs17176643 was different between the NSCL/P and
control groups and this difference was marginally significant
following Benjamini-Hochberg corrections (P=0.075). The dominant
model and Cochran-Armitage trend test implied that the common A
variant may be a risk factor of NSCL/P in a Chinese population. Lee
et al indicated that a haplotype with three SNPs (rs2073247,
rs17104928 and rs17176643) showed a significantly increased
association with NSCL/P (17).
Interactions between IRF6, MSX1 and PAX9 are
important for the development of dentition (27). However, interactions between IRF6
and PAX9 do not contribute to human tooth agenesis (28). The relationship between these
interactions and NSCL/P remains unknown. In the present study, a
logistic regression model was used to explore gene-gene
interactions and their involvement in NSCL/P. The model was based
on the combinations of two SNPs, which were significantly or
marginally different between the NSCL/P and control groups. The
combinations of rs2073485 (GG) and rs17176643 (aC+CC), rs2235371
(CC) and rs17176643 (aC+CC), and rs2236909 (aG+aa) and rs17176643
(aC+CC), significantly increased the risk of NSCL/P, which suggests
that these gene-gene interactions are critical in susceptibility to
NSCL/P in our study population. The underlying molecular mechanisms
of interaction between IRF6 and PAX9 requires further
clarification.
In conclusion, regardless of the limitations of a
small sample size, the present study demonstrated an association of
rs2073485, rs2235371, rs2236909 and rs861020 in the IRF6 gene with
NSCL/P in Chinese patients. Haplotype analysis of the gene
supported these findings. We also identified that gene-gene
interactions between IRF6 and PAX9 may be important in the
susceptibility to NSCL/P. Additional studies are required to
clarify the associations between IRF6, MSX1 and PAX9 with
NSCL/P.
Acknowledgements
The authors thank all the participants who donated
samples for this study. This study has been supported by the
National Natural Science Foundation of China (grant no.
30901569).
Abbreviations:
|
NSCL/P
|
non-syndromic cleft lip with or
without cleft palate
|
|
IRF6
|
interferon regulatory factor 6
|
|
MSX1
|
muscle segment homeobox 1
|
|
PAX9
|
paired box gene 9
|
|
HWE
|
Hardy-Weinberg equilibrium
|
|
LD
|
linkage disequilibrium
|
|
MAF
|
minor allele frequency
|
|
OR
|
odds radio
|
|
CI
|
confidence interval
|
References
|
1
|
Dixon MJ, Marazita ML, Beaty TH and Murray
JC: Cleft lip and palate: understanding genetic and environmental
influences. Nat Rev Genet. 12:167–178. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vieira AR: Genetic and environmental
factors in human cleft lip and palate. Front Oral Biol. 16:19–31.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zucchero TM, Cooper ME, Maher BS, et al:
Interferon regulatory factor 6 (IRF6) gene variants and the risk of
isolated cleft lip or palate. N Engl J Med. 351:769–780. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu T, Liang KY, Hetmanski JB, et al:
Evidence of gene-environment interaction for the IRF6 gene and
maternal multivitamin supplementation in controlling the risk of
cleft lip with/without cleft palate. Hum Genet. 128:401–410. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Scapoli L, Palmieri A, Martinelli M, et
al: Strong evidence of linkage disequilibrium between polymorphisms
at the IRF6 locus and nonsyndromic cleft lip with or without cleft
palate, in an Italian population. Am J Hum Genet. 76:180–183. 2005.
View Article : Google Scholar
|
|
6
|
Park JW, McIntosh I, Hetmanski JB, et al:
Association between IRF6 and nonsyndromic cleft lip with or without
cleft palate in four populations. Genet Med. 9:219–227. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brito LA, Bassi CF, Masotti C, et al: IRF6
is a risk factor for nonsyndromic cleft lip in the Brazilian
population. Am J Med Genet A. 158A:2170–2175. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi J, Song T, Jiao X, Qin C and Zhou J:
Single-nucleotide polymorphisms (SNPs) of the IRF6 and TFAP2A in
non-syndromic cleft lip with or without cleft palate (NSCLP) in a
northern Chinese population. Biochem Biophys Res Commun.
410:732–736. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pan Y, Ma J, Zhang W, et al: IRF6
polymorphisms are associated with nonsyndromic orofacial clefts in
a Chinese Han population. Am J Med Genet A. 152A:2505–2511. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nugent P and Greene RM: MSX-1 gene
expression and regulation in embryonic palatal tissue. In Vitro
Cell Dev Biol Anim. 34:831–835. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nakatomi M, Wang XP, Key D, et al: Genetic
interactions between Pax9 and Msx1 regulate lip development and
several stages of tooth morphogenesis. Dev Biol. 340:438–449. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Salahshourifar I, Halim AS, Wan Sulaiman
WA, et al: Contribution of MSX1 variants to the risk of
non-syndromic cleft lip and palate in a Malay population. J Hum
Genet. 56:755–758. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang YQ, Ma J, Ma M, et al: Association
between MSX1 variants and oral clefts in Han Chinese in western
China. DNA Cell Biol. 30:1057–1061. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Suazo J, Santos JL, Jara L and Blanco R:
Parent-of-origin effects for MSX1 in a Chilean population with
nonsyndromic cleft lip/palate. Am J Med Genet A. 152A:2011–2016.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Perry GH, Verrelli BC and Stone AC:
Molecular evolution of the primate developmental genes MSX1 and
PAX9. Mol Biol Evol. 23:644–654. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ichikawa E, Watanabe A, Nakano Y, et al:
PAX9 and TGFB3 are linked to susceptibility to nonsyndromic cleft
lip with or without cleft palate in the Japanese: population-based
and family-based candidate gene analyses. J Hum Genet. 51:38–46.
2006. View Article : Google Scholar
|
|
17
|
Lee JK, Park JW, Kim YH and Baek SH:
Association between PAX9 single-nucleotide polymorphisms and
nonsyndromic cleft lip with or without cleft palate. J Craniofac
Surg. 23:1262–1266. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sull JW, Liang KY, Hetmanski JB, et al:
Maternal transmission effects of the PAX genes among cleft
case-parent trios from four populations. Eur J Hum Genet.
17:831–839. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Matalova E, Fleischmannova J, Sharpe PT
and Tucker AS: Tooth agenesis: from molecular genetics to molecular
dentistry. J Dent Res. 87:617–623. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Slayton RL, Williams L, Murray JC, et al:
Genetic association studies of cleft lip and/or palate with
hypodontia outside the cleft region. Cleft Palate Craniofac J.
40:274–279. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zucchero TM, Cooper ME, Maher BS, et al:
Interferon regulatory factor 6 (IRF6) gene variants and the risk of
isolated cleft lip or palate. N Engl J Med. 351:769–780. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jia ZL, Li Y, Li L, et al: Association
among IRF6 polymorphism, environmental factors, and nonsyndromic
orofacial clefts in western china. DNA Cell Biol. 28:249–257. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang Y, Wu J, Ma J, et al: Association
between IRF6 SNPs and oral clefts in West China. J Dent Res.
88:715–718. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pegelow M, Peyrard-Janvid M, Zucchelli M,
et al: Familial non-syndromic cleft lip and palate - analysis of
the IRF6 gene and clinical phenotypes. Eur J Orthod. 30:169–175.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tongkobpetch S, Siriwan P and Shotelersuk
V: MSX1 mutations contribute to nonsyndromic cleft lip in a Thai
population. J Hum Genet. 51:671–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Butali A, Mossey PA, Adeyemo WL, et al:
Genetic studies in the Nigerian population implicate an MSX1
mutation in complex oral facial clefting disorders. Cleft Palate
Craniofac J. 48:646–653. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Boeira BR Jr and Echeverrigaray S:
Dentistry and molecular biology: a promising field for tooth
agenesis management. Tohoku J Exp Med. 226:243–249. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vieira AR, Cooper ME, Marazita ML, Oriolo
IM and Castilla EE: Interferon regulatory factor 6 (IRF6) is
associated with oral-facial cleft in individuals that originate in
South America. Am J Med Genet A. 143A:2075–2078. 2007. View Article : Google Scholar : PubMed/NCBI
|