Introduction
Progestagen-associated endometrial protein (PAEP) is
a secreted glycoprotein, which was first isolated from human
placenta, amniotic fluid, decidua of pregnancies and seminal plasma
(1). It has been demonstrated to
be important in a number of physiological processes, such as
embryonic implantation, immunotolerance, contraception and gland
differentiation. In recent years, several studies have demonstrated
that PAEP is abnormally expressed in various types of tumors, such
as endometrial carcinoma, ovarian cancer, breast cancer (2–5),
lung cancer (6) and melanoma
(7). Transfection of PAEP cDNA
into the MCF-7 breast cancer cell line results in marked changes in
cell growth behavior, with the suppression of proliferation and
formation of acinar structures (8), suggesting that PAEP inhibits tumor
cell growth and promotes cell differentiation as a tumor
suppressive factor. Song et al(9) demonstrated that PAEP is also involved
in neovascularization during tumor growth (9). However, our previous study presumed
that PAEP is a tumor promoter in melanoma (10), suggesting contradictory results as
compared to other studies.
Melanogenesis is a result of the malignant
transformation of neural crest-derived melanocytes (11), and melanoma is one of the most
aggressive forms of human cancer. Once metastasized, it is
difficult to treat and is associated with high mortality rates.
PAEP has been shown to be highly expressed in melanoma tissues and
the majority of the melanoma cell lines tested in the literature
thus far. Therefore, the present study aimed to establish melanoma
cell lines with low PAEP gene expression to investigate their
cytological and genetic functions in tumorigenesis and tumor
development.
In the present study, RNA interference (RNAi)
technology was utilized to silence PAEP gene expression, using
transfection with PAEP-specific small interfering RNA (siRNA) or
infection with lentiviral vector-mediated small hairpin RNA (shRNA)
to lead to the transient or stable knockdown of PAEP gene
expression in melanoma cells. The cell lines obtained from this
study may be utilized to clarify the function of the PAEP gene.
Materials and methods
Cell lines and culture
Four cell lines (MCC69B, 624-MEL, 624.38-Mel and
FEMX-I lines) originally procured from melanoma patients were used.
MCC69B cells were isolated from a distant metastasis, the 624-Mel
and 624.38-Mel metastatic melanoma cell lines were obtained from
the National Cancer Institute (NIH). FEMX-I, provided by Oystein
Fodstand from Norwegian Radium Hospital (Oslo, Norway), originated
from a lymph node metastasis in a patient, uniquely and selectively
produced extrapulmonary metastases following intravenous (i.v.)
injection of cells prepared from xenografts into adult, nude mice
(12). All the melanoma cells were
maintained in RPMI-1640 culture media (Hyclone, Thermo Scientific,
Beijing, China) supplemented with 10% fetal bovine serum (Hyclone,
Thermo Scientific).
Design of siRNA
SiGLO green is a fluorescent marker that is used to
monitor infection efficiency. In the present study, four duplex
siRNAs targeting PAEP mRNA were designed. The target sequences were
as follows: siPAEP9, TCA ACT ATA CGG TGG CGA A; siPAEP10, GGA AGA
GCC GUG CCG UUU UU; siPAEP11, CCA CGC UGC UCG AUA CUG AUU; and
siPAEP12, ACA GCU GUG UUG AGA AGA AUU. The four duplex PAEP siRNAs,
one siControl non-targeting pool and siGLO green transfection
indicator were synthesized by Thermo Scientific Dharmacon
(Lafayette, CO, USA).
siRNA knockdown
Transfection conditions were optimized by
transfecting melanoma cells with siGLO of varying concentrations
followed by analysis with fluorescence microscopy. For knockdown
experiments, 624.38-Mel and MCC69B melanoma cells
(1–2×105) were cultured in a 6-well plate in complete
medium in a 5% CO2 humidified atmosphere at 37°C
overnight. Gene-specific siRNAs or non-specific siControl were then
transfected into tumor cells using DharmaFECT1 (Dharmacon,
Lafayette, CO, USA) according to the manufacturer’s instructions.
After 8 h, the supernatant was removed and the cells were washed
with phosphate-buffered saline (PBS) and maintained in complete
medium for 24 h. The cells were incubated in serum-free RPMI-1640
medium. To assess the knockdown efficiency, transfected cells were
collected for subsequent reverse transcription-polymerase chain
reaction (RT-PCR) and western blot analysis, 48 and 72 h following
transfection.
Construction of lentiviral vectors
To obtain stable PAEP shRNA melanoma cells, the
lentiviral shRNA vector system was selected and three PAEP shRNAs
(gene target sequences: shPAEP1, 5′-AAG ATC AAC TAT ACG GTG G-3′;
shPAEP2, 5′-AAG AGC CGT GCC GTT TCT A-3′; shPAEP3, 5′-ATA AAC CCT
TGG AGC ATG A-3′) were screened. The lentiviral particles
constructed by Thermo Scientific Dharmacon contained one TurboGFP
(Evrogen, Moscow, Russia) reporter gene, one puromycin resistance
gene and one of three PAEP shRNA sequences.
Optimization of polybrene concentration
for enhanced transduction
Polybrene (hexadimethrine bromide) is a cationic
polymer that increases the viral infection of mammalian cells by
neutralizing the charge repulsion between the virus and cell
surface. As it is cytotoxic to cells, the concentration of
polybrene was optimized prior to lentiviral infection. Melanoma
cells (624-Mel, 624.38-Mel and FEMX-I) were seeded into a 96-well
plate at a density of 1×104 cells/well and cultured in a
incubator in a 5% CO2 atmosphere at 37°C. Following
overnight culture, the culture medium was replaced with fresh
medium containing polybrene at different concentrations (0–10
μg/ml), and the cells were returned to the incubator for 24 h. Cell
proliferation was determined using a cell proliferation kit
(Pregene, Beijing, China) and the optical density (OD) of each well
was determined at 490 nm with a microplate reader (SpectraMax M5;
Molecular Devices, Sunnyvale, CA, USA). Assays were performed in
triplicate at each concentration.
Determination of puromycin
concentrations
The puromycin resistance gene in the lentiviral
vector allows for the generation of stable cell lines by drug
selection. Once the optimal concentration had been identified,
transduced cells were selected and propagated. Melanoma cells
(624-Mel, 624.38-Mel and FEMX-I) were seeded in a 96-well plate at
a density of 1×104 cells/well and cultured in 5%
CO2 at 37°C overnight. Puromycin (American
Bioanalytical, Natick, MA, USA) was then added to each well at
different concentrations, and the plate was returned to the
incubator. Every 2 days, the cells were provided with fresh medium
supplemented with puromycin of different concentrations. The OD of
each well at different time-points was determined at 490 nm as
previously described.
Multiplicity of infection (MOI)
optimization of lentiviral particles
MOI, the ratio of lentiviral particles to cells, was
optimized. Melanoma cells (624.38-Mel and FEMX-I) were seeded in a
96-well plate at a density of 1×104 cells/well and
incubated in a 5% CO2 atmosphere at 37°C overnight. The
cells were then infected with non-targeting shControl lentiviral
particles at MOI ratios of 10, 50, 100 and 150 plaque-forming units
(pfu)/cell in the presence of polybrene. After 48 h, the cells were
observed using a Nikon Eclipse TE-2000U microscope (Nikon
Instruments Inc., Melville, NY, USA) and collected for flow
cytometric analysis.
Transient and stable transfection of
melanoma cells with shRNA lentivirus
To assess the abilities of three PAEP shRNA
lentiviral particles to silence PAEP gene expression, melanoma
cells (624-Mel, 624.38-Mel and FEMX-I) were seeded in a 96-well
plate at a density of 1×104 cells/well and were
incubated in a 5% CO2 atmosphere at 37°C overnight. The
cells were then infected with each of the three PAEP shRNA
lentiviral particles at a MOI of 100 pfu/cell in the presence of
polybrene. Shglyceraldehyde 3-phosphate dehydrogenase (GAPDH) and
non-targeting shControl lentiviral particles were used as positive
and negative controls, respectively. After 24 h, transduced cells
were collected for qPCR analysis.
To obtain stable PAEP knockdown cell lines, melanoma
cells were infected with a low MOI of lentiviral particles
expressing PAEP shRNA. Melanoma cells (624-Mel, 624.38-Mel) were
seeded in a 96-well plate at a density of 1×104
cells/well and were incubated in 5% CO2 at 37°C
overnight. The cells were then infected with PAEP shRNA3 lentiviral
particles at a MOI of 10 pfu/cell in the presence of polybrene.
After 24 h, the supernatant was replaced with complete medium
supplemented with 1.5 μg/ml puromycin. Following puromycin
screening and propagation, transduced cells were collected for qPCR
and western blot analysis.
RT-PCR and qPCR
Total RNA from transduced cells was extracted using
TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA).
RNA concentration and purity were determined using a UV-2550
spectrophotometer (Shimadzu, Kyodo, Japan). Total RNA (5 μg) was
reverse-transcribed into cDNA using a reverse transcription kit
(Takara Biotechnology, Dalian, China). The resultant cDNA was
amplified using the primers: Forward: 5′-AGG TTG GCA GGG ACC TGG
CAC TC-3′ and reverse: 5′-ACG GCA CGG CTC TTC CAT CTG TT-3′ for
PAEP; and forward: 5′-ACA CTG TGC CCA TCT ACG AGG-3′ and reverse:
5′-AGG GGC CGG ACT CGT CAT ACT-3′ for β-actin. PCR cycling
conditions for PAEP were as follows: Initial denaturation at 95°C
for 1 min followed by 26 cycles of 95°C for 10 sec, 60°C for 15
sec, 72°C for 45 sec and 72°C for 7 min. PCR cycling conditions for
β-actin were as follows: Initial denaturation at 94°C for 2 min
followed by 26 cycles of 94°C for 30 sec, 60°C for 30 sec, 72°C for
30 sec and 72°C for 7 min. The amplified products were analyzed
using 1.5% agarose gel electrophoresis.
qPCR using gene-specific primers [unlabeled;
PAEP-Hs00171462_m1 and GAPDH-Hs99999905_m1] and TaqMan MGB
probes (6-FAM dye-labeled) purchased from Applied Biosystems
(Foster City, CA, USA) was conducted in triplicate on a BioRad iQ5
multicolor real-time PCR detection system (Bio-Rad, Hercules, CA,
USA). Transcript levels were normalized to GAPDH.
Western blot analysis
A total of 30 μg of each protein sample from the
cell lysate or serum-free culture medium was loaded onto a sodium
dodecyl sulfate-polyacrylamide gel and electrophoresed.
Immunostaining was accomplished with the incubation of a PAEP
antibody (2 μg/ml; Invitrogen, South San Francisco, CA, USA)
followed by incubation with a secondary antibody conjugated to
horseradish peroxidase (Jackson Immuno Research Labs, West Grove,
PA, USA). Visualization was performed with enhanced
chemiluminescence detection reagents (Thermo Fisher Scientific,
Rockford, IL, USA) using a Fujifilm Luminescent Image Analyzer
LAS-3000 (Fujifilm, Valhalla, NY, USA). The blot was analyzed using
multi-gauge v3.1 software (Fujifilm).
Mass spectrometry
To observe changes in the secreted protein profiles
of melanoma samples following PAEP-silencing, mass spectral
analysis was conducted on a Thermo Fisher Orbitrap mass
spectrometer (Thermo Fisher, Waltham, MA, USA) with an Agilent 1200
Series Nanoflow LC (Agilent Technologies, Santa Clara, CA, USA) for
sample introduction, as previously described (13–15).
Prior to analysis, secreted proteins isolated from PAEP-knockdown
and non-targeting knockdown 624.38-Mel cells were digested with
trypsin in the presence of a reducing agent. Analysis data were
then obtained at 60,000 resolution in the Orbitrap, while MS/MS
spectra were obtained in parallel within the linear ion trap (LTQ).
Analyses were performed in 3 h LC/MS runs and the data converted to
search files and searched against the Mascot search engine (Matrix
Science, available at www.matrixscience.com). The data were searched as a
combined sample set (3 injection set) to produce the data shown.
Mascot search data (Matrix Science, Boston, MA, USA) were
automatically aligned between sets and the protein profiles were
compared.
Statistical analysis
Statistical analysis was performed with the SAS
statistical software (SAS Institute, Inc., Cary, NC, USA).
Numerical data were expressed as the mean ± SD. Statistical
differences between groups were assessed using analysis of variance
and Dunnett’s t-tests for each group. P<0.05 was considered to
indicate a statistically significant difference.
Results
siRNA effectively silences PAEP gene
expression
siRNA, as an inducer or RNAi, is a useful tool
utilized to assess gene function. In the present study this
technology was applied to knock down PAEP gene expression.
Transfection conditions were optimized by transfecting melanoma
cells with siGLO of varying concentrations followed by fluorescence
microscopy and flow cytometric analysis. Transfection efficiencies
in 624.38-Mel and MCC69B cells were 88.5 and 78%, respectively, at
siGLO green and DharmaFECT1 concentrations of 100 nmol/l and 2
μl/ml, respectively (Fig. 1A).
Four duplex siRNA sequences were analyzed to identify the
sequence(s) that inhibited PAEP gene expression most efficiently in
the melanoma cell lines. qPCR results showed that siRNAs 10, 11 and
12 were more effective than siRNA 9 in MCC69B and 624.38-Mel cell
lines. Moreover, the mixture of PAEP-specific siRNA (siPAEP)-9–12
resulted in the greatest overall gene silencing of PAEP. Therefore,
siRNAs 10, 11 and 12 were pooled to silence PAEP expression in
subsequent experiments.
 | Figure 1Optimization of siRNA and shRNA
transfection conditions. (A) SiGLO green is used as an indicator to
optimize transfection. Flow cytometric analysis shows that when the
concentration of siGLO green was 100 nmol/l, the transfection
efficiencies of 624.38-Mel and MCC69B cells were 88.5 and 78%,
respectively. (B) The expression of GFP in 624.38-Mel and FEMX-I
cells shown by the TurboGFP reporter following infection of the two
cell lines at an MOI range of 0–150 pfu/cell. Flow cytometric
analysis showed that the infection efficiencies for these two cell
lines were ~50% at a MOI of 100. Infection efficiency increased
when the MOI was increased. (C) Under a fluorescent microscope,
624.38-Mel cells were shown to be efficiently infected by all five
vectors at a MOI of 100 (original magnification, ×20). (D)
Knockdown efficiency was detected by qPCR. Non-targeting shRNA
served as a standard, and GAPDH was chosen as a positive control.
Of the three lentiviral PAEP shRNAs tested, shPAEP3 was the most
effective, silencing PAEP expression by 90%. GFP, green fluorescent
protein; MOI, multiplicity of infection; PAEP,
progestagen-associated endometrial protein; shRNA, small hairpin
RNA; siRNA, small interfering RNA; GAPDH, glyceraldehyde
3-phosphate dehydrogenase. |
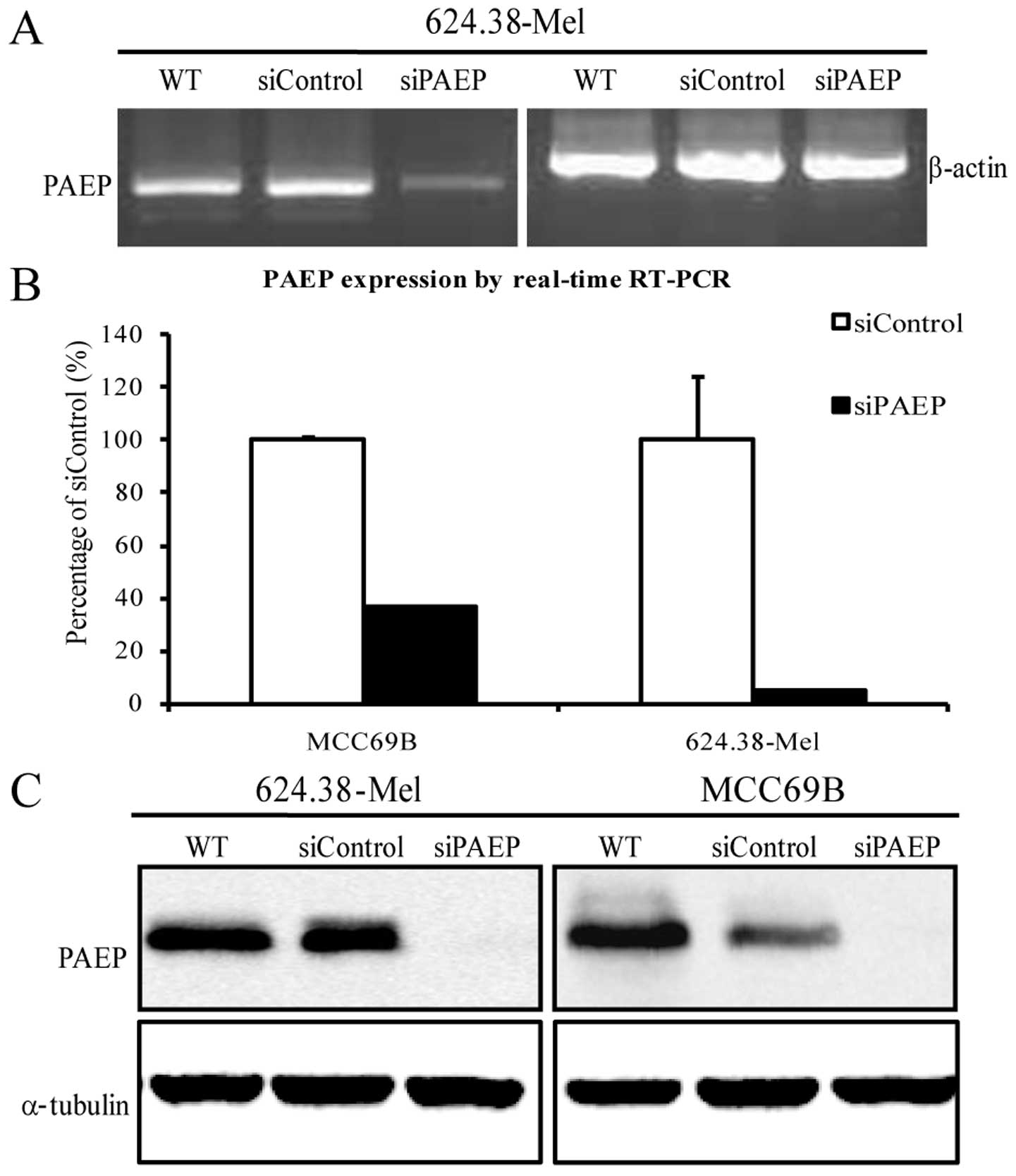
By utilizing the siPAEP10–12 pool to silence PAEP
expression in MCC69B and 624.38-Mel cells, RT-PCR and qPCR results
showed that PAEP mRNA was significantly downregulated (Fig. 2A and B; P<0.05, Dunnet’s
t-test). Moreover, the level of PAEP protein in the two cell lines
decreased by 90% following siRNA transfection (Fig. 2C).
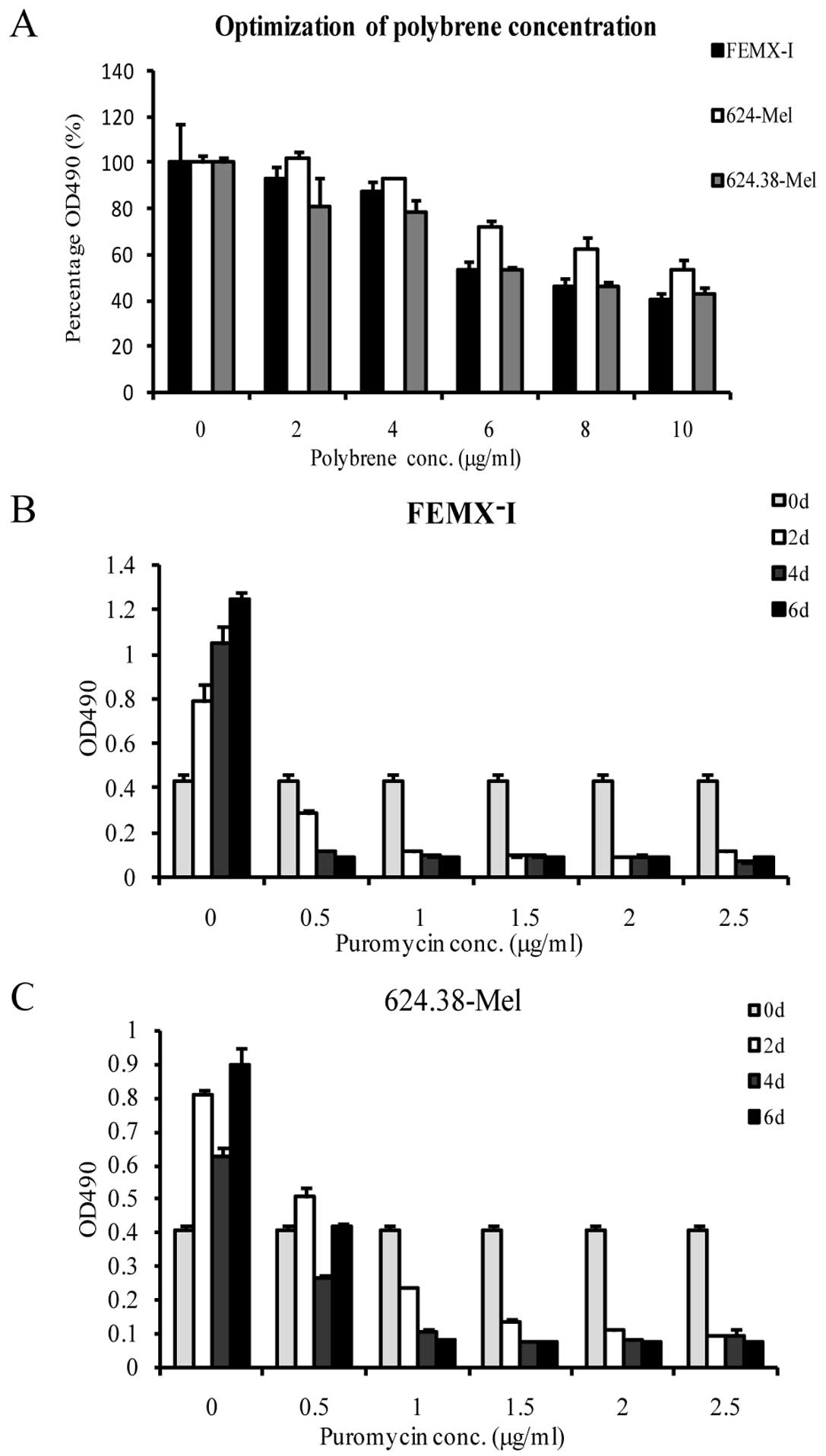
Lentiviral vector mediated shRNA
temporarily silences PAEP gene expression
This study screened for and optimized the conditions
of lentiviral vector-mediated shRNA transfection in the melanoma
cell lines by testing various concentrations of polybrene and
puromycin, the MOI of PAEP shRNA lentiviral particles and the
ability of three shRNA molecules to effectively silence PAEP gene
expression. MTS assay results demonstrated the optimal polybrene
concentration to be 2 μg/ml (Fig.
3A). Similarly, a puromycin concentration of 1.5 μg/ml was
selected as suitable to screen target cells in subsequent
experiments (Fig. 3B and C). The
melanoma cell lines were efficiently infected by lentiviral
particles at a MOI of 100 pfu/cell (Fig. 1B).
As only a minority of constructs elicit efficient
gene knockdown, studies typically prepare several candidate
constructs. Thus, the present study screened three PAEP shRNA
molecules to identify the most effective one. GAPDH and
non-targeting shRNA were used as positive and negative controls,
respectively. Utilizing siGLO as a visual indicator of successful
transfection, fluorescent microscopy demonstrated that 624.38-Mel
cells were efficiently infected by all five tested vectors at a MOI
of 100 pfu/cell (Fig. 1C).
Identical results were observed in 624-Mel and FEMX-I cells. Of the
three lentiviral PAEP shRNAs tested, shPAEP3 exhibited the greatest
ability to silence PAEP in each of the cell lines (90% expression
decrease) (Fig. 1D). Therefore,
shPAEP3 was selected to establish stable PAEP shRNA cell lines.
Notably, the target site of shPAEP3 is near the poly A tail on the
PAEP cDNA sequence.
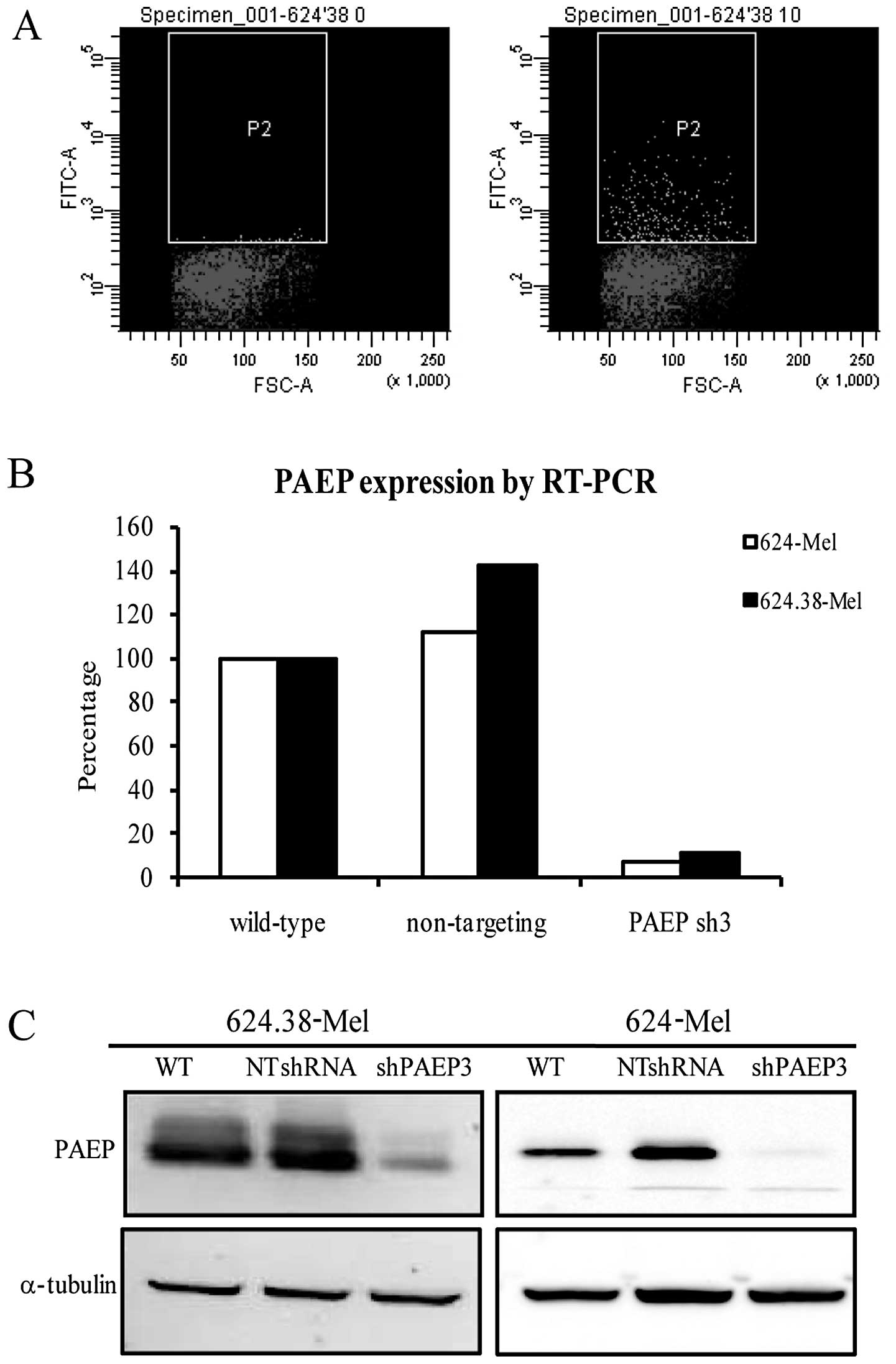
Stable PAEP knockdown cell lines were
successfully established
To obtain stable cell lines with a marked decrease
of PAEP gene expression, 624.38-Mel and 624-Mel cells were infected
with lentiviral particles at a low MOI (10 pfu/cell) and achieved
~3% infection efficiency (Fig.
4A). Following regular culture and puromycin screening, the
cells and serum-free supernatant were collected for qPCR and
western blot analysis. The results indicated that the expression of
the PAEP gene in the shPAEP3 group was knocked down by 90% at the
mRNA level and >80% at the protein level (Fig. 4B and C). Following puromycin
screening and expansion in culture, two shPAEP3 and four shControl
clones were derived from the 624.38-Mel and 624-Mel cells.
PAEP knockdown results in alteration of
certain secreted proteins
PAEP knockdown by shRNA resulted in an increase of
certain secreted proteins and a decrease in others in the culture
medium (Tables I and II). Elevated proteins included GDP
dissociation inhibitor 2, transgelin-2 and thrombospondin-1.
Decreased proteins included secreted phosphoprotein-1 (SPP-1), heat
shock 90 kDa protein and laminin-α1.
 | Table IProtein expression upregulated by PAEP
shRNA in 624.38-Mel cells. |
Table I
Protein expression upregulated by PAEP
shRNA in 624.38-Mel cells.
| Abbreviation | Protein | Fold changea | Involvement in
cancer |
|---|
| GDI-2 | GDP dissociation
inhibitor-2 | 16.84 | Inhibits tumor cell
invasion and metastasis in stomach cancer |
| Tagln-2 | Transgelin-2 | ndb | Tumor supression
factor in ovarian cancer |
| MMP-2 | Matrix
metalloproteinase-2 | 6.58 | Unknown |
| TSP-1 | Thrombospondin-1 | 5.05 | Induces endothelial
cell apoptosis, thus inhibits tumor angiogenesis |
 | Table IIProtein expression downregulated by
PAEP shRNA in 624.38-Mel cells. |
Table II
Protein expression downregulated by
PAEP shRNA in 624.38-Mel cells.
| Abbreviation | Protein | Fold changea | Involvement in
cancer |
|---|
| SPP-1 | Secreted
phosphoprotein-1 | ndb | Promotes tumor cell
migration and inhibits cell apoptosis |
| LN-α1 | Laminin-α1 | ndb | Activates tumor
migration |
| HSP-90 | Heat shock 90 kDa
protein | 2.38 | Inhibits cell
apoptosis |
| Vgf | VGF nerve growth
factor | 2.04 | Unknown |
Discussion
Melanoma is the most lethal form of skin cancer.
When diagnosed early, melanoma may be cured by surgical resection.
However, metastatic malignant melanoma is largely refractory to
existing therapies and has a very poor prognosis, with a median
survival rate of six months and a five-year survival rate of <5%
(16).
Thus a large number of studies have been conducted
with the aim of identifying a correlation between gene alteration
and tumor development. Our previous study identified a list of
specific genes that were overexpressed and underexpressed
throughout the various time-points of melanoma tumor progression
(17). PAEP is one of the genes
that was demonstrated to exhibit a higher level of expression in
freshly procured melanoma tissues and daughter cell lines (7). The gene has also been demonstrated to
be differentially involved in certain tumor (18,19).
Several studies have suggested that PAEP inhibits tumor cell growth
and promotes cell differentiation as a tumor suppressive factor;
however, other authors have demonstrated that PAEP increases the
invasion of endometrial adenocarcinoma cells (20) and melanoma (10). Therefore, the function of PAEP gene
expression in tumorigenesis and tumor progression remains unclear.
The silencing of intact wild-type gene expression, a valuable
method of functional discovery, is required to be utilized to
characterize PAEP in cancer systems.
Since the identification of RNAi in
Caenorhabditis elegans in 1998, this mechanism has been
demonstrated to be conserved in a wide variety of species,
including insects, plants and mammals. siRNA, an important inducer
of RNAi, is thus a useful tool to assess gene function. Formation
of siRNA occurs when dicer binds to dsRNA and digests it into
duplexes of 21–23 nucleotides. These, in turn, are incorporated
into the RNA-induced silencing complex, which has been suggested to
eliminate one of the strands and thus initiate a cyclical process
as the siRNA associates with novel target molecules (21,22).
In the present study, this technique was used to silence PAEP
expression. Four duplex siRNAs were selected using the PAEP mRNA
sequence and significantly suppressed PAEP expression by as much as
80% at the post-transcriptional and translational levels.
As siRNA only binds to its respective mRNA
counterpart in the cytoplasm, its inhibitory effect is transient.
Alternatively, there is another method by which to prolong
inhibition. In mammals, shRNA may be expressed by using several
expression vectors. Studies have demonstrated that adenovirus and
lentivirus vectors are useful tools for delivering a target gene
into the nucleus. However, adenoviral-mediated gene expression is
not maintained for long, and adenovirus vectors may induce an
immune response in the host (23–25).
By contrast, the lentiviral vector has several useful
characteristics for RNAi experiments, including broad host tropism
and stable gene transduction to dividing and non-dividing cells,
permitting stable depletion of target genes (26). Accordingly, a lentiviral vector
system was used to deliver and express PAEP shRNA in the melanoma
cells in order to establish stable cell lines. Following
optimization of the transfection methods, a stable PAEP shRNA cell
line with low MOI was successfully established. qPCR confirmed that
PAEP gene expression in shPAEP stable transfectants was reduced by
>80%, and western blot analysis validated these results by
showing a reduction in PAEP protein of >75%.
Moreover, it was demonstrated that knockdown by
PAEP-specific shRNA resulted in the alteration of the secreted
protein profiles in the melanoma cell lines, as determined by
protein changes in the culture medium. TSP-1, one of the proteins
highly secreted upon PAEP-silencing, is known to induce endothelial
cell apoptosis, thus inhibiting tumor angiogenesis, and is
considered to be a tumor suppressive factor (27–29).
It is therefore possible that PAEP promotes tumor cell growth at
least in part by downregulating TSP-1 expression. The presence of
SPP-1, a putative tumor oncogene (30–32),
in the medium was lost following the knockdown of PAEP. Thus,
secretion of SPP-1 is directly related to the presence of PAEP. Our
previous study demonstrated that silencing the PAEP gene expression
decreased the migration and invasion of melanoma cells and
inhibited tumor growth in a human xenograft model (10). These results suggested that PAEP
may act as a tumor-promoting factor influencing the biological
behavior of melonom and knockdown of PAEP gene was essential for
this observation. PAEP is regarded as an immunosuppressive factor
in embryonic implantation, and may hamper immunological responses
in the tumor microenvironment. In our subsequent study, the
PAEP-silenced cell models established in the present study may be
used to further investigate this hypothesis. Considering the
significance of silencing the PAEP gene, this study thoroughly
demonstrated and characterized the procedure of gene knockdown and
may be used for other gene function studies in cancer systems.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81071709) and the
Chinese Major Infectious Disease Research Projects (grant no.
2012ZX10001003).
References
|
1
|
Seppälä M, Taylor RN, Koistinen H,
Koistinen R and Milgrom E: Glycodelin: a major lipocalin protein of
the reproductive axis with diverse actions in cell recognition and
differentiation. Endocr Rev. 23:401–430. 2002.
|
|
2
|
Jeschke U, Kuhn C, Mylonas I, Schulze S,
Friese K, Mayr D, Speer R, et al: Development and characterization
of monoclonal antibodies for the immunohistochemical detection of
glycodelin A in decidual, endometrial and gynaecological tumour
tissues. Histopathology. 48:394–406. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Richter C, Baetje M, Bischof A, Makovitzky
J, Richter DU, Gerber B, Briese V, et al: Expression of the
glycodelin A gene and the detection of its protein in tissues and
serum of ovarian carcinoma patients. Anticancer Res. 27:2023–2025.
2007.PubMed/NCBI
|
|
4
|
Hautala LC, Greco D, Koistinen R,
Heikkinen T, Heikkilä P, Aittomäki K, Blomqvist C, et al:
Glycodelin expression associates with differential tumour phenotype
and outcome in sporadic and familial non-BRCA1/2 breast cancer
patients. Breast Cancer Res Treat. 128:85–95. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Seppälä M, Koistinen H, Koistinen R,
Hautala L, Chiu PC and Yeung WS: Glycodelin expression in
reproductive endocrinology and hormone-related cancer. Eur J
Endocrinol. 160:121–133. 2009.
|
|
6
|
Kunert-Keil C, Steinmüller F, Jeschke U,
Gredes T and Gedrange T: Immunolocalization of glycodelin in human
adenocarcinoma of the lung, squamous cell carcinoma of the lung and
lung metastases of colonic adenocarcinoma. Acta Histochem.
113:798–802. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ren S, Liu S, Howell PM Jr and Riker AI:
Identification of a putative oncogene in human melanoma:
progestagen-associated endometrial protein. Ann Surg Oncol.
15(Suppl 2): 102008.
|
|
8
|
Hautala LC, Koistinen R, Seppälä M, Bützow
R, Stenman UH, Laakkonen P and Koistinen H: Glycodelin reduces
breast cancer xenograft growth in vivo. Int J Cancer.
123:2279–2284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song M, Ramaswamy S, Ramachandran S,
Flowers LC, Horowitz IR, Rock JA and Parthasarathy S: Angiogenic
role for glycodelin in tumorigenesis. Proc Natl Acad Sci USA.
98:9265–9270. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ren S, Liu S, Howell PM Jr, Zhang G,
Pannell L, Samant R, et al: Functional characterization of the
progestagen-associated endometrial protein gene in human melanoma.
J Cell Mol Med. 14:1432–1442. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gray-Schopfer V, Wellbrock C and Marais R:
Melanoma biology and new targeted therapy. Nature. 445:851–857.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fodstad O, Kjønniksen I, Aamdal S, Nesland
JM, Boyd MR and Pihl A: Extrapulmonary, tissue-specific metastasis
formation in nude mice injected with FEMX-I human melanoma cells.
Cancer Res. 48:4382–4388. 1988.
|
|
13
|
Pellitteri-Hahn MC, Warren MC, Didier DN,
Winkler EL, Mirza SP, Greene AS and Olivier M: Improved mass
spectrometric proteomic profiling of the secretome of rat vascular
endothelial cells. J Proteome Res. 5:2861–2864. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mbeunkui F, Metge BJ, Shevde LA and
Pannell LK: Identification of differentially secreted biomarkers
using LC-MS/MS in isogenic cell lines representing a progression of
breast cancer. J Proteome Res. 6:2993–3002. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mitra A, Fillmore RA, Metge BJ, Rajesh M,
Xi Y, King J, Ju J, et al: Large isoform of MRJ (DNAJB6) reduces
malignant activity of breast cancer. Breast Cancer Res. 10:R222008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cummins DL, Cummins JM, Pantle H,
Silverman MA, Leonard AL and Chanmugam A: Cutaneous malignant
melanoma. Mayo Clin Proc. 81:500–507. 2006. View Article : Google Scholar
|
|
17
|
Riker AI, Enkemann SA, Fodstad O, et al:
The gene expression profiles of primary and metastatic melanoma
yields a transition point of tumor progression and metastasis. BMC
Med Genomics. 1:132008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Scholz C, Toth B, Barthell E, Mylonas I,
Weissenbacher T, Friese K and Jeschke U: Glycodelin expression in
correlation to grading, nodal involvement and steroid receptor
expression in human breast cancer patients. Anticancer Res.
30:1599–1603. 2010.PubMed/NCBI
|
|
19
|
Mandelin E, Lassus H, Seppälä M, Leminen
A, Gustafsson JA, Cheng G, Bützow R and Koistinen R: Glycodelin in
ovarian serous carcinoma: association with differentiation and
survival. Cancer Res. 63:6258–6264. 2003.PubMed/NCBI
|
|
20
|
Uchida H, Maruyama T, Ono M, Ohta K, et
al: Histone deacetylase inhibitors stimulate cell migration in
human endometrial adenocarcinoma cells through up-regulation of
glycodelin. Endocrinology. 148:896–902. 2007. View Article : Google Scholar
|
|
21
|
Verdel A, Jia S, Gerber S, Sugiyama T,
Gygi S, Grewal SI and Moazed D: RNAi-mediated targeting of
heterochromatin by the RITS complex. Science. 303:672–676. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yao MC, Fuller P and Xi X: Programmed DNA
deletion as an RNA-guided system of genome defense. Science.
300:1581–1584. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Naldini L, Blömer U, Gage FH, Trono D and
Verma IM: Efficient transfer, integration, and sustained long-term
expression of the transgene in adult rat brains injected with a
lentiviral vector. Proc Natl Acad Sci USA. 93:11382–11388. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Naldini L, Blömer U, Gallay P, Ory D,
Mulligan R, Gage FH, Verma IM and Trono D: In vivo gene delivery
and stable transduction of nondividing cells by a lentiviral
vector. Science. 272:263–267. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sumimoto H and Kawakami Y: Lentiviral
vector-mediated RNAi and its use for cancer research. Future Oncol.
3:655–664. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sumimoto H and Kawakami Y: The RNA
silencing technology applied by lentiviral vectors in oncology.
Methods Mol Biol. 614:187–199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lawler PR and Lawler J: Molecular basis
for the regulation of angiogenesis by thrombospondin-1 and -2. Cold
Spring Harb Perspect Med. 2:a0066272012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jia L and Waxman DJ: Thrombospondin-1 and
pigment epithelium-derived factor enhance responsiveness of KM12
colon tumor to metronomic cyclophosphamide but have disparate
effects on tumor metastasis. Cancer Lett. 330:241–249. 2013.
View Article : Google Scholar
|
|
29
|
Kim NH, Kim SN, Seo DW, Han JW and Kim YK:
PRMT6 overexpression upregulates TSP-1 and downregulates MMPs: its
implication in motility and invasion. Biochem Biophys Res Commun.
432:60–65. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Senger DR, Perruzzi CA and Papadopoulos A:
Elevated expression of secreted phosphoprotein 1 (osteopontin, 2ar)
as a consequence of neoplastic transformation. Anticancer Res.
9:1291–1299. 1989.PubMed/NCBI
|
|
31
|
Das R, Philip S, Mahabeleshwar GH, Bulbule
A and Kundu GC: Osteopontin: it’s role in regulation of cell
motility and nuclear factor kappa B-mediated urokinase type
plasminogen activator expression. IUBMB Life. 57:441–447. 2005.
|
|
32
|
Wu Y, Jiang W, Wang Y, Wu J, Saiyin H,
Qiao X, Mei X, et al: Breast cancer metastasis suppressor 1
regulates hepatocellular carcinoma cell apoptosis via suppressing
osteopontin expression. PLoS One. 7:e429762012. View Article : Google Scholar : PubMed/NCBI
|