Introduction
Lung cancer is the most common form of malignancy
and a major determinant of the overall cancer-related mortality
worldwide (1). Based on the
biology, therapy and prognosis, lung cancers are divided into two
predominant classes: non-small cell lung cancer (NSCLC) and small
cell lung cancer (SCLC). NSCLCs consist of three types,
adenocarcinoma (ADC), squamous cell carcinoma (SC) and large cell
carcinoma (2).
Lung ADC, an epithelial cancer of glandular origin,
is the most prevalent of these lung cancer diagnoses. In addition,
due to the recent advances in computed tomographic technology, the
number of patients diagnosed with small-sized lung ADC as well as
stage I lung ADC has increased. To improve the prognosis of ADC
patients, the identification of suitable markers is required to
select patients with a poor prognosis who may benefit from adjuvant
therapy subsequent to surgery (3,4).
Gene amplification is a predominant genomic force
contributing to the development of numerous solid tumors, including
ADC, and providing an important resource for identifying the
location of candidate oncogenes (5). Previous genome-wide analyses for copy
number changes in cancer cells have identified various chromosomal
loci that are amplified in lung ADCs (5–7).
However, as copy number alterations in lung ADC genomes are
complex, target genes often remain unclear in amplified chromosomal
segments. In addition, the clinical significance of gene
amplification in early-stage lung ADC also remains to be
elucidated. Thus, in the present study, the copy number changes of
14 early-stage lung ADCs were determined, with the aim of
identifying novel high level alterations and candidate genes that
may be important in ADC progression.
Materials and methods
Preparation of patient samples
Fourteen lung ADCs were observed from patients
undergoing surgery as a primary treatment, without previous
radiation or chemotherapy. The original diagnostic material of all
ADC patients was reviewed to verify the previous histopathological
diagnosis and staging according to the World Health Organization
classification system. The stage of disease was based on the
tumor-node-metastasis (TNM) classification using the UICC (Union
Internationale Contre Le Cancer) staging system. No patients had
received pre-operative chemotherapy or radiation. This study was
reviewed and approved by the Institutional Review Board of the
Chungnam National University Hospital (Daejeon, Korea). Written
informed consent was obtained from each patient according to the
institutional regulations of the Chungnam National University
Hospital. The demographic and pathological data, including age,
gender and the tumor stage were obtained by a review of the medical
records.
Array-comparative genomic hybridization
(CGH) analysis
Microarray-CGH was performed on the MacArray™ Karyo
4000 K BAC-chip (Macrogen, Seoul, Korea) (8–11),
consisting of 4,046 human bacterial artificial chromosomes (BACs)
applied in duplicate at a resolution of 1 Mbp as described in our
previous studies (12,13). Briefly, all clones were two-end
sequenced using an ABI Prism 3700® DNA analyzer (Applied
Biosystems, Foster City, CA, USA) and their sequences were blasted
[using basic local alignment search tool (BLAST); http://blast.ncbi.nlm.nih.gov/Blast.cgi]. Mapping of
large insert clones was conducted according to the genomic location
in the UCSC Genome Bioinformatics database [http://genome.ucsc.edu; Build 36, version Mar. 2006
(hg18)].
Preparation of DNA targets, labeling, hybridization,
washing, staining and scanning was conducted according to the
manufacturer’s instructions (Macrogen, Seoul, Korea) (8–13).
Briefly, arrays were pre-hybridized with salmon sperm DNA to block
repetitive sequences in the BACs. A total of 500 ng normal male DNA
(reference) and digested tumor DNA (test) were labeled with
Cy5-dCTP and Cy3-dCTP, respectively, by randomly primed labeling
(Array CGH Genomic Labeling System; Invitrogen, Carlsbad, CA, USA).
The labeled probe and human Cot-I DNA (Invitrogen) were mixed and
dissolved in hybridization solution.
Statistical analysis
To adjust for effects due to the variation between
the red and green dyes, Lowess normalization was applied. The ratio
of the red to green channels of each clone was calculated and
log2 transformed. The spot quality criteria were set as
foreground to background >3.0 and the standard deviation of
triplicates <0.2. Breakpoint detection and status assignment of
the genomic regions were performed using GLAD software (14). The R 2.2.1 package of the
Bioconductor Project (http://www.bioconductor.org) was used for the
detection of the frequency of gain or loss, and for statistical
analysis. The median of the signal ratio (test signal/reference
signal) of each triplicate spot was defined as a gain or a loss
when it was >0.25 or <−0.25, respectively. High-level
amplification of clones was defined when their intensity ratios
were >1.0 in log2 scale and vice versa for homozygous
deletion. The threshold value was determined empirically as a value
3-fold greater than that of the standard deviations calculated from
30 normal males and females in hybridization experiments. The
Benjamini-Hochberg false discovery rate (FDR) was applied for
multiple testing correction for the high number of false-positive
calls.
Results
Whole genome array analysis of ADC
cases
To clarify the critical genetic markers associated
with ADC pathogenesis, high-resolution array-CGH was conducted on
14 ADC cases. A broad range of aberrations were detected, such as
deletions and/or gains of various sizes. All patients (100.0%) in
this genomic profile showed multiple segmental alterations,
including single copy as well as high level gains and losses. A
detailed overview of the clinicopathological data of the 14 ADCs is
shown in Table I. Although entire
chromosomal arm changes appeared occasionally, the majority of copy
number alterations in ADCs were localized regional changes.
Notably, large copy number gains involving chromosomes 5p, 7p, 20q,
1p and 16p (>35% of patients) were more prevalent than copy
number losses in the cases. The delineation of the most frequently
gained chromosomal regions and possible target genes in the ADCs is
listed in Table II.
 | Table IA detailed overview of
clinicopathological data of the 14 early-stage lung
adenocarcinomas. |
Table I
A detailed overview of
clinicopathological data of the 14 early-stage lung
adenocarcinomas.
| Case no. | Gender | Age (years) | TNM
classification | Tumor stage | Smoking status |
|---|
| 1 | M | 61 | T2N0M0 | 1B | Former smoker |
| 2 | F | 50 | T2N2M0 | 3A | Current smoker |
| 3 | F | 47 | T2N0M0 | 1B | Former smoker |
| 4 | M | 66 | T1N0M0 | 1A | Current smoker |
| 5 | M | 65 | T2N2M0 | 3A | Current smoker |
| 6 | F | 61 | T1N0M0 | 1A | Never smoked |
| 7 | F | 56 | T2N1M0 | 2B | Never smoked |
| 8 | M | 72 | T1N0M0 | 1A | Former smoker |
| 9 | M | 61 | T2N0M0 | 2A | Current smoker |
| 10 | F | 70 | T2N2M0 | 3A | Former smoker |
| 11 | M | 60 | T3N1M0 | 3A | Current smoker |
| 12 | M | 70 | T3N1M0 | 3A | Current smoker |
| 13 | M | 75 | T3N1M0 | 3A | Current smoker |
| 14 | F | 69 | T1N0M0 | 1A | Never smoked |
 | Table IIMost frequently gained regions of
overlap detected by microarray comparative genomic hybridization in
early-stage lung adenocarcinomas and the candidate genes. |
Table II
Most frequently gained regions of
overlap detected by microarray comparative genomic hybridization in
early-stage lung adenocarcinomas and the candidate genes.
| BAC clone | Chromosome
location | Gene contained in
clones | BAC size (bp) | Cases with copy
number gainsa (%) |
|---|
| BAC91_J20 | 5p15.33 | SLC6A19,
SLC6A18, TERT | 115,211 | 79 |
| BAC151_L22 | 5p15.33 | Cep72,
TPPP | 89,632 | 71 |
| BAC170_A22 | 7p22.3 | MGC11257,
LOC393076, GPR146, GPR30,
LOC402518 | 84,220 | 62 |
| BAC1_I06 | 7p11.2 | CALM1P2 | 116,026 | 57 |
| BAC15_B08 | 7p11.2 | EGFR | 100,083 | 57 |
| BAC107_K11 | 20q13.33 | ARFGAP1,
KIAA1510, CHRNA4, KCNQ2 | 147,123 | 54 |
| BAC137_F15 | 7p14.1 | TRGJP1,
TRGV11, TRGVB, TRGV10, TRGV9,
TRGVA, TRGV8, TRGV7, TRGV6,
TRGV5P, TRGV5, TRGV4 | 80,960 | 50 |
| BAC147_B17 | 7p22.3 | MAD1L1,
LOC402663, LOC442696, LOC442609 | 85,873 | 50 |
| BAC218_N01 | 7p15.2 | HOXA4,
HOXA5, HOXA6, HOXA7, HOXA9,
HOXA10, HOXA11, HOXA13 | 111,540 | 43 |
| BAC183_C18 | 7p14.1 | LOC222103,
TRGJP2, TRGC1, TRGJ1, TRGJP,
TRGJP1, TRGV11, TRGVB, TRGV10,
TRGV9, TRGVA, TRGV8, TRGV7,
TRGV6, TRGV5P, TRGV5 | 98,051 | 43 |
| BAC38_N15 |
1p36.33-1p36.32 | SKI,
FLJ13941 | 85,791 | 43 |
| BAC62_J14 | 7p21.1 | HDAC9 | 78,272 | 43 |
| BAC97_B23 | 7p22.3 | OC442586,
LOC442589, LOC442282, FAM20C,
LOC442651 | 94,921 | 43 |
| BAC239_F18 | 16p13.3 | LOC441443,
LOC389753 | 72,701 | 43 |
| BAC142_O10 | 7p22.3 | MAD1L1,
LOC442699, LOC442592, LOC442654,
LOC442593, LOC442594, LOC442595,
LOC442700 | 144,899 | 43 |
| BAC122_H21 | 7p12.2 | GRB10 | 85,692 | 43 |
| BAC139_D05 | 7p21.2 | ETV1 | 87,963 | 36 |
| BAC161_H20 | 7p14.3 | PDE1C | 99,714 | 36 |
| BAC178_O13 | 7p11.2 |
LOC442681 | 92,874 | 36 |
| BAC65_K10 | 7p21.1 | LOC442511,
MEOX2 | 90,760 | 36 |
| BAC46_P02 | 7p13 | CCM2,
KIAA0363, TBRG4, RAMP3 | 91,839 | 36 |
Copy number alterations on the short arm
of chromosome 7 in ADCs
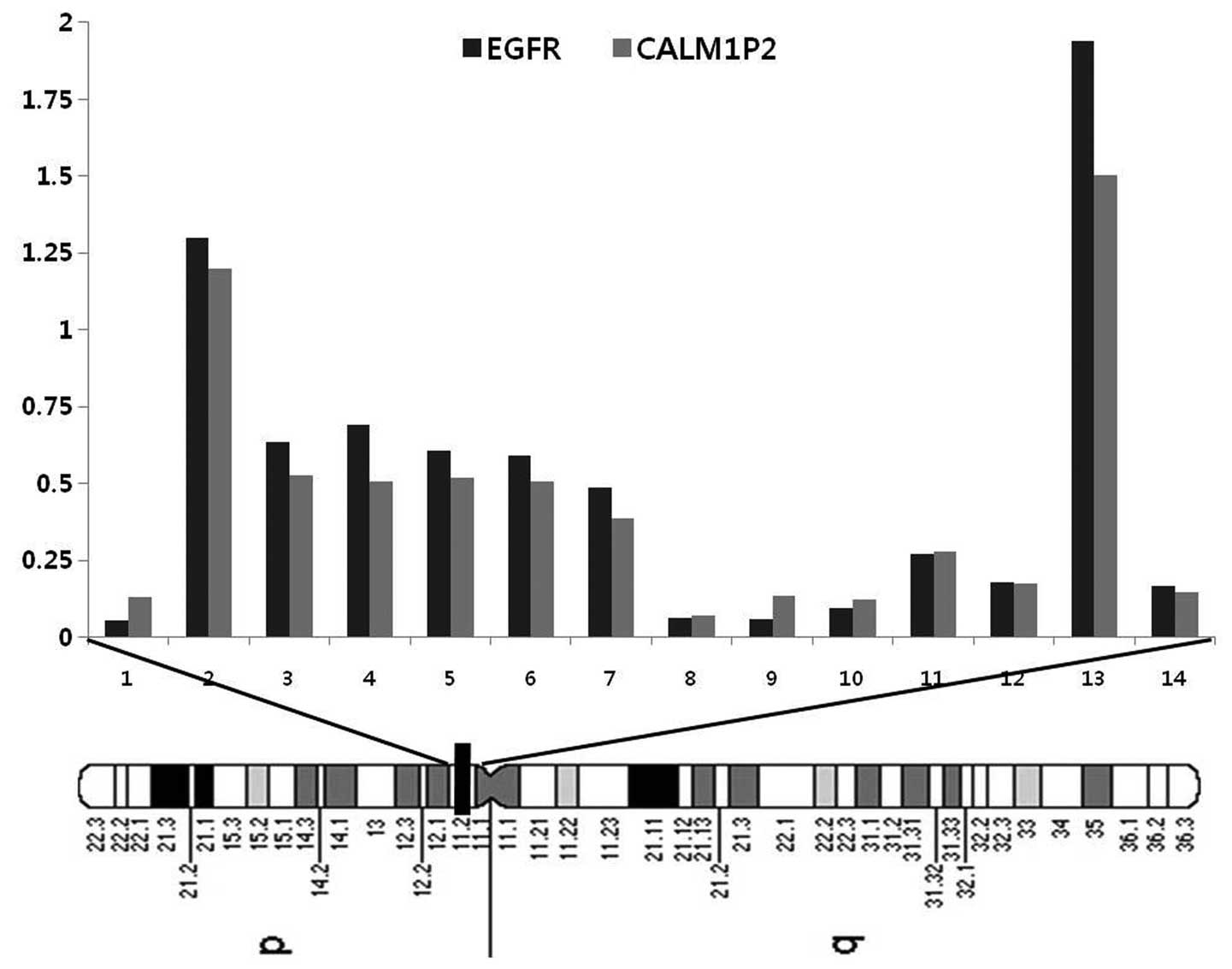
Array-CGH analysis revealed several copy number
changes in the ADC cases. Initially, the analysis focused on the
short arm of chromosome 7, the most frequently affected regions in
the ADC cases (85.7%, 12/14). More specifically, three distinct
regions of amplifications were identified in 28.6% (4/14) of the
cases. In addition, three minimal overlapping regions were defined
on chromosome 7p, (7p22.3-p21.1, 7p15.2-p14.1) and 7p12.3-p11.2.
The minimal common region of chromosome 7p was identified, by
array-CGH, to be located between BAC41_H11 and BAC178_O13 (position
92.8–123.6 kb).
The first locus of amplification was located
distally on 7p22.3-p21.1 regions (82.4–123.6 kb). According to the
information archived by human genome database (http://genome.ucsc.edu/), it is flanked by the BAC
clones between BAC130_G18 and BAC113_E07 and contains 63 possible
target genes (4.1 Mb segment). These terminal gains were often
large and were located to the ETS translocation variant 1
(ETV1) gene. Notably, a high-frequency of single copy number
gains (>0.25 log2 ratio) and high-level gains
(>0.5 log2 ratio) from the 7p22.3-p21.1 region were
detected in 71.4% (10/14) and 35.7% (5/14) of the cases,
respectively. The most frequently gained clone was BAC170_A22 at
the 7p22.3 region (57.1%, 8/14), which is located in the
MGC11257, LOC393076, GPR146, GPR30 and
LOC402518 genes.
The second candidate locus spanned 91.8–118.2 kb in
the 7p15.2-p14.1 regions, encompassed 15 target clones and was
identified as exhibited copy number gains in 7 of 14 cases (50.0%).
ADC cases with 7p14.1-p15.2 gains, displayed a varying degree of
copy number increases predominantly from 7p14.1 (50%, 7/14), 7p15.2
(42.9%, 6/14) and 7p14.3 (35.7%, 5/14). It was flanked by the BAC
clones between BAC137_F15 and BAC218_N01, and encompassed 57 genes
(http://genome.ucsc.edu/). Notably, a 111.5 kb
high-level amplification of the 7p15.2 region contained
HOXA4, HOXA5, HOXA6, HOXA7,
HOXA9, HOXA10, HOXA11 and HOXA13 genes
in one case.
The third region was flanked by BAC157_N08 and
BAC178_O13, and mapped at the 7p11.2-p12.3 regions (76.0–92.9 kb).
A high frequency of single copy number gains (>0.25
log2 ratio) from the 7p11.2-p12.3 regions was observed
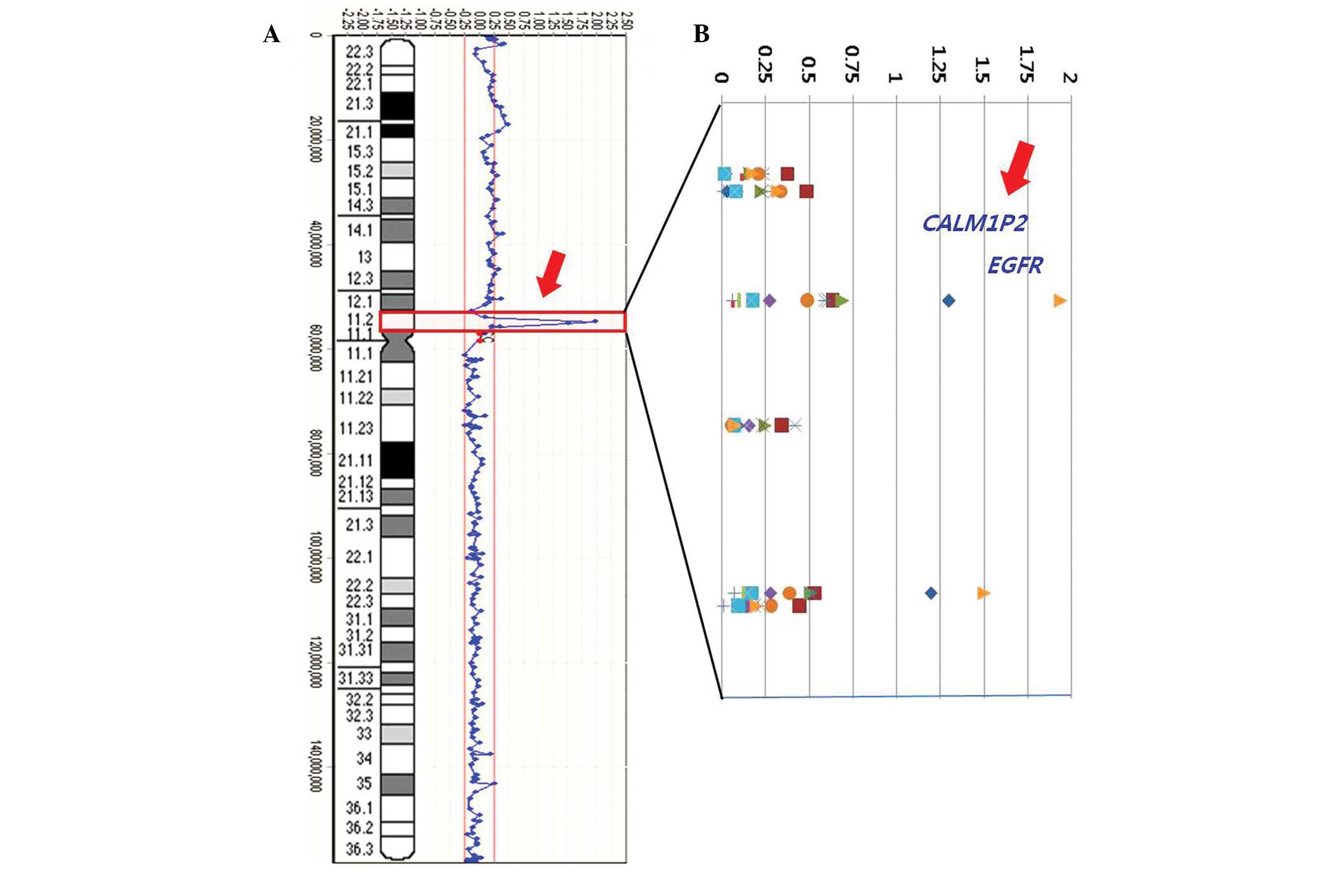
in 57.1% (8/14) of the cases. In addition, two amplified (>1
log2 ratio) loci on the 7p11.2 region were identified in
14.3% of the cases. One locus contained contiguous amplified clones
covering a region of ~100.1 kb and comprised the oncogenic variant
of the epidermal growth factor receptor (EGFR) gene in 2 of 14 ADCs
(14.3%), with the highest level of amplification in case 13
(Fig. 2).
Furthermore, a candidate target gene for
CALM1P2 was identified in 14.3% (2/14) of cases at the
7p11.2 region. To the best of our knowledge, the pathogenesis of
the CALM1P2 gene has not been previously reported in ADCs.
As increased amplification of the EGFR gene has been
previously observed in ADCs, the present study aimed to determine
whether there was a correlation between EGFR and the newly
identified CALM1P2 gene. Notably, co-amplifications were
demonstrated between EGFR and CALM1P2 genes in 100%
(2/2, case 2 and 13; Fig. 1). An
example of an individual profile at the 7p11.2 region in the 14 ADC
cases is presented in Fig. 1.
High-level amplifications are clearly observed in cases 2 and 13.
Fig. 2A represents the frequency
of the copy number changes on chromosome 7 and a weighted frequency
(%) diagram with high-level amplifications at the 7p11.2 region
from the 14 ADC cases is shown in Fig.
2B.
Discussion
Array-CGH is a successful and valuable tool for the
analysis of chromosome copy-number alterations in human cancer and
may be suitable for individualized diagnostic, prognostic and
therapeutic decision-making (12).
In this study, genome wide array-CGH was conducted to
comprehensively characterize genome copy number aberrations
associated with early-stage ADC.
The most noteworthy observation in this study was
the high frequency of copy number gains at chromosome 7p, in 85.7%
(12/14) of the cases. The short arm of chromosome 7 is implicated
as being involved in the initiation and/or progression of ADC and
has been suggested to include critical cancer related genes in ADC
(16–18). Job et al(15) demonstrated that the high frequency
of copy number gains on chromosome 7p contained CARD11,
ETV1 and IKZF1 genes in 78–92% of the ADCs observed.
Single nucleotide polymorphism array analysis determined the
high-level amplifications on chromosome 7p (>10%) in small-sized
ADCs and lung ADC cell lines (3).
Previously, frequent copy number gains on chromosome 7p in ADC
cases in non-smokers has also been observed (16). In conclusion, chromosome 7p appears
to harbor multiple tumor-related genes that may be implicated in
ADC pathogenesis.
Three distinct amplified (>1 log2
ratio) loci from the 7p22.3-11.2 region were identified in 28.6% of
the cases. The first loci located on 7p22.3 contained
FAM20C, LOC442586, LOC442589, LOC442282
and LOC442651 genes. The FAM20 family of secreted
proteins consists of three members (FAM20A, FAM20B
and FAM20C) which have recently been linked to developmental
disorders (17).
The second amplification region of 7p15.2 contained
HOXA4, HOXA5, HOXA6, HOXA7,
HOXA9, HOXA10, HOXA11 and HOXA13 genes.
Homeobox (HOX) genes encode homeodomain-containing transcription
factors critical for development, differentiation and homeostasis.
Their dysregulation has been implicated in various types of cancer,
including lung adenocarcinoma. Abe et al(18) determined that the expression levels
of HOXA5 and A10 in adenocarcinoma (and HOXA1,
A5, A10 and C6 in squamous cell carcinoma of
the lung) were significantly higher than those in the non-cancerous
tissues. It was suggested that the disordered patterns of
HOX gene expression were involved in the development of
non-small cell lung cancer and in the histological changes (such as
adenocarcinoma and squamous cell carcinoma) of the lung. A previous
study by Marra et al(19)
observed the involvement of the HOX B13 gene in several
tumors of the urogenital system. In non-muscle invasive bladder
transitional cancer, nuclear HOX B13 expression showed
significant correlation with higher Gleason grade, clinical stage
of the tumor and a poor survival outcome, thus determining its
potential prognostic value.
HOX genes have also been demonstrated to be a
hallmark of numerous hematological malignancies (20). The dysregulation of HOX
genes is correlated with a number of hematological malignancies,
including acute myeloid leukemia (AML) and acute lymphoid leukemia,
where they have been shown to support the immortalization of
leukemic cells as chimeric partners in fusion genes and when
overexpressed in their wild-type form (21). Furthermore, overexpression of
individual Hox proteins expanded various bone marrow populations
in vitro, leading to myeloproliferation and in certain cases
inhibition of differentiation and AML in vivo(22). A high concentration of HOXA9
gene product in leukemic blasts has been shown to be an adverse
prognostic parameter and HOXA9 expression was associated
with a certain state of myeloid differentiation (23). These results suggest that
HOX genes represent important prognostic and predictive
markers for solid tumors and may be rational targets for
therapeutic approaches for the poor prognosis leukemia subset.
Additional studies are required to further investigate the
mechanism and clinical significance of these results.
The third amplification region was located distally
in the 7p22.3 chromosomal region and this locus contained the
oncogenic variant of the epidermal growth factor receptor gene
(EGFR) in 7.1% of the cases. The involvement of the
EGFR gene as the driver of the 7p11.2 amplicon is well
established in ADC cases (24–27).
Liu et al(24) determined that EGFR gene
mutation rates were significantly greater in patients with
adenocarcinoma (35.5 versus 9.9% non-adenocarcinoma) and there was
a correlation between EGFR gene mutation and gene
amplification, particularly in early-stage adenocarcinoma.
Moreover, Reinmuth et al(25) demonstrated that EGFR gene
mutations are frequently observed in ADC with bronchioloalveolar
differentiation and may be linked to chromosomal imbalances. In a
study by Sholl et al(26),
EGFR amplification demonstrated a unique association with
exon 19 deletion mutations and represented distinct
clinicopathological features associated with a significantly
worsened prognosis in ADC patients. Furthermore, Yoshizawa et
al(27) observed that
EGFR mutations were significantly associated with
adenocarcinoma in situ, minimally invasive adenocarcinoma
and lepidic-and papillary-predominant adenocarcinoma, suggesting
that EGFR mutations may aid in the prediction of patient
prognosis and selection of those who require adjuvant chemotherapy.
These results and the results of the present study suggested that
the EGFR mutation may be an early event in the pathogenesis
of lung ADC and may facilitate aggressive behavior of the
tumor.
In addition, a potential oncogenic variant of
CALM1P2 was identified in 14.3% (2/14) of cases from the
7p11.2 region. To the best of our knowledge, the involvement of the
CALM1P2 gene in the pathogenesis of ADC has not been
previously described; however, genetic mutations of CALM
genes are observed in other types of cancer (28,29).
Toutenhoofd et al(28)
concluded that the CALM gene family is differentially active
at the transcriptional level in teratoma cells and that the 5′
untranslated regions are required to recover full promoter
activation. Furthermore, Stanislaus et al(29) suggested that the CALM1 and PLCG2
signaling pathways are the two potential targets for gene knockdown
in doxorubicin- and paclitaxel-based chemotherapy of cervical
cancer. As a gain of amplification of the EGFR gene has been
described previously in ADCs, the present study aimed to determine
whether there was a correlation between the EGFR gene and
the newly identified amplified CALM1P2 gene. Notably,
co-amplification was demonstrated between the EGFR and
CALM1P2 genes in 100% (2/2) of cases.
The present study established critical regions on
the 7p chromosome implicated in ADC. The present results warrant
future studies to identify the putative oncogenes at 7p to gain a
better understanding of the molecular pathogenesis of early-stage
lung ADC. The genomic analysis allowed the proposition of novel
candidate genes that may be associated with the pathogenesis of
early-stage lung adenocarcinoma. The newly identified target genes
may contribute to ADC pathogenesis as well as provide novel targets
for therapeutic intervention in early-stage ADC pending functional
validation.
Acknowledgements
This study was funded by the research fund of Korea
Nazarene University in 2013 (2013-0302).
References
|
1
|
Parkin DM, Bray FI and Devesa SS: Cancer
burden in the year 2000. The global picture. Eur J Cancer. 37(Suppl
8): S4–S66. 2001.PubMed/NCBI
|
|
2
|
Balsara BR, Sonoda G, du Manoir S,
Siegfried JM, Gabrielson E and Testa JR: Comparative genomic
hybridization analysis detects frequent, often high-level,
overrepresentation of DNA sequences at 3q, 5p, 7p, and 8q in human
non-small cell lung carcinomas. Cancer Res. 57:2116–2120. 1997.
|
|
3
|
Iwakawa R, Kohno T, Kato M, Shiraishi K,
Tsuta K, Noguchi M, Ogawa S and Yokota J: MYC amplification as a
prognostic marker of early-stage lung adenocarcinoma identified by
whole genome copy number analysis. Clin Cancer Res. 15:1481–1489.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Greulich H: The genomics of lung
adenocarcinoma: opportunities for targeted therapies. Genes Cancer.
1:1200–1210. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao X, Weir BA, LaFramboise T, Lin M,
Beroukhim R, Garraway L, Beheshti J, Lee JC, Naoki K, Richards WG,
et al: Homozygous deletions and chromosome amplifications in human
lung carcinomas revealed by single nucleotide polymorphism array
analysis. Cancer Res. 1:5561–5570. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tonon G, Wong KK, Maulik G, Brennan C,
Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, et
al: High-resolution genomic profiles of human lung cancer. Proc
Natl Acad Sci USA. 102:9625–9630. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li X, Wan L, Shen H, Geng J, Nie J, Wang
G, Jia N, Dai M and Bai X: Thyroid transcription factor-1
amplification and expressions in lung adenocarcinoma tissues and
pleural effusions predict patient survival and prognosis. J Thorac
Oncol. 7:76–84. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hwang KT, Han W, Cho J, Lee JW, Ko E, Kim
EK, Jung SY, Jeong EM, Bae JY, Kang JJ, et al: Genomic copy number
alterations as predictive markers of systemic recurrence in breast
cancer. Int J Cancer. 15:1807–1815. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Choe J, Kang JK, Bae CJ, Lee DS, Hwang D,
Kim KC, Park WY, Lee JH and Seo JS: Identification of origin of
unknown derivative chromosomes by array-based comparative genomic
hybridization using pre- and postnatal clinical samples. J Hum
Genet. 52:934–942. 2007. View Article : Google Scholar
|
|
10
|
Kim JI, Ju YS, Park H, Kim S, Lee S, Yi
JH, Mudge J, Miller NA, Hong D, Bell CJ, et al: A highly annotated
whole-genome sequence of a Korean individual. Nature. 20:1011–1015.
2009.PubMed/NCBI
|
|
11
|
Chochi Y, Kawauchi S, Nakao M, Furuya T,
Hashimoto K, Oga A, Oka M and Sasaki K: A copy number gain of the
6p arm is linked with advanced hepatocellular carcinoma: an
array-based comparative genomic hybridization study. J Pathol.
217:677–684. 2009. View Article : Google Scholar
|
|
12
|
Kang JU, Koo SH, Kwon KC, Park JW and Kim
JM: Identification of novel candidate target genes, including
EPHB3, MASP1 and SST at 3q26.2–q29 in squamous cell carcinoma of
the lung. BMC Cancer. 9:2372009.PubMed/NCBI
|
|
13
|
Kang JU and Koo SH: ORAOV1 is a probable
target within the 11q13.3 amplicon in lymph node metastases from
gastric adenocarcinoma. Int J Mol Med. 29:81–87. 2012.PubMed/NCBI
|
|
14
|
Willenbrock H and Fridlyand J: A
comparison study: applying segmentation to array CGH data for
downstream analyses. Bioinformatics. 21:4084–4091. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Job B, Bernheim A, Beau-Faller M,
Camilleri-Broët S, Girard P, Hofman P, Mazières J, Toujani S,
Lacroix L, Laffaire J, et al: Genomic aberrations in lung
adenocarcinoma in never smokers. PLoS One. 5:e151452010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thu KL, Vucic EA, Chari R, Zhang W,
Lockwood WW, English JC, Fu R, Wang P, Feng Z, MacAulay CE, et al:
Lung adenocarcinoma of never smokers and smokers harbor
differential regions of genetic alteration and exhibit different
levels of genomic instability. PLoS One. 7:e330032012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vogel P, Hansen GM, Read RW, Vance RB,
Thiel M, Liu J, Wronski TJ, Smith DD, Jeter-Jones S and Brommage R:
Amelogenesis imperfecta and other biomineralization defects in
Fam20a and Fam20c null mice. Vet Pathol. 49:998–1017. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Abe M, Hamada J, Takahashi O, Takahashi Y,
Tada M, Miyamoto M, Morikawa T, Kondo S and Moriuchi T: Disordered
expression of HOX genes in human non-small cell lung cancer. Oncol
Rep. 15:797–802. 2006.PubMed/NCBI
|
|
19
|
Marra L, Cantile M, Scognamiglio G,
Perdonà S, La Mantia E, Cerrone M, Gigantino V, Cillo C, Caraglia
M, Pignata S, et al: Deregulation of HOX B13 expression in urinary
bladder cancer progression. Curr Med Chem. 20:833–839.
2013.PubMed/NCBI
|
|
20
|
Bach C, Buhl S, Mueller D, García-Cuéllar
MP, Maethner E and Slany RK: Leukemogenic transformation by HOXA
cluster genes. Blood. 115:2910–2918. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Alharbi RA, Pettengell R, Pandha HS and
Morgan R: The role of HOX genes in normal hematopoiesis and acute
leukemia. Leukemia. 27:1000–1008. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Eklund E: The role of Hox proteins in
leukemogenesis: insights into key regulatory events in
hematopoiesis. Crit Rev Oncog. 16:65–76. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Golub TR, Slonim DK, Tamayo P, Huard C,
Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri
MA, et al: Molecular classification of cancer: class discovery and
class prediction by gene expression monitoring. Science.
286:531–537. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu H, Li Y, Chen G, Wang J, Li Y, Wang Y,
Wei S, Zhu D, Qiu X, Wang W, et al: Detection and its clinical
significance of EGFR gene mutation and gene amplification in 187
patients with non-small cell lung cancer. Zhongguo Fei Ai Za Zhi.
12:1219–1228. 2009.(In Chinese).
|
|
25
|
Reinmuth N, Jauch A, Xu EC, Muley T,
Granzow M, Hoffmann H, Dienemann H, Herpel E, Schnabel PA, Herth
FJ, et al: Correlation of EGFR mutations with chromosomal
alterations and expression of EGFR, ErbB3 and VEGF in tumor samples
of lung adenocarcinoma patients. Lung Cancer. 62:193–201. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sholl LM, Yeap BY, Iafrate AJ,
Holmes-Tisch AJ, Chou YP, Wu MT, Goan YG, Su L, Benedettini E, Yu
J, et al: Lung adenocarcinoma with EGFR amplification has distinct
clinicopathologic and molecular features in never-smokers. Cancer
Res. 69:8341–8348. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoshizawa A, Sumiyoshi S, Sonobe M,
Kobayashi M, Fujimoto M, Kawakami F, Tsuruyama T, Travis WD, Date H
and Haga H: Validation of the IASLC/ATS/ERS lung adenocarcinoma
classification for prognosis and association with EGFR and KRAS
gene mutations: analysis of 440 Japanese patients. J Thorac Oncol.
8:52–61. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Toutenhoofd SL, Foletti D, Wicki R, Rhyner
JA, Garcia F, Tolon R and Strehler EE: Characterization of the
human CALM2 calmodulin gene and comparison of the transcriptional
activity of CALM1, CALM2 and CALM3. Cell Calcium. 23:323–338. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stanislaus A, Bakhtiar A, Salleh D, Tiash
S, Fatemian T, Hossain S, Akaike T and Chowdhury EH: Knockdown of
PLC-gamma-2 and calmodulin 1 genes sensitizes human cervical
adenocarcinoma cells to doxorubicin and paclitaxel. Cancer Cell
Int. 12:302012. View Article : Google Scholar
|