Introduction
A safe and efficient gene transfer system has always
been the aim of gene therapy. The clinical application of viral
vectors is limited due to the potential safety problems, although
these vectors provide high gene transfection efficiency and the
possibility of stable gene expression. Over the past three decades,
numerous non-viral carriers have been studied and produced to
replace viral vectors. However, the transfection efficiency was not
desirable due to extracellular and intracellular obstacles during
gene delivery (1,2). The cytoplasmic and nuclear membranes
are two of the greatest barriers (3,4). To
improve the transfection efficiency, various approaches have been
investigated for entering the cell and nuclear membrane barriers
(4–6).
As a novel non-viral gene delivery system,
ultrasound- targeted microbubble destruction (UTMD) appears to be a
promising technology. Previously, numerous studies demonstrated
improved gene transfer in the presence of ultrasound and
microbubbles (7–9). In addition, as a physical method, the
ultrasound-microbubble technology showed superiority over other
non-viral gene carriers with the ability to target gene delivery to
a specific area. This possibility provides extensive application
prospects of the technique for in vivo studies (10). Therefore, in the present study,
UTMD technology was utilized to promote the entry of plasmid DNA
(pDNA) into the cytoplasm.
The poor nuclear import rate is another critical
challenge for non-viral gene delivery methods. Previous studies
showed that directly micro-injecting pDNA into the cytoplasm only
resulted in ~3% nuclear import and low expression (11). Therefore, breaking through the
nuclear membrane barrier and promoting nuclear import are crucial
for gene transfer. In the early stage, numerous nuclear
localization signals (NLSs), such as the SV40 large T antigen, have
been used to promote gene import into the nucleus of non-dividing
cells (12); however, the gene
transfection efficiency was not as high as expected. Nuclear factor
κB (NFκB), which contains a natural NLS and shuttles between the
cytoplasm and nucleus, has been found to facilitate the nuclear
import of pDNA (13).
In the present study, a specific DNA-targeting
sequence (five optimal repeats of the NFκB binding motif) was
designed to embed into pDNA-containing therapeutic genes. The NFκB
binding motif is recognized by and binds to NFκB, which activates
the nuclear-protein guided intracellular trafficking of the pDNA
and may improve the nuclear intake and transfection efficiency. To
further increase the transfection efficiency, the NFκB binding
motif was combined with UTMD. It was hypothesized that UTMD
enhances the cellular uptake of pDNA and the NFκB binding motif
promotes the nuclear import of pDNA. Thus, the transfection
efficiency would be markedly increased by combining these two
approaches.
Materials and methods
hSDF-1α plasmid construction
The primers for cloning human SDF-1α (hSDF-1α) were
synthesized by Invitrogen Life Technologies (Shanghai, China). The
primers were as follows: Forward: 5′-ATGAACGCCAAGGTCGTGGTCG-3′;
reverse: 5′-TCACATCTTGAACCTCTTGTTT-3′. Total mRNA from human
fibroblasts was extracted using TRIzol reagent (Invitrogen Life
Technologies, New York, NY, USA) and reverse
transcription-polymerase chain reaction (RT-PCR) was used to obtain
the hSDF-1α DNA. Following this, pcDNA3.1(−) vectors (Invitrogen
Life Technologies, New York, NY, USA) were extracted using
NheI/EcoRI restriction enzymes and the RT-PCR
products were inserted into the same restriction sites using T4-DNA
ligase.
hSDF-1α-NFκB plasmid construction
The 5X NFκB fragment was as follows:
5′-CTGGGGACTTTCCAG
CTGGGGACTTTCCAGCTGGGGACTTTCCAGCTGGGG ATTTCCAGCTGGGGACTTTCCAGCT-3′ (each
underlined section represents one NFκB binding motif and five 10-bp
NFκB sites were separated by the 5-bp optimized spacer AGCTG to
ensure the best structural fit with NFκB). The fragment was
designed, as described previously (13) and added to the hSDF-1α primers. The
hSDF-1α-NFκB DNA was obtained as described above. Following
digestion using BamHI/HindIII restriction enzymes,
the hSDF-1α-NFκB DNA was cloned into the same restriction site of
the pcDNA3.1(−) vector using T4-DNA ligase (Fig. 1).
Ligation products were transformed into the
E.coli strain DH5a for amplification and then isolated and
purified using a Qiagen Endofree Plasmid kit (Qiagen, Hilden,
Germany).
Plasmid labeling
Plasmids were labeled with Cy3 using the Label IT
Tracker Intracellular Nucleic Acid Localization kits (Mirus Bio
LLC., Madison, WI, USA), according to the manufacturer’s
instructions. The reagent:plasmid weight ratio was 1:2 and the
mixture was purified by ethanol precipitation and then dissolved to
a concentration of 1 mg/ml.
Cell culture
Human umbilical vein endothelial cells (HUVECs) were
supplied by Doctor Jinyue Hu, Central Laboratory of Renmin
Hospital, Wuhan University (Wuhan, China). The HUVECs were cultured
in endothelial cell medium (ECM, ScienCell Research Laboratories,
Carlsbad, CA, USA) containing 10% fetal bovine serum and 1%
endothelial cell growth supplement. The cells were maintained in
10-cm culture dishes at 37°C in a humidified 5% CO2
atmosphere. Confluent cells were trypsinized and seeded at a
density of 2×105 cells per well (2 ml ECM) in 6-well
plates (Corning Inc., Corning, NY, USA) 24 h prior to
transfection.
Microbubble preparation
A commercially available second-generation contrast
agent, SonoVue (Bracco, Geneva, Switzerland), was used in this
study. Following the addition of 5 ml normal saline solution and
vigorous oscillation, a suspension containing ~2×108
microbubbles per ml was obtained. The microbubbles were filled with
sulfur hexafluoride gas and encapsulated by a thin and flexible
monolayer of phospholipids. The diameters of the microbubbles were
0.8–10 μm and the majority were 2–5 μm.
Ultrasound- and microbubble-mediated
plasmid transfection
Gene transfection was performed when the cells in
the 6-well plates were 60–70% confluent. The culture medium was
replaced with 4 ml OptiMEM® medium (Gibco, Grand Island,
NY, USA) suspension containing 10 μg Cy3-labeled plasmid and
various quantities of microbubbles per well prior to transfection.
Subsequent to this, ultrasound irradiation was performed on the
cell and plasmid-microbubbles suspension using the UGT2007
ultrasound irradiation machine (Ultrasonic Research Institute of
Chongqing Medical University, Chongqing, China) with various
acoustic intensities and exposure times. The transducer was wrapped
with a thin sterile latex sheath and then immersed into the
suspension for irradiation (Fig.
2).
To optimize the ultrasound- and microbubble-mediated
transfection parameters, a series of variables were tested as
follows: i) Microbubble concentration: 0, 1×105,
1×106, 1×107 and 1×108 per ml. The
acoustic intensity and exposure time were fixed at 1.0
W/cm2 and 30 sec, respectively. ii) Acoustic intensity:
0, 0.5, 1.0, 1.5 and 2.0 W/cm2. The microbubble
concentration and exposure time were fixed at 1×107
microbubbles per ml and 30 sec, respectively. iii) Exposure time:
10, 30, 45, 60 and 120 sec, with a 20% duty cycle. The microbubble
concentration and acoustic intensity were fixed at 1×107
microbubbles per ml and 1.0 W/cm2, respectively.
The optimal parameters were determined using the
criteria of high cell viability and ideal pDNA uptake.
Transfections were performed using the determined optimal
ultrasound-microbubble transfection condition.
Cell viability
HUVECs were seeded in 96-well plates
(5×103 cells and 100 μl culture medium, per well). The
transfections were performed in exactly the same manner as
described above. Following incubation at 37°C for 24 h, 10 μl Cell
Counting kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan) was
added to each well. Following incubation at 37°C for 2 h, the
absorbance was measured at 450 nm using an automatic microplate
reader (Lambda 1050; Perkin Elmer, Waltham, MA, USA). Four
duplicate experiments were performed to reduce random error. Cell
viability was calculated as:
(ODsample/ODcontrol) × 100%, where OD is the
optical density.
Cellular uptake of pDNA
To detect the cellular uptake of the plasmid, HUVECs
were incubated at 37°C for 4 h subsequent to being transfected with
Cy3-labeled pDNA. Following this, the transfected HUVECs were
washed three times with phosphate-buffered saline (PBS; Gibco, Life
Technologies, New York, NY, USA) and harvested using trypsin. The
number of fluorescent cells was detected using flow cytometry
(Beckman Coulter Inc., Brea, CA, USA) and the cellular uptake
efficiency was calculated as the percentage of Cy3-positive
cells.
Nuclear uptake of pDNA
The cells were seeded on sterile coverslips that
were placed on the bottom of 6-well plates. Transfection was
performed with Cy3-labeled phSDF-1α or phSDF-1α-NFκB plasmids using
the same method as described when the cells were 50% confluent. The
transfected cells were incubated for an additional 4 h in the
absence or presence of 20 ng/ml recombinant human IL-1β (PeproTech,
Rocky Hill, NJ, USA) and washed six times with PBS. Cells were then
fixed with 4% paraformaldehyde. DAPI (Beyotime Biotechnology Inc.,
Nantong, China) was added to clearly stain the nucleus and the
cells were washed with PBS six times prior to observation. The
subcellular localization of the Cy3-labeled plasmids was observed
using fluorescence microscopy (BX61; Olympus, Inc., Tokyo, Japan).
The fluorescence intensity (FL) of the whole cell
(FLcell) and the nucleus (FLnucleus) was
quantified using ImageJ 1.46 software (http://rsb.info.nih.gov/ij). The nuclear uptake
efficiency of the pDNA was calculated as
(FLnucleus/FLcell) × 100%.
RT-PCR
The mRNA expression of the hSDF-1α gene was
semi-quantitatively analyzed using an RT-PCR kit (Thermo
Scientific, Waltham, MA, USA). The primers were as follows:
Forward: 5′-TCAGCCTGAGCT ACAGATGC-3′ and reverse:
5′-CTTTAGCTTCGGGTCAA TGC-3′ for hSDF-1α; forward:
5′-CAAGGTCATCCATGA CAACTTTG-3′ and reverse: 5′-GTCCACCACCCTGTTGCT
GTAG-3′ for glyceraldehyde 3-phosphate dehydrogenase (GAPDH). PCR
products were electrophoresed on a 1% agarose gel and stained with
ethidium bromide. GAPDH was used to normalize the cDNA from
different samples. The relative expression of hSDF-1α
(hSDF-1α/GAPDH) was measured using a gel imaging analysis system
(Geliance 200; Perkin Elmer).
Western blot analysis and enzyme-linked
immunosorbent assay (ELISA)
Qualitative and quantitative analysis of the hSDF-1α
protein expression was performed using western blot analysis and
ELISA. Protein samples were extracted from the medium and the cell
lysis product and then separated using 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis. The separated proteins
were transferred to polyvinylidene fluoride membranes and incubated
with anti-hSDF-1α antibody (Abcam Inc., Cambridge, MA, USA) at 4°C
overnight. The membranes were blocked with 5% non-fat dry milk and
incubated with horseradish peroxidase-coupled secondary antibodies
for 1 h at room temperature. The membranes were washed and exposed
to X-ray to detect the expression bands. The quantity of SDF-1α
protein was measured using an hSDF-1α ELISA kit (R&D Systems,
Minneaopolis, MN, USA) according to the manufacturer’s
instructions. The 96-well polystyrene microplate was coated with a
rabbit polyclonal anti-SDF-1α antibody, and recombinant human
SDF-1α was used as the standard. The results are expressed as the
quantity of hSDF-1α/mg protein.
Statistical analysis
Data are expressed as the mean ± SEM. For analysis
of the difference between the two groups, the Student’s t-test was
performed. For multiple group comparisons, analysis of variance was
performed, followed by the Student-Newman-Keuls q-test. P<0.05
was considered to indicate a statistically significant
difference.
Results
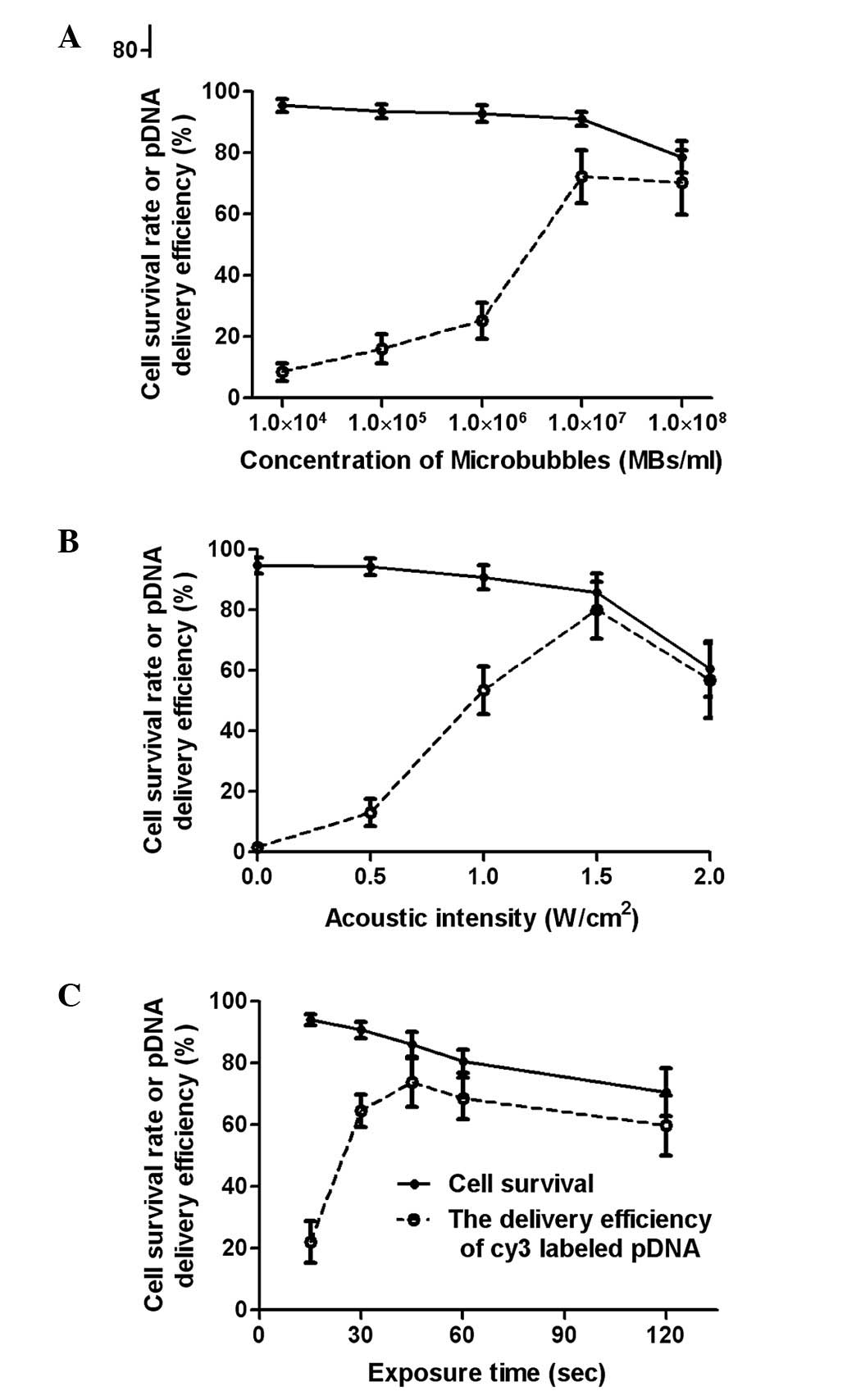
Optimization of ultrasound exposure
parameters
The ultrasound exposure parameters were optimized to
increase the efficiency of the pDNA cellular uptake while
minimizing the cytotoxicity mediated by the ultrasound and
microbubbles.
The gene delivery efficiency increased with the
microbubble concentration but decreased when the microbubble
concentration was higher than 1×107 microbubbles/ml. The
cell survival rate did not significantly differ as microbubble
concentration increased from 0 to 1×107/ml; however, a
notable decrease occurred when the concentration exceeded
1×107/ml microbubbles (Fig.
3A). The gene delivery efficiency peaked at 1.5
W/cm2, and a marked reduction of the cell survival rate
occurred at the same acoustic intensity (Fig. 3B). When the exposure time was
between 10 and 45 sec, gene delivery efficiency increased and
reached the maximum level of 74%. The gene delivery efficiency was
reduced with further increases in the ultrasound exposure time due
to a significant increase in cell death.
For the criteria of cell viability and efficient
gene delivery, the optimal exposure parameters in this study were
determined to be a microbubble concentration of
1×107/ml, acoustic intensity of 1.5 W/cm2 and
exposure time of 45 sec with a 20% duty cycle.
Ultrasound and microbubbles enhanced
cellular uptake of pDNA
As shown in Fig.
4A, when HUVECs were treated with UTMD using the optimal
exposure condition described above, the ratio of fluorescent cells
was 81%, which was significantly higher than that for the
transfection with only ultrasound or with microbubbles alone (11
and 2%, respectively). The cell viability was >85% under the
optimal exposure conditions. These results indicated that the
ultrasound exposure combined with microbubbles was an effective and
safe method for gene delivery.
 | Figure 4Cellular and nuclear import of pDNA.
(A) Efficiency of the cellular uptake of the pDNA with MB, US and
the combination of MB with US. (B) Efficiency of the nuclear import
of the pDNA with or without the NFκB binding motif in the absence
or presence of IL-1β. pDNA, plasmid DNA; MB, microbubbles, US,
ultrasound, NFκB, nuclear factor-κB; IL-1β, interleukin-1β; FL,
fluorescence intensity; NS, not significant. |
NFκB binding motif promoted the nuclear
import of pDNA
When activated by IL-1β, the nuclear uptake of pDNA
in the HUVECs transfected with phSDF-1α-NFκB was 6.5 times greater
than that in the HUVECs transfected with phSDF-1α, whereas there
was no significant difference between the two groups without IL-1β
stimulation (Fig. 4B). The results
suggested that the NFκB binding motif significantly promoted the
nuclear import of the plasmid but was unable to do so without the
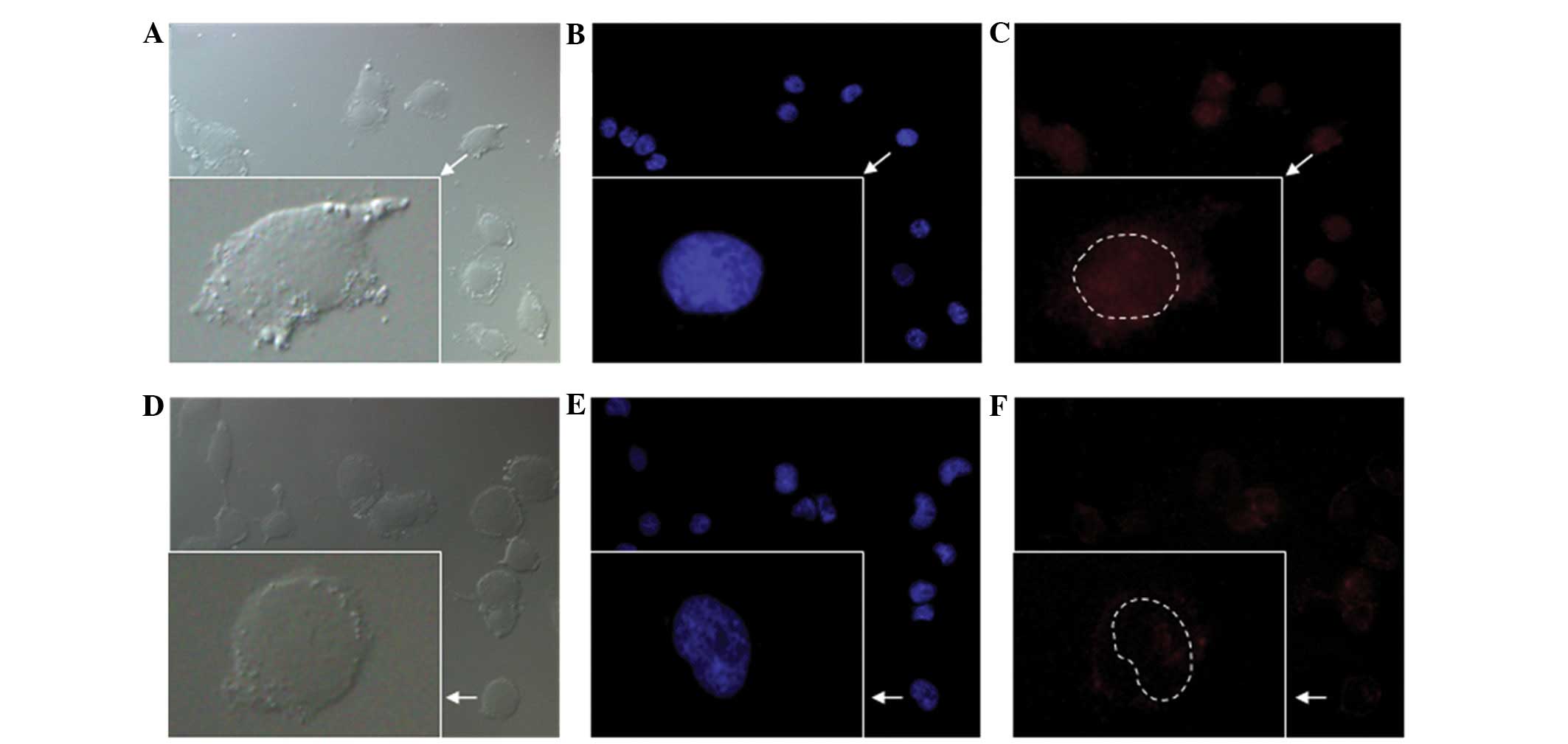
stimulation by IL-1β. Fig. 5 shows
the subcellular localization of Cy3-labeled phSDF-1α-NFκB and
phSDF-1α plasmids. The fluorescence intensity in the nucleus of the
phSDF-1α-NFκB group was markedly higher than that of the phSDF-1α
group.
Increased hSDF-1α mRNA expression
HUVECs were transfected with pcDNA3.1 (control),
phSDF-1α or phSDF-1α-NFκB and activated by IL-1β stimulation for 24
h. Subsequent to this, hSDF-1α mRNA expression was determined using
the semi-quantitative RT-PCR. Fig.
6A shows the gene expression of hSDF-1α and GAPDH. In the
pcDNA3.1 (control) group, a weak gene expression was observed as
normal vascular endothelial cells were only able to express a small
quantity of SDF-1α following activation by IL-1β. Fig. 6B shows the relative expression of
the hSDF-1α mRNAs normalized by GAPDH. The relative expression of
mRNA in the phSDF-1α-NFκB group was significantly higher than that
of the phSDF-1α group.
Increased hSDF-1α protein expression
Consistent with the mRNA expression, the SDF-1α
protein level was significantly higher in the phSDF-1α-NFκB group
than in the remaining two groups. Western blot analysis data
demonstrated that the relative expression of the hSDF-1α protein in
the phSDF-1α-NFκB group was ~3 times as high as that of the
phSDF-1α group. Moreover, the quantitative ELISA results suggested
that the expression of the hSDF-1α protein in the phSDF-1α-NFκB
group increased 4-fold compared with that in the phSDF-1α group.
(Fig. 7)
Discussion
Poor transfection efficiency is a great challenge
for non-viral gene therapy. The cytoplasmic and nuclear membrane
barriers, which seriously restrict the import of exogenous genes,
are two key reasons for this inefficiency (14). In the present study, UTMD was
combined with the NFκB binding motif to promote the passage of an
exogenous gene through the cytoplasmic and nuclear membrane
barriers. The results showed that the cytoplasmic and nuclear
uptake of the exogenous pDNA significantly increased and the
expression of the targeted gene was ~4-times greater than that of
the control.
Over the past two decades, UTMD has been recognized
as a valuable method for gene delivery. Various microbubble and
ultrasound parameters were investigated to improve the transfection
efficiency, although the mechanism has not been clearly defined
(15–16). Numerous studies have suggested that
the following mechanisms are mostly likely: microbubbles, serving
as cavitation nuclei, facilitate cavitation during ultrasound
exposure. The microjet and the rupture of microbubbles resulting
from cavitation releases large quantities of energy, creating
transient and reversible nanopores on the cell membrane, the
creation of these pores is termed as ‘sonoporation’ and has been
demonstrated by scanning electron microscopy in previous studies
(17). Thus, exogenous pDNA or
other molecules enter cells through the nanopores without resulting
in irreversible cell damage. In the present study, commercially
available SonoVue lipid microbubbles and optimized ultrasound
exposure parameters (microbubble concentration of
1×107/ml; acoustic intensity of 1.5 W/cm2 and
exposure time of 45 sec) were applied for gene delivery. The
results showed that ultrasound only marginally increased the gene
delivery efficiency (expressed as the percentage of fluorescent
cells). However, when UTMD was applied, the gene delivery
efficiency was significantly improved. The synergistic bioeffect
was significantly higher than the ultrasound bioeffect or the
microbubble bioeffect alone. When transfection was performed under
the optimal ultrasound exposure parameters, Cy3-labeled pDNA was
detected in ~81% of cells while the cell viability was >85%.
This result indicated that UTMD-mediated gene delivery was safe and
effective.
Although the entry of exogenous genes into the
cytoplasm is essential for transfection, its efficiency is
unsatisfactory. The high cytoplasmic import of a gene does not
result in high expression. The exogenous pDNA in the cytoplasm is
required to enter the nucleus for transcription, rendering the
nuclear membrane an insurmountable obstacle for pDNA (18).
The nuclear membrane is a double membrane on the
surface of the nucleus and numerous nuclear pore complexes (NPCs)
are distributed on it. NPCs are key in nucleocytoplasmic exchange
in eukaryotes and support two modes of transport. Small particles
(particles <9 nm, proteins <40 kDa and DNA<300 bp) pass
through the NPCs by passive diffusion and the transportation of
large molecules is mediated by nuclear localization signal (NLS)
peptides (19). NLS interact with
importin-α and -β and then form a complex, which is docked to NPCs
and shuttled to the inner membrane of the nucleus (20–21).
The size of pDNA (usually ~5,000 bp) renders it highly unlikely to
transverse the NPC channels as a free molecule.
To aid pDNA circumvent the nuclear entry bottleneck,
considerable efforts have been made to mimic the process of
NLS-mediated nuclear import of large molecules. In the early stage,
NLS peptides were directly attached to nucleic acid molecules to
promote the nuclear uptake of pDNA (22–23).
However, the result was unsatisfactory due to the strong
interaction between the positively charged peptide and the
negatively charged pDNA, which modified the physicochemical
properties of the NLS peptide and the pDNA. Certain nucleotide
sequences that interacted with endogenous NLS-containing proteins
were inserted into pDNA and this addition proved to be beneficial
for nuclear import (24). However,
the benefit from the addition of this device in terms of gene
transfection efficiency was also not as high as expected.
Previously, NFκB was identified to exhibit a natural NLS, allowing
it to pass through the NPCs and facilitate nuclear transport.
In the present study, a specific DNA targeting
sequence (five optimal repeats of NFκB binding motif) was designed
and inserted into pDNA to promote nuclear import. Plasmids
containing the NFκB binding motif specifically attach to NFκB and
form a plasmid-NFκB complex. NFκB usually exists as an inactive
heterodimer with the IκB protein (the inhibitor protein of NFκB) in
the cytoplasm. When cells are stimulated by a specific signal, such
as IL-1β, the IκB protein rapidly degrades, activating NFκB. The
NLS of NFκB is then recognized by a heterodimeric protein complex
importin-α/-β. Importin-α interacts directly with the NLS and
importin-β docks the plasmid-NFκB complex to the perinuclear space
allowing it to pass through the NPCs (25). As shown in Fig. 5, the majority of the Cy3-labeled
phSDF-1α plasmids were localized in the cytoplasmic space rather
than in the nucleus, while the phSDF-1α-NFκB plasmids were
predominanltly localized in the nucleus. The quantity of plasmids
containing the NFκB binding motif in the nucleus was 6.5-times that
of the NFκB-free plasmids following stimulation by IL-1β. This
indicated that the NFκB binding motif significantly promoted the
entry of the plasmid into the nucleus from the cytoplasm. However,
without IL-1β stimulation, no significant difference was observed
between the NFκB-attached plasmids and the NFκB-free plasmids in
the nucleus, suggesting that the NFκB binding motif cannot function
independently of IL-1β.
In addition, the protein expression of the SDF-1α
gene was detected. The SDF-1α protein expressed by HUVECs
transfected with phSDF-1α-NFκB was ~4-times greater than that when
transfected with phSDF-1α. This result was consistent with the
increase of nuclear import, although the expression efficiency was
not as high as the efficiency of nuclear import (4- versus
6.5-fold).
In the present study, SDF-1α was selected as the
target gene and HUVECs as the transfection object as numerous
studies have demonstrated that transfecting the SDF-1α plasmid into
vascular endothelial cells results in the release of SDF-1α. This
release facilitated the targeting of circulating CXCR4-positive
cells and other stem cells to the site of injury, thus initiating
organ regeneration and repair (26–27).
Therefore, this study demonstrates a safe and efficient method for
promoting SDF-1α gene transfection, which may be beneficial for
numerous types of injuries and diseases.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 302-163534). The authors
would like to thank Doctor Jinyue Hu (Central Laboratory of Renmin
Hospital, Wuhan University, Wuhan, China) for providing the
HUVECs.
References
|
1
|
Elsabahy M, Nazarali A and Foldvari M:
Non-viral nucleic acid delivery: key challenges and future
directions. Curr Drug Deliv. 8:235–244. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jafari M, Soltani M, Naahidi S,
Karunaratne DN and Chen P: Nonviral approach for targeted nucleic
acid delivery. Curr Med Chem. 19:197–208. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mudhakir D and Harashima H: Learning from
the viral journey: how to enter cells and how to overcome
intracellular barriers to reach the nucleus. AAPS J. 11:65–77.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lentacker I, Vandenbroucke RE, Lucas B,
Demeester J, De Smedt SC and Sanders NN: New strategies for nucleic
acid delivery to conquer cellular and nuclear membranes. J Control
Release. 132:279–288. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pichon C, Billiet L and Midoux P: Chemical
vectors for gene delivery: uptake and intracellular trafficking.
Curr Opin Biotechnol. 21:640–645. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kawazu T, Kanzaki H, Uno A, Azuma H and
Nagasaki T: HVJ-E/importin-β hybrid vector for overcoming
cytoplasmic and nuclear membranes as double barrier for non-viral
gene delivery. Biomed Pharmacother. 66:519–524. 2012.PubMed/NCBI
|
|
7
|
Burke CW, Suk JS, Kim AJ, Hsiang YH,
Klibanov AL, Hanes J and Price RJ: Markedly enhanced skeletal
muscle transfection achieved by the ultrasound-targeted delivery of
non-viral gene nanocarriers with microbubbles. J Control Release.
162:414–421. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cool SK, Geers B, Lentacker I, De Smedt SC
and Sanders NN: Enhancing nucleic acid delivery with ultrasound and
microbubbles. Methods Mol Biol. 948:195–204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Geis NA, Katus HA and Bekeredjian R:
Microbubbles as a vehicle for gene and drug delivery: current
clinical implications and future perspectives. Curr Pharm Des.
18:2166–2183. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou Y, Yang K, Cui J, Ye JY and Deng CX:
Controlled permeation of cell membrane by single bubble acoustic
cavitation. J Control Release. 157:103–111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miller AM, Munkonge FM, Alton EW and Dean
DA: Identification of protein cofactors necessary for
sequence-specific plasmid DNA nuclear import. Mol Ther.
17:1897–1903. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Prasad TK and Rao NM: The role of plasmid
constructs containing the SV40 DNA nuclear-targeting sequence in
cationic lipid-mediated DNA delivery. Cell Mol Biol Lett.
10:203–215. 2005.PubMed/NCBI
|
|
13
|
Gonçalves C, Ardourel MY, Decoville M,
Breuzard G, Midoux P, Hartmann B and Pichon C: An optimized
extended DNA kappa B site that enhances plasmid DNA nuclear import
and gene expression. J Gene Med. 11:401–411. 2009.PubMed/NCBI
|
|
14
|
Pérez-Martínez FC, Guerra J, Posadas I and
Ceña V: Barriers to non-viral vector-mediated gene delivery in the
nervous system. Pharm Res. 28:1843–1858. 2011.PubMed/NCBI
|
|
15
|
Xie A, Belcik T, Qi Y, et al:
Ultrasound-mediated vascular gene transfection by cavitation of
endothelial-targeted cationic microbubbles. JACC Cardiovasc
Imaging. 5:1253–1262. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cavalli R, Bisazza A, Trotta M, Argenziano
M, Civra A, Donalisio M and Lembo D: New chitosan nanobubbles for
ultrasound-mediated gene delivery: preparation and in vitro
characterization. Int J Nanomedicine. 7:3309–3318. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mehier-Humbert S, Bettinger T, Yan F and
Guy RH: Plasma membrane poration induced by ultrasound exposure:
implication for drug delivery. J Control Release. 104:213–222.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Glover DJ, Leyton DL, Moseley GW and Jans
DA: The efficiency of nuclear plasmid DNA delivery is a critical
determinant of transgene expression at the single cell level. Gene
Med. 12:77–85. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Allen TD, Cronshaw JM, Bagley S, Kiseleva
E and Goldberg MW: The nuclear pore complex: mediator of
translocation between nucleus and cytoplasm. J Cell Sci.
113:1651–1659. 2000.PubMed/NCBI
|
|
20
|
Lange A, Mills RE, Lange CJ, Stewart M,
Devine SE and Corbett AH: Classical nuclear localization signals:
definition, function, and interaction with importin alpha. J Biol
Chem. 282:5101–5105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
van Gaal EV, Oosting RS, van Eijk R, et
al: DNA nuclear targeting sequences for non-viral gene delivery.
Pharm Res. 28:1707–1722. 2011.PubMed/NCBI
|
|
22
|
Cartier R and Reszka R: Utilization of
synthetic peptides containing nuclear localization signals for
nonviral gene transfer systems. Gene Ther. 9:157–167. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Martin ME and Rice KG: Peptide-guided gene
delivery. AAPS J. 9:E18–E29. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Munkonge FM, Amin V, Hyde SC, et al:
Identification and functional characterization of cytoplasmic
determinants of plasmid DNA nuclear import. J Biol Chem.
284:26978–26987. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jeong JH, Kim SH, Christensen LV, Feijen J
and Kim SW: Reducible poly (amido ethylenimine)-based gene delivery
system for improved nucleus trafficking of plasmid DNA. Bioconjug
Chem. 21:296–301. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ghadge SK, Mühlstedt S, Ozcelik C and
Bader M: SDF-1α as a therapeutic stem cell homing factor in
myocardial infarction. Pharmacol Ther. 129:97–108. 2011.
|
|
27
|
Wen J, Zhang JQ, Huang W and Wang Y:
SDF-1α and CXCR4 as therapeutic targets in cardiovascular disease.
Am J Cardiovasc Dis. 2:20–28. 2012.
|