Introduction
Coronary heart disease (CHD) is a severe condition
in which plaque builds up inside the coronary arteries. Over time,
plaque hardens and narrows the coronary arteries, eventually
leading to myocardic infarction and mortality. Triglycerides (TGs)
are the major components of plaque and aberrant levels of TGs
significantly correlate with the risk of CHD (1). Increased TGs contribute to the
development of hypertriglyceridemia and metabolic syndrome
(2,3), which are associated with
cardiovascular events (2).
Although a small number of candidate genes have been identified for
the risk of CHD, it is estimated that 95% of genetic factors remain
unidentified in elucidating the pathogenesis of this complex
disease (4).
Apolipoprotein A-V (ApoA-V) is a key regulator of TG
levels (5,6) and apolipoprotein A5 (APOA5)
gene variants, including -1131T>C (rs662799) and S19W
(rs3135506). These genetic variants have been significantly
associated with TG levels (7–9) and
the risk of CHD (10). TG levels
have been shown to be significantly higher in -1131C controls
compared with -1131T controls (7,8). In
addition, the -1131C allele frequency of the APOA5 gene in
the early-onset CHD group (43.2%) has been observed to be
significantly higher than that in a control group (33.0%) (10). In the S19W variant, the minor
allele 19W was found to be rare in the Chinese (0–4.7%) (11) in contrast to that of 15% of the
Latin American populations (12).
Despite the low minor allele frequency, a positive correlation was
found between the 19W allele and CHD in the Chinese population
(8,12). In addition, another variant,
553G>T (rs2075291), was shown as a risk factor for CHD in the
Han Chinese population (13,14).
However, conflicting results of APOA5 variants were observed
in a number of other studies (6,15,16).
Studies showed that S19W and -1131T>C were not associated with
the risk of CHD in Italian (6) and
Brazilian populations (15). The
19W allele was not identified as a risk factor for stroke in
Hungarian populations (16). Since
previous epidemiological studies indicate that there is an ethnic
difference in the APOA5 gene variants, a meta-analysis of
the available data was necessary to investigate the role of the
APOA5 gene in the risk of CHD.
The aim of the current study was to assess whether
the APOA5 gene variants, -1131T>C, S19W and 553G>T,
are associated with CHD in the populations studied and to evaluate
the contribution of APOA5 gene variants to CHD in various
ethnic populations by meta-analysis.
Materials and methods
Sample collection
A total of 819 unrelated individuals were recruited
for the case-control study. These included 290 CHD cases, 198
non-CHD controls and 331 healthy controls. CHD cases were patients
with >50% coronary artery occlusion of one or more major
coronary arteries (17) or a
history of prior angioplasty or coronary artery bypass surgery.
Non-CHD participants were selected from inpatients who had <50%
occlusion in the major coronary artery (18) and did not have any atherosclerotic
vascular disease. In addition, 331 apparently healthy individuals
from the Ximen Community residents in Ningbo were recruited as
healthy controls. CHD cases and non-CHD controls were collected
from the Lihuili Hospital (Ningbo, China). CHD cases and non-CHD
controls had been examined by standardized coronary angiography
according to Seldinger’s method (19) and assessed by at least two
independent cardiologists. Subjects were excluded from this study
if the individual had congenital heart disease, cardiomyopathy,
liver or renal disease or cancer. Blood samples were stored at
−80°C until analysis was performed and were treated by the same
investigators. The study was approved by the Ethical Committee of
Lihuili Hospital in Ningbo (Zhejiang, China) and informed written
consent was obtained from all subjects.
Single nucleotide polymorphism (SNP)
genotyping
Human genomic DNA was prepared from peripheral blood
samples using the Lab-Aid 820 nucleic acid extraction automatic
analyzer (Zeesan Biotech, Xiamen, China) and was quantified using
the Quant-iT™ PicoGreen® dsDNA assay kit (Molecular
Probes, Inc., Eugene, OR, USA). Amplification was performed on the
Geneamp® PCR System 9700 Dual 384-Well Sample Block
Module (Applied Biosystems, Foster City, CA, USA) for polymerase
chain reaction (PCR). Primers for the single base extension
reaction are shown in Table I. The
distinct mass of the extended primer indicates different SNP
alleles. PCR conditions included an initial denaturation stage at
94°C for 15 sec, followed by 45 cycles at 94°C for 20 sec, 56°C for
30 sec and primer extension at 72°C for 1 min and a final extension
for 3 min at 72°C. Primer extension for genotyping was performed on
the Sequenom MassARRAY iPLEX® platform (Sequenom, San
Diego, CA, USA) according to the manufacturer’s instructions
(20). The primer extension
reaction included an initial denaturation stage at 94°C for 30 sec,
followed by 40 cycles of amplification, including 94°C for 5 sec,
52°C for 5 sec and 80°C for 5 sec, and 5 cycles of amplification,
including 52°C for 5 sec and 80°C for 5 sec, and a final extension
for 3 min at 72°C. Following purification, the products were
subjected to MALDI-TOF mass spectrometry for SNP genotyping using a
SpectroCHIP array (Sequenom, San Diego, CA, USA). To verify the
repeatability and stability of the experiment, 5% of random samples
and 18 control samples, including 9 negative and 9 positive
controls, were used for quality control.
 | Table IPrimer sequences for single base
extension reaction. |
Table I
Primer sequences for single base
extension reaction.
| SNP | Name | Primer | Sequence
(5′-3′) |
|---|
| rs662799 | -1131T>C | 1st-P |
ACGTTGGATGGCCCTGCGAGTGGAGTTCA |
| 2nd-P |
ACGTTGGATGACTCTGAGCCCCAGGAACT |
| UEP_SEQ |
GGGTGAACTGGAGCGAAAGT |
| rs3135506 | S19W | 1st-P |
ACGTTGGATGTGGTCTGGCTGAAGTAGTCC |
| 2nd-P |
ACGTTGGATGTGATTACCTAGTCCCTCTCC |
| UEP_SEQ |
TAGGCCCTCTCCACAGCGTTTT |
| rs2075291 | 553G>T | 1st-P |
ACGTTGGATGTTGGGCTTTGCTGCAGGGAC |
| 2nd-P |
ACGTTGGATGATGGGTGGAAGAGCTCTTTG |
| UEP_SEQ |
GCTCTTTGAAGCGGC |
Retrieval of published studies
A search of the studies on APOA5 gene
variants and CHD was conducted in electronic databases, including
the Chinese National Knowledge infrastructure, PubMed, Embase,
SpringerLink and ScienceDirect, between 2001 and 2012. Specific
combinations of keywords were used for the following Medical
Subject Heading terms, including ‘coronary heart disease’,
‘coronary artery disease’ or ‘myocardial infarction’ combined with
‘APOA5’, ‘apolipoprotein A5’, ‘C56G’, ‘S19W’, ‘-1131T>C’
or ‘553G>T’ and ‘single nucleotide polymorphism’, ‘SNP’ or
‘genetic association’. All studies were considered eligible if they
aimed to investigate the correlation between APOA5 and the
risk of CHD. For meta-analysis, studies with one of the following
conditions were excluded: i) Studies lacking controls, ii) a lack
of detailed main allele or genotype information and iii) duplicate
publications.
Statistical analysis
Departure of Hardy-Weinberg equilibrium (HWE) was
analyzed by Arlequin program version 3.5 (21). The allele frequencies and genotype
distribution between CHD patients and each of the 2 control groups
were compared by CLUMP16 software (Department of Psychological
Medicine, Institute of Psychiatry, Denmark Hill, London, UK) with
10,000 Monte Carlo simulations (22). Linkage disequilibrium of
APOA5 gene variants was measured by an online calculator
(http://www.oege.orgsoftware/cubex/).
Haplotype frequencies were inferred by Arlequin program version 3.5
based on the expectation-maximization algorithm. The odds ratio
(OR) with 95% confidence interval (95% CI) was calculated by an
online tool (http://faculty.vassar.edu/lowry/odds2x2.html). The
statistical power of the study was calculated by the PS power and
sample size calculation software version 3.0.43 (23). The correlation between the variants
and the severity of CHD was analyzed by the R statistics software
(University of Auckland, Auckland, New Zealand). Severity grade of
CHD was defined by the number of major coronary arteries with
>50% occlusion. Meta-analysis was performed using the Review
Manager version 5.1 (The Cochrane Collaboration, The Nordic
Cochrane Centre, Copenhagen, Denmark). Heterogeneity of the studies
in the meta-analysis was assessed with the Q and I2
tests. Publication bias was presented using funnel plots by Review
Manager 5.1. The type I error rate was set at 0.05. Two-tailed
P<0.05 was considered to indicate a statistically significant
difference.
Results
Case-control study in Han Chinese
populations
SNP S19W was monomorphic in samples of the current
study and thus, discarded from further analysis. SNPs -1131T>C
and 553G>T were consistent with HWE (P>0.05). As shown in
Table II, no significant
differences in the 2 SNPs were observed between CHD cases and each
of the 2 control groups (P>0.05). Haplotypes of the 2 SNPs were
associated with the risk of CHD (data not shown). Under the
dominant inheritance model, -1131C was observed to be a CHD risk
factor (P=0.030; OR, 1.422; 95% CI, 1.036–1.952; Table III). Further breakdown analysis
by gender did not produce significant results between the 2
variants and the risk of CHD (data not shown).
 | Table IIFrequencies of the genotype and
allele for SNPs. |
Table II
Frequencies of the genotype and
allele for SNPs.
| A, 553G>T |
|---|
|
|---|
| Total | Genotype (n) | χ2 | P-value, df=2 | HWE | Allele (n) | χ2 | P-value, df=1 | OR (95% CI) |
|---|
|
|
|---|
| GG | GT | TT | G | T |
|---|
| CHD cases,
n=290 | 258 | 31 | 1 | | | 1.000 | 547 | 33 | | | |
| Non-CHD controls,
n=198 | 169 | 29 | 0 | 2.357 | 0.261 | 0.605 | 367 | 29 | 1.056 | 0.352 | 0.764
(0.456–1.279) |
| Healthy controls,
n=331 | 299 | 31 | 1 | 0.312 | 0.799 | 0.567 | 629 | 33 | 0.305 | 0.614 | 1.150
(0.700–1.888) |
|
| B, -1131T>C |
|
| Total | Genotype (n) | χ2 | P-value, df=2 | HWE | Allele (n) | χ2 | P-value, df=1 | OR (95% CI) |
|
|
| AA | AG | GG | A | G |
|
| CHD cases,
n=290 | 134 | 124 | 32 | | | 0.685 | 392 | 188 | | | |
| Non-CHD controls,
n=198 | 106 | 75 | 17 | 2.675 | 0.257 | 0.476 | 287 | 109 | 2.656 | 0.107 | 1.263
(0.954–1.672) |
| Healthy controls,
n=331 | 182 | 117 | 32 | 4.808 | 0.090 | 0.051 | 481 | 181 | 3.809 | 0.054 | 1.275
(0.999–1.626) |
 | Table IIISignificant differences in genotype
distributions under the dominant model. |
Table III
Significant differences in genotype
distributions under the dominant model.
| CHD cases vs.
non-CHD controls | CHD cases vs.
healthy controls |
|---|
|
|
|
|---|
| Dominant model | OR (95% CI) | P-value, df=1 | OR (95% CI) | P-value, df=1 |
|---|
| Total |
| rs2075291 (GT + TT
vs. GG) | 0.723
(0.422–1.239) | 0.266 | 1.159
(0.691–1.945) | 0.599 |
| rs662799 (AG + GG
vs. AA) | 1.341
(0.934–1.927) | 0.118 | 1.422
(1.036–1.952) | 0.030 |
| Male |
| rs2075291 (GT + TT
vs. GG) | 0.638
(0.325–1.249) | 0.211 | 1.720
(0.677–4.370) | 0.295 |
| rs662799 (AG + GG
vs. AA) | 1.343
(0.834–2.163) | 0.229 | 1.499
(0.903–2.488) | 0.126 |
| Female |
| rs2075291 (GT + TT
vs. GG) | 0.787
(0.305–2.031) | 0.643 | 0.936
(0.406–2.159) | 1.000 |
| rs662799 (AG + GG
vs. AA) | 1.515
(0.834–2.753) | 0.178 | 1.664
(0.998–2.774) | 0.054 |
Correlation between the 2 variants and
the severity of CHD
The severity of CHD was defined by the number of
coronary arteries with >50% coronary artery occlusion. A
logistic regression test was performed between the 2 variants and
the severity of CHD in all cases and in a gender-stratified manner.
The results indicated that 553G>T correlated with CHD severity
in males (Table IV; P=0.032);
however, following Bonferroni’s correction, this result was not
statistically significant.
 | Table IVLogistic regression analysis of
association of SNPs and the serious extent of CHD disease. |
Table IV
Logistic regression analysis of
association of SNPs and the serious extent of CHD disease.
| Parameters | Non-CHD
controls | One artery | Two arteries | ≥Three
arteries | rs2075291 | rs662799 |
|---|
| Total | 198 | 106 | 65 | 119 | 0.091 | 0.283 |
| Male | 101 | 77 | 49 | 84 | 0.032 | 0.568 |
| Female | 97 | 29 | 16 | 35 | 0.898 | 0.219 |
Inclusion of case-control studies for
meta-analysis
A total of 23 association studies between
APOA5 gene variants and the risk of CHD were retrieved from
the online databases. Among them, 9 studies were excluded from the
current meta-analysis as they focused on other APOA5
variants (24–27) or did not present sufficient
information on genotype or allele frequencies or OR values
(9,28–31).
A total of 14 studies were included in the meta-analysis:
-1131T>C, 6,848 cases and 5,452 controls; S19W, 7,644 cases and
10,610 controls; and 553G>T, 4,450 cases and 5,068 controls.
Meta-analysis of the association studies
between -1131T>C and the risk of CHD
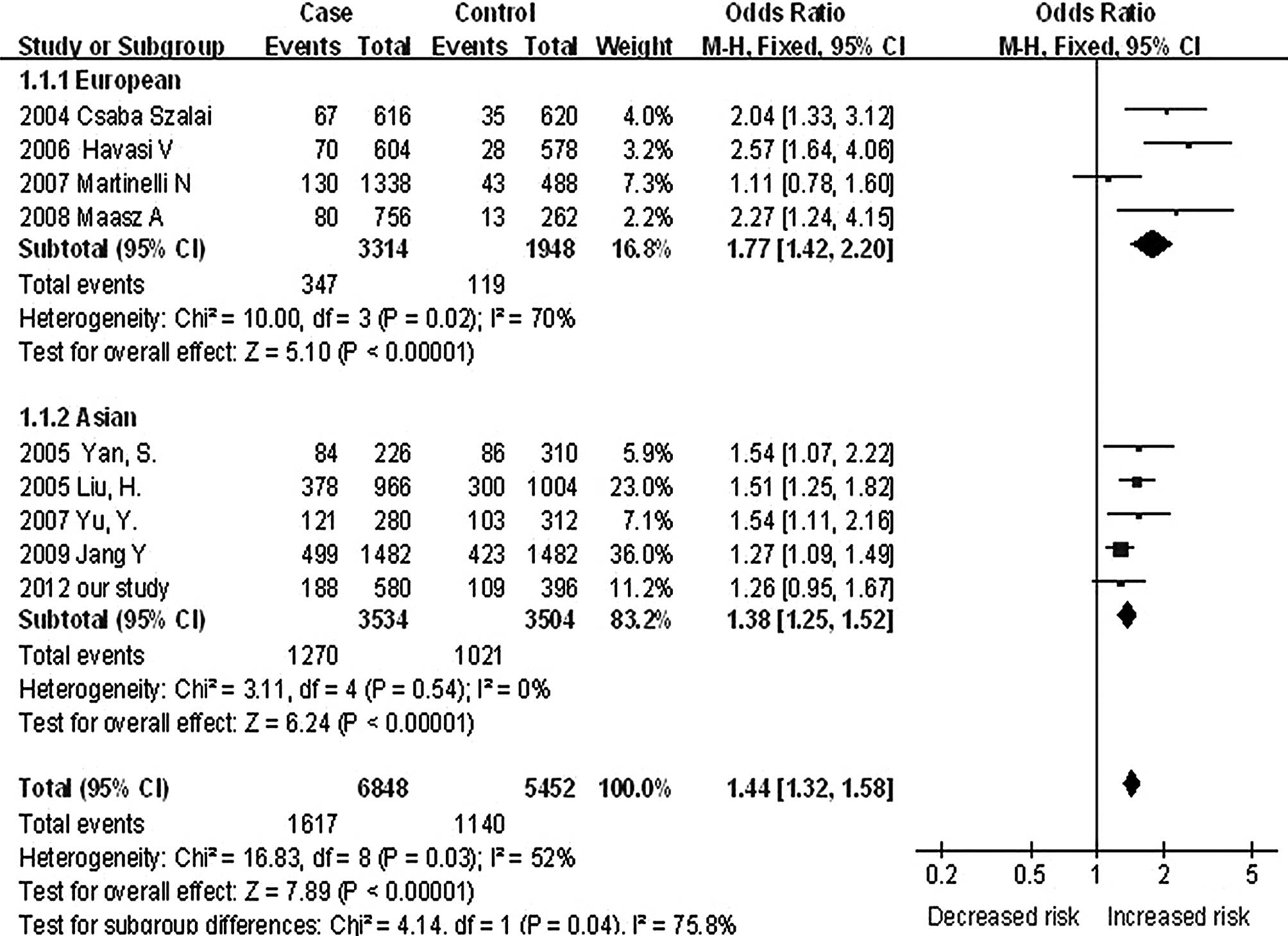
As shown in Fig. 1,
the case-control study (CHD cases vs. non-CHD controls) and 8 other
studies (6–8,10,32–35)
were included in the meta-analysis. Significant heterogeneity was
observed among the 9 studies (I2, 52%;
χ2,16.83; df, 8; P=0.03). Due to the high heterogeneity
of these studies, the groups were divided into 2 ethnic subgroups,
European and Asian. A significant heterogeneity was observed among
Europeans (6,7,33,34)
(I2, 70%; χ2, 10.00; df, 3; P=0.02) in
contrast to minimal heterogeneity among Asians (8,10,32,35)
(I2, 0%; χ2, 3.11; df, 4; P=0.54). A
significant association between -1131T>C and CHD risk was
observed in European individuals (OR, 1.77; 95% CI, 1.42–2.20;
P<0.0001) and Asian subgroups (OR, 1.38; 95% CI, 1.25–1.52;
P<0.0001). A significant difference was observed between the 2
subgroups (I2, 75.8%; χ2, 4.14; df, 1;
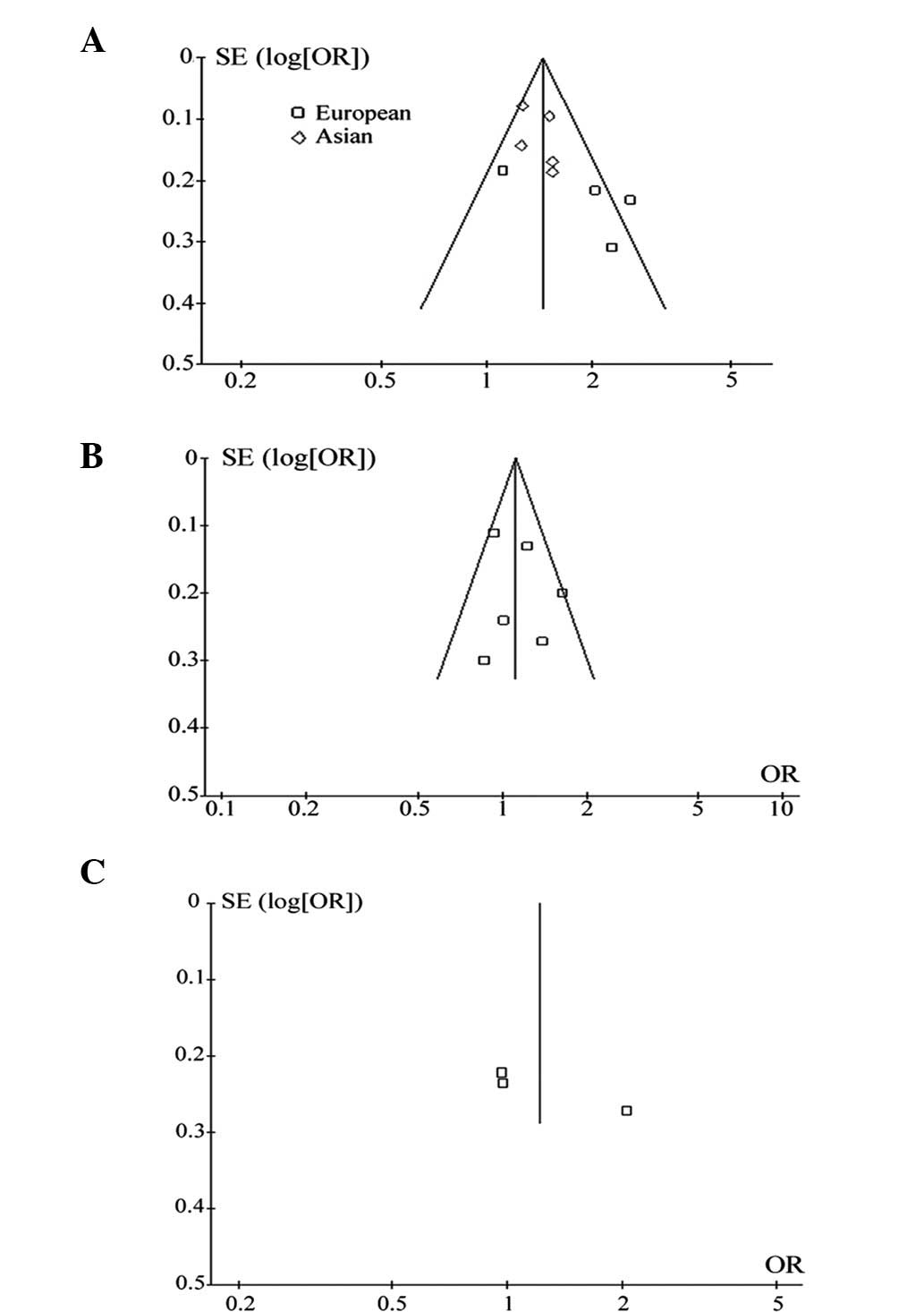
P=0.04). Funnel plot analysis did not reveal publication bias
(Fig. 2A).
Meta-analysis of association studies
between 553G>T and the risk of CHD
Using the fixed effect analysis model, the
meta-analysis of 553G>T showed a moderate heterogeneity among
the 2 case-control studies (14,36)
and the current study (I2, 64%; χ2, 5.48; df,
2; P=0.06). Due to the moderate heterogeneity, the random effects
analysis model was selected for meta-analysis (Fig. 3). The results showed that 553G>T
had no significant association with CHD (P=0.40; OR, 1.22; 95% CI,
0.77–1.91). Funnel plot analysis did not reveal publication bias
(Fig. 2B).
Meta-analysis of the association studies
between S19W and the risk of CHD
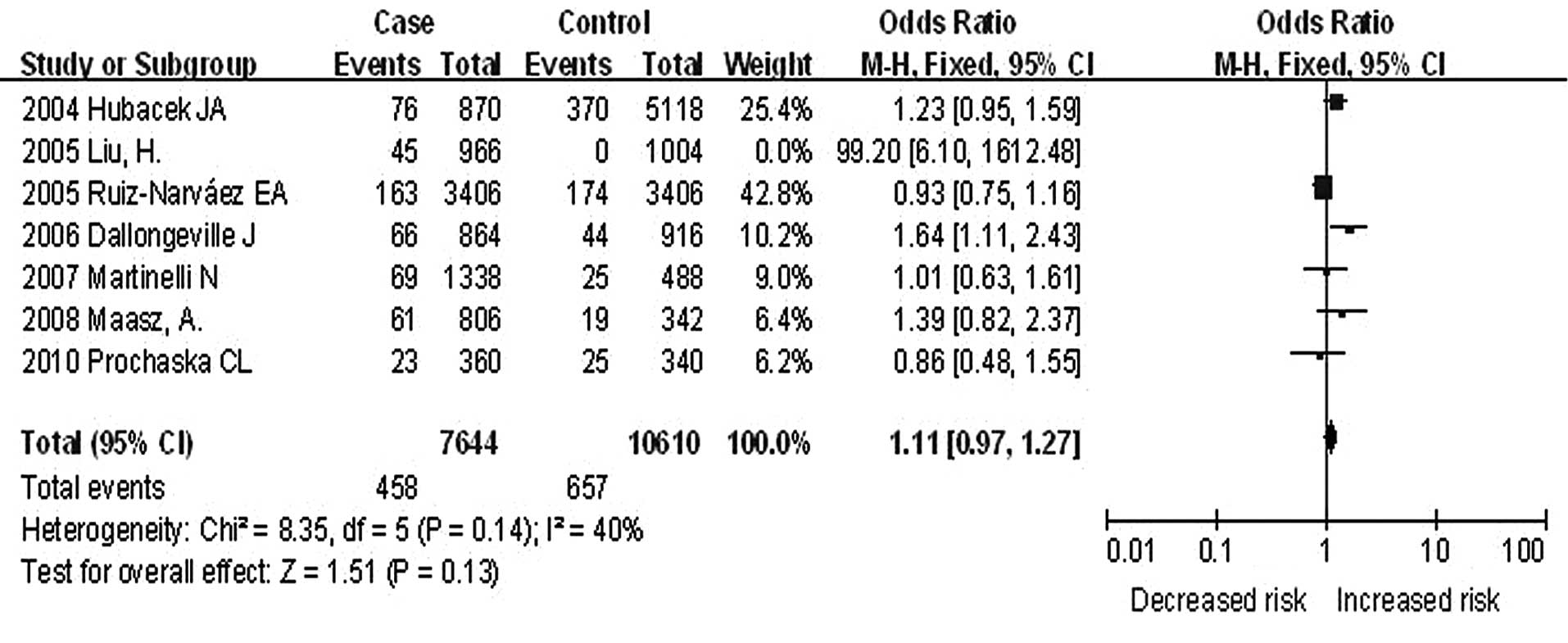
Fig. 4 shows the
results of meta-analysis of the associations between S19W and CHD.
Since S19W was monomorphic in the samples, only 7 studies were
included in the meta-analysis (6,8,15,16,36–38).
Under the random effects model, a significantly higher
heterogeneity was observed (I2, 70%; χ2,
19.98; df, 6, P=0.003). An outlier OR-value was observed in 1 study
(8) (OR, 99.20; 95% CI,
6.10–1612.48). Following exclusion of the outlier, another
meta-analysis was performed and a lower heterogeneity was observed
with fixed effect analysis model (Fig.
4; I2, 40%; χ2, 8.35; df, 5; P=0.14). The
results showed that S19W had no significant association with CHD
(OR, 1.11; 95% CI, 0.97–1.27; P=0.13). Funnel plot analysis did not
reveal publication bias (Fig.
2C).
Discussion
The APOA5 gene codes for a 366-amino acid
protein, apoA-V, which enhances lipoprotein lipase (LPL) activity
(24). Loss of LPL activity
interferes with the ability of apoA-V to interact with lipids and
lipoproteins, including TGs, very low density lipoproteins and high
density lipoproteins (39,40). Elevated plasma TGs are a known risk
factor for CHD (41,42) and apoA-V is a major risk factor of
CHD as it activates TG hydrolysis in the blood (43). APOA5 gene variants have been
identified as the genetic determinants of TG concentration
(9). Since discrepancies exist in
previous epidemiological studies on the association of APOA5
gene variants with CHD, the current study investigated a
case-control study in specific populations and meta-analysis of the
available case-control data was performed to clarify the role of
APOA5 gene variants in CHD.
SNP -1131T>C is located in the proximal promoter
of the APOA5 gene and is associated with elevated TG levels
and hyperinsulinemia (2). A number
of studies have found that the -1131T>C gene is significantly
associated with CHD in Chinese populations (8,10,32).
However, the association between -1131T>C and CHD in the
European population remains controversial (6,7,33,34).
Three independent studies (7,33,34)
observed that patients carrying the -1131CT>C gene had higher TG
levels and a significantly increased risk of coronary events.
However, Martinelli et al demonstrated that 2 APOA5
variants, including -1131T>C, which are independent predictors
of TGs (6), were not associated
with CHD (6). The current
meta-analysis of 9 studies among 12,300 individuals indicates that
-1131CT>C is a risk factor for CHD (pooled OR, 1.44; 95% CI,
1.32–1.58; P<0.00001). An ethnic difference in the prevalence of
-1131T>C was observed between the Asian and the European studies
(I2, 5.8%; P=0.04). The frequency of -1131CT>C in
non-CHD controls and healthy controls was 0.380 and 0.376,
respectively, which is close to 0.267 in HapMap-HCB and 0.291 in
HapMap-JPT (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=662799).
A lower frequency of -1131C was observed in Europeans (0.017 in
HapMap-CEU). This is in agreement with the current heterogeneity
test results in the meta-analysis.
SNP 553G>T is a rare APOA5 gene variant that has
been studied in Han Chinese populations (13,14,29,36).
SNP 553G>T has been found to correlate with serum levels of TG
and total cholesterol in Han Chinese individuals in Xinjiang, China
(29). A significant association
between 553G>T and CHD in Han Chinese populations, by 2 separate
groups, has been found in Taiwan (P<0.001) and Nanjing (P=0.017)
(13,14). In the present study, the previous
positive association between 553G>T and CHD was not observed. A
meta-analysis of 3 studies among 9,518 individuals indicates that
the 553G>T gene is not associated with CHD risk (P=0.40; OR,
1.22; 95% CI, 0.77–1.91). However, a correlation between 553G>T
and the severity of CHD was observed in males by a logistic
regression analysis (P=0.032). Further investigation of the
contribution of 553G>T to the progression of CHD is
required.
The minor allele frequency of S19W in Chinese
populations was significantly different from that in Caucasians.
The 19W allele is rare in HapMap-CHB (0%), thus providing an
explanation as to why S19W was monomorphic in the samples,
including 290 CHD cases, 198 non-CHD controls and 331 healthy
controls. Liu et al observed a 4.7% prevalence of 19W in the
CHD cases, while in Chinese populations, 19W was not observed (0%)
(8). However, another case-control
study in Chinese populations did not observe 19W in cases and
controls and thus, hypothesized a negative association between SNP
19W and CHD (11,44). The allele frequency of 19W was 0.1%
in Chinese Singaporean populations (12). However, 19W was more common in
Europeans (HapMap-CEU: 5.8%). The allele frequencies of 19W in
Malay and the Asian-Indian populations were 1.7 and 3.1%,
respectively, while in Latin-American populations, allele frequency
was 15% (12). The current
meta-analysis of 6 studies (6,15,16,36–38)
among 18,254 individuals found no significant association between
CHD and S19W (P=0.13; OR, 1.11; 95% CI, 0.97–1.27).
In summary, the current case-control study shows
that the -1131CT>C gene is a CHD risk factor in the populations
studied and this association was further supported by
meta-analysis. The case-control study has <80% statistical power
(the strongest power observed for -1131T>C was 58.8%). An
improved case-control investigation with larger sample sizes and a
balanced gender structure is required in the future.
Acknowledgements
The research was supported by the grants from the
National Natural Science Foundation of China (31100919), Natural
Science Foundation of Zhejiang Province (LR13H020003), K.C. Wong
Magna Fund in Ningbo University, Ningbo Social Development Research
Projects (2012C50032), Advanced Key Scientific and Technological
Programs of NingBo (2011C51001), Fund of NingBo Science and
Technology Innovation Team (2011B82015), Natural Science Foundation
of the Zhejiang Province (LY13H020008), Ningbo Personnel Training
Project (first level), and the Project of Ningbo Medicine and
Science (2009A02).
Abbreviations:
|
SNP
|
single nucleotide polymorphism
|
|
CHD
|
coronary heart disease
|
|
HWE
|
Hardy-Weinberg equilibrium
|
|
GWAS
|
genome-wide association studies
|
|
APOA5
|
apolipoprotein A5
|
|
TG
|
triglyceride
|
|
OR
|
odds ratio
|
|
CI
|
confidence interval
|
References
|
1
|
Carey VJ, Bishop L, Laranjo N, Harshfield
BJ, Kwiat C and Sacks FM: Contribution of high plasma triglycerides
and low high-density lipoprotein cholesterol to residual risk of
coronary heart disease after establishment of low-density
lipoprotein cholesterol control. Am J Cardiol. 106:757–763. 2010.
View Article : Google Scholar
|
|
2
|
Maász A, Kisfali P, Horvatovich K, et al:
Apolipoprotein A5 T-1131C variant confers risk for metabolic
syndrome. Pathol Oncol Res. 13:243–247. 2007.PubMed/NCBI
|
|
3
|
Evans D, Aberle J and Beil FU:
Resequencing the apolipoprotein A5 (APOA5) gene in patients with
various forms of hypertriglyceridemia. Atherosclerosis.
219:715–720. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Peden JF and Farrall M: Thirty-five common
variants for coronary artery disease: the fruits of much
collaborative labour. Hum Mol Genet. 20:R198–R205. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pennacchio LA, Olivier M, Hubacek JA,
Cohen JC, Cox DR, Fruchart JC, Krauss RM and Rubin EM: An
apolipoprotein influencing triglycerides in humans and mice
revealed by comparative sequencing. Science. 294:169–173. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martinelli N, Trabetti E, Bassi A, Girelli
D, Friso S, Pizzolo F, Sandri M, Malerba G, Pignatti PF, Corrocher
R and Olivieri O: The -1131 T>C and S19W APOA5 gene
polymorphisms are associated with high levels of triglycerides and
apolipoprotein C-III, but not with coronary artery disease: an
angiographic study. Atherosclerosis. 191:409–417. 2007.
|
|
7
|
Szalai C, Keszei M, Duba J, Prohászka Z,
Kozma GT, Császár A, Balogh S, Almássy Z, Fust G and Czinner A:
Polymorphism in the promoter region of the apolipoprotein A5 gene
is associated with an increased susceptibility for coronary artery
disease. Atherosclerosis. 173:109–114. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu H, Zhang S, Lin J, et al: Association
between DNA variant sites in the apolipoprotein A5 gene and
coronary heart disease in Chinese. Metabolism. 54:568–572. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Triglyceride Coronary Disease Genetics
Consortium and Emerging Risk Factors Collaboration. Sarwar N,
Sandhu MS, Ricketts SL, et al: Triglyceride-mediated pathways and
coronary disease: collaborative analysis of 101 studies. Lancet.
375:1634–1639. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu Y, Xue L and Zhao CY: Study on
polymorphism in the apolipoprotein A5 gene in patients with
premature coronary heart disease. Beijing Da Xue Xue Bao.
39:576–580. 2007.(In Chinese).
|
|
11
|
Li X, Yao C, Song M, Ma J, Ning H and Hu
B: The relationship between the polymorphism of apolipoprotein A5
gene 56C>G and the level of serum triglyceride. Ningxia Yi Xue
Za Zhi. 9:643–645. 2006.(In Chinese).
|
|
12
|
Lai CQ, Tai ES, Tan CE, Cutter J, Chew SK,
Zhu YP, Adiconis X and Ordovas JM: The APOA5 locus is a strong
determinant of plasma triglyceride concentrations across ethnic
groups in Singapore. J Lipid Res. 44:2365–2373. 2003. View Article : Google Scholar
|
|
13
|
Kao JT, Wen HC, Chien KL, Hsu HC and Lin
SW: A novel genetic variant in the apolipoprotein A5 gene is
associated with hypertriglyceridemia. Hum Mol Genet. 12:2533–2539.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang Y, Sun P, Guo D, Ferro A, Ji Y, Chen
Q and Fan L: A genetic variant c.553G>T in the apolipoprotein A5
gene is associated with an increased risk of coronary artery
disease and altered triglyceride levels in a Chinese population.
Atherosclerosis. 185:433–437. 2006.PubMed/NCBI
|
|
15
|
Prochaska CL, Picheth G, Anghebem-Oliveira
MI, Costantini CO, de Souza EM, Pedrosa FO and Scartezini M: The
polymorphisms -1131T>C and the S19W of the APOA5 gene are not
associated with coronary artery disease in a Brazilian population.
Clin Chem Lab Med. 48:419–422. 2010.
|
|
16
|
Maász A, Kisfali P, Szolnoki Z, Hadarits F
and Melegh B: Apolipoprotein A5 gene C56G variant confers risk for
the development of large-vessel associated ischemic stroke. J
Neurol. 255:649–654. 2008.PubMed/NCBI
|
|
17
|
No authors listed. Nomenclature and
criteria for diagnosis of ischemic heart disease. Report of the
Joint International Society and Federation of Cardiology/World
Health Organization task force on standardization of clinical
nomenclature. Circulation. 59:607–609. 1979. View Article : Google Scholar
|
|
18
|
Nikus KC: Chapter 4: Coranory angiography.
Multimodal Imaging: Principles and Clinical Applications: Section
I: Current methods and their applications for cardiovascular
multimodal imaging. Pahlm O and Wagner GS: McGraw-Hill Medical; New
York, NY: pp. 752011
|
|
19
|
Higgs ZC, Macafee DA, Braithwaite BD and
Maxwell-Armstrong CA: The Seldinger technique: 50 years on. Lancet.
366:1407–1409. 2005.PubMed/NCBI
|
|
20
|
Gabriel S, Ziaugra L and Tabbaa D: SNP
genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc
Hum Genet. (Suppl 60): 2.12.1–2.12.18. 2009.
|
|
21
|
Excoffier L and Lischer HE: Arlequin suite
ver 3.5: a new series of programs to perform population genetics
analyses under Linux and Windows. Mol Ecol Resour. 10:564–567.
2010. View Article : Google Scholar
|
|
22
|
Sham PC and Curtis D: Monte Carlo tests
for associations between disease and alleles at highly polymorphic
loci. Ann Hum Genet. 59(Pt 1): 97–105. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dupont WD and Plummer WD Jr: Power and
sample size calculations. A review and computer program. Control
Clin Trials. 11:116–128. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Aberle J, Evans D, Beil FU and Seedorf U:
A polymorphism in the apolipoprotein A5 gene is associated with
weight loss after short-term diet. Clin Genet. 68:152–154. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu HK, Wang CT, Zhang SZ, et al:
Association of APOA5 gene single nucleotide polymorphism with
levels of lipids and coronary heart disease in Chinese. Zhonghua Yi
Xue Yi Chuan Xue Za Zhi. 21:335–338. 2004.(In Chinese).
|
|
26
|
Yuan S, Ma YT, Xie X, Yang YN, Fu ZY, Ma
X, Li XM, Xiang Y, Liu F and Chen BD: Association of apolipoprotein
A5 gene polymorphism with coronary heart disease in Uygur
population of Xinjiang. Zhonghua Yi Xue Yi Chuan Xue Za Zhi.
28:73–77. 2011.(In Chinese).
|
|
27
|
Hubacek JA, Wang WW, Skodová Z, Adámková
V, Vráblík M, Horínek A, Stulc T, Ceska R and Talmud PJ: APOA5
Ala315>Val, identified in patients with severe
hypertriglyceridemia, is a common mutation with no major effects on
plasma lipid levels. Clin Chem Lab Med. 46:773–777. 2008.PubMed/NCBI
|
|
28
|
Eichenbaum-Voline S, Olivier M, Jones EL,
et al: Linkage and association between distinct variants of the
APOA1/C3/A4/A5 gene cluster and familial combined hyperlipidemia.
Arterioscler Thromb Vasc Biol. 24:167–174. 2004. View Article : Google Scholar
|
|
29
|
Yuan S, Ma YT, Xie X, Yang YN, Fu ZY, Ma
X, Li XM, Liu F and Chen BD: Association between apolipoprotein A5
gene polymorphism and coronary heart disease in the Han population
from Xinjiang. Zhonghua Liu Xing Bing Xue Za Zhi. 32:51–54.
2011.(In Chinese).
|
|
30
|
Song KH, Yu SG, Cha S and Kim JY:
Association of the Apolipoprotein A5 gene -1131T>C polymorphism
with serum lipids in Korean subjects: impact of Sasang
constitution. Evid Based Complement Alternat Med.
2012:5983942012.
|
|
31
|
Ramakrishnan L, Sachdev HS, Sharma M, et
al: Relationship of APOA5, PPARγ and HL gene variants with serial
changes in childhood body mass index and coronary artery disease
risk factors in young adulthood. Lipids Health Dis. 10:682011.
|
|
32
|
Yan SK, Cheng XQ, Song YH, Xiao XH, Bi N
and Chen BS: Apolipoprotein A5 gene polymorphism -1131T→C:
association with plasma lipids and type 2 diabetes mellitus with
coronary heart disease in Chinese. Clin Chem Lab Med. 43:607–612.
2005.
|
|
33
|
Havasi V, Szolnoki Z, Talián G, et al:
Apolipoprotein A5 gene promoter region T-1131C polymorphism
associates with elevated circulating triglyceride levels and
confers susceptibility for development of ischemic stroke. J Mol
Neurosci. 29:177–183. 2006. View Article : Google Scholar
|
|
34
|
Maasz A, Kisfali P, Jaromi L, Horvatovich
K, Szolnoki Z, Csongei V, Safrany E, Sipeky C, Hadarits F and
Melegh B: Apolipoprotein A5 gene IVS3+G476A allelic variant confers
susceptibility for development of ischemic stroke. Circ J.
72:1065–1070. 2008.
|
|
35
|
Jang Y, Paik JK, Hyun YJ, Chae JS, Kim JY,
Choi JR, Lee SH, Shin DJ, Ordovas JM and Lee JH: The apolipoprotein
A5 -1131T>C promoter polymorphism in Koreans: association with
plasma APOA5 and serum triglyceride concentrations, LDL particle
size and coronary artery disease. Clin Chim Acta. 402:83–87.
2009.
|
|
36
|
Ruiz-Narváez EA, Yang Y, Nakanishi Y,
Kirchdorfer J and Campos H: APOC3/A5 haplotypes, lipid levels, and
risk of myocardial infarction in the Central Valley of Costa Rica.
J Lipid Res. 46:2605–2613. 2005.PubMed/NCBI
|
|
37
|
Hubacek JA, Skodová Z, Adámková V, Lánská
V and Poledne R: The influence of APOAV polymorphisms (T-1131>C
and S19>W) on plasma triglyceride levels and risk of myocardial
infarction. Clin Genet. 65:126–130. 2004.
|
|
38
|
Dallongeville J, Cottel D, Montaye M,
Codron V, Amouyel P and Helbecque N: Impact of APOA5/A4/C3 genetic
polymorphisms on lipid variables and cardiovascular disease risk in
French men. Int J Cardiol. 106:152–156. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dorfmeister B, Zeng WW, Dichlberger A, et
al: Effects of six APOA5 variants, identified in patients with
severe hypertriglyceridemia, on in vitro lipoprotein lipase
activity and receptor binding. Arterioscler Thromb Vasc Biol.
28:1866–1871. 2008. View Article : Google Scholar
|
|
40
|
Johansen CT, Kathiresan S and Hegele RA:
Genetic determinants of plasma triglycerides. J Lipid Res.
52:189–206. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lim S, Park YM, Sakuma I and Koh KK: How
to control residual cardiovascular risk despite statin treatment:
Focusing on HDL-cholesterol. Int J Cardiol. 166:8–14. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hokanson JE and Austin MA: Plasma
triglyceride level is a risk factor for cardiovascular disease
independent of high-density lipoprotein cholesterol level: a
meta-analysis of population-based prospective studies. J Cardiovasc
Risk. 3:213–219. 1996. View Article : Google Scholar
|
|
43
|
Merkel M and Heeren J: Give me A5 for
lipoprotein hydrolysis! J Clin Invest. 115:2694–2696.
2005.PubMed/NCBI
|
|
44
|
Li G, Wang JY, Yan S, Zhang LJ, Xue H,
Zeng WW, Wu G and Chen BS: The relationship between the
polymorphism of apolipoprotein A5 gene 56C→G and the levels of
serum triglyceride. Zhonggua Dong Mai Ying Hua Za Zhi. 5:574–576.
2004.(In Chinese).
|