Introduction
Hepatocellular carcinoma (HCC) is the sixth most
common type of cancer worldwide and the third most frequent cause
of cancer-related mortality (1).
Chronic viral infections and alcohol exposure are the predominant
risk factors of hepatocarcinogenesis (2). Hepatocarcinogenesis exhibits a
multistage course, from chronic liver disease, dysplastic nodules
and early through to advanced HCC, in which numerous cancer-related
genes involved (3). A number of
genes may be used as diagnostic or prognostic molecular markers of
HCC (4). For example, glypican-3
(GPC3) is an important diagnostic marker for early HCC (5). α-fetoprotein (AFP) is a potential
predictor of survival and tumor recurrence for AFP-producing HCC
(6); however, it has been
discontinued as a marker for HCC diagnosis (7). GPC3 and AFP may be transcriptionally
regulated by zinc-fingers and homeoboxes protein 2 (ZHX2) (8–10),
indicating that ZHX2 acts as an important transcriptional regulator
in HCC. Our previous study demonstrated that ZHX2 inhibits the
proliferation of HCC cells and the growth of xenograft tumors in
mice (11).
ZHX2 is a member of the ZHX family, which also
includes ZHX1 and ZHX3. The three members share a number of common
features, including expression in various tissues and localization
in the cell nucleus. These ZHX family members contain two
C2H2 zinc-finger motifs and four to five
homeodomains, appear to be transcriptional repressors, interact
with the α subunit of nuclear factor-Y (NF-YA) and form homodimers
or form heterodimers with other family members (12–17).
In addition, ZHX1 was observed to bind to DNA methyltransferases
(DNMT) 3B (18). NF-YA is a
subunit of NF-Y, which was shown to be involved in tumorigenesis
(19). In addition, DNMT3B and
ZHX2 are important in hepatocarcinogenesis (11,20).
Data from these studies indicate that ZHX1 may be involved in
tumorigenesis, including in hepatocarcinogenesis.
In the current study, the correlation between ZHX1
and HCC was investigated; thus, a recombinant eukaryotic expression
plasmid, pcDNA3-ZHX1, was constructed and transfected into
SMMC-7721 cells. The potential biological activity of ZHX1 in HCC
was also investigated.
Materials and methods
Reagents
SYBR®-Green Polymerase Chain Reaction
(PCR) Master mix, TRIzol reagent and Lipofectamine™ 2000 were
purchased from Invitrogen Life Technologies (Carlsbad, CA, USA).
ReverTra Ace quantitative PCR (qPCR) RT kit and KOD DNA polymerase
were obtained from Toyobo (Shanghai, China). 2X Taq DNA
polymerase, TIANgel Midi Purification kit and TIANprep Mini Plasmid
kit were obtained from Tiangen Biotech Co., Ltd. (Beijing, China).
DNA markers, pEASY-T1 vector and Escherichia coli
DH5α were purchased from Beijing TransGen Biotech Co., Ltd.
(Beijing, China). Restriction enzymes and T4 DNA ligase were from
Takara Bio, Inc. (Shiga, Japan). RPMI-1640 and fetal calf serum
(FCS) were obtained from Gibco-BRL (Carlsbad, CA, USA). Polyclonal
rabbit anti-ZHX1 antibody was from Abcam (Cambridge, MA, USA) and
the monoclonal mouse anti-β-actin antibody was from Sigma-Aldrich
(St. Louis, MO, USA). Cell counting kit-8 (CCK-8) was purchased
from Beyotime Institute of Biotechnology (Shanghai, China). The
SMMC-7721 liver cancer cell line was obtained from the Cell Bank of
Shanghai Institute of Cell Biology, Chinese Academy of Science
(Shanghai, China). PCR primers were synthesized by the Shanghai
Biosune Biotechnology Co., Ltd., (Shanghai, China).
Patients
This study was approved by the Ethics Committee of
the Provincial Hospital affiliated to Shandong University for
Clinical Investigation. The study included 12 patients with HCC who
underwent surgery for radical resection. Patients were excluded if
they had received any other therapy prior to the surgery. Specimens
of cancer liver tissue were obtained from the patients during
surgery, when written consent had been obtained. Specimens of
adjacent cirrhotic liver tissues were obtained as controls during
the same time period. Tissues were stored at −70ºC for qPCR. For
each patient, the diagnosis was confirmed by a paraffin
section.
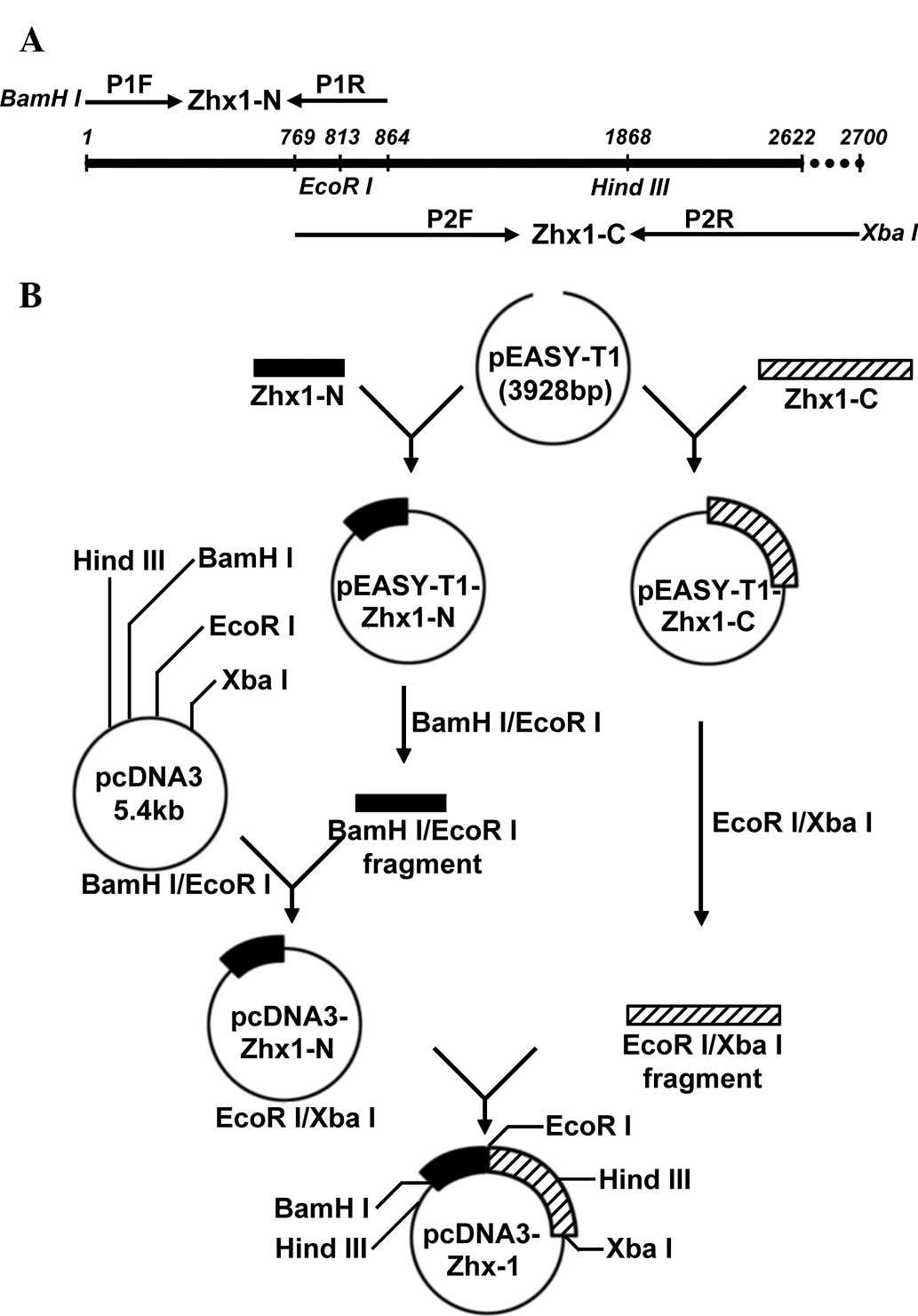
Primer design and gene amplification
The full length of the human ZHX1 (hZHX1) gene
coding sequence (CDS) is 2622 bp (GenBank accession no.
NM_001017926.2), which is difficult to obtain from one PCR
amplification. To acquire full-length hZHX1 complementary DNA
(cDNA), two pairs of primers were designed as follows: P1F
5′-CGCggatccATG GCAAGCAGGCGAAAAT-3′, which contained a BamHI
site and P1R, 5′-GGTGGGAATGCTATTAACAGGGATTAAG-3′ for the N-terminal
fragment of ZHX1 (ZHX1-N); P2F, 5′-CTTCCTGGATTGGCACAGGTGAT-3′ and
P2R, 5′-GCt ctagaCAAAGATTGGGCAAATTCACAG-3′, which contained an
XbaI site for the C-terminal fragment of ZHX1 (ZHX1-C).
Lowercase letters indicate restrictions sites. ZHX1-N and ZHX1-C
overlapped and contained an endogenous EcoRI site (Fig. 1A).
cDNA was synthesized from total RNA, which was
isolated from frozen paraneoplastic liver tissues. Using the cDNA
as a template, ZHX1-N and ZHX1-C gene fragments were amplified with
P1F/P1R and P2F/P2R, respectively. PCR mixtures were prepared in a
total volume of 50 μl, including 5.0 μl of 10X buffer, 5.0 μl dNTP
mixture (2 mM), 3.0 μl MgSO4 (25 mM), 1.5 μl of each
paired primer (10 μM), 2.0 μl template cDNA, 1.0 μl KOD DNA
polymerase (1.0 U/μl) and double distilled water. Parameters for
PCR were as follows: 5 min pre-denaturation at 94ºC; 35 cycles of
denaturation at 94ºC for 45 sec, annealing at 58ºC for 45 sec and
extension at 72ºC for 1 min (ZHX1-N) or 2 min (ZHX1-C), followed by
a final 10-min extension at 72ºC. To obtain poly(A) tails, the PCR
products were mixed with an equal volume of 2X Taq DNA
polymerase in a water bath at 72ºC for 30 min. The PCR products
were subjected to agarose gel electrophoresis and purified using
the TIANgel Midi Purification kit, according to the manufacturer’s
instructions.
TA cloning and sequencing
ZHX1-N and ZHX1-C were respectively cloned into the
pEASY-T1 vector by TA strategy resulting in pEASY-T1-ZHX1-N and
pEASY-T1-ZHX1-C. DNA sequencing was used to analyze the two ZHX1
gene fragments.
Generation of recombinant plasmid
pcDNA3-ZHX1
To obtain the recombinant expression vector
containing the full length ZHX1 gene, BamHI/EcoRI
fragments of pEASY-T1-ZHX1-N were subcloned into pcDNA3 to produce
pcDNA3-ZHX1-N. ZHX1-C was obtained by EcoRI and XbaI
digestion with pEASY-T1-ZHX1-C and subcloned into
EcoRI/XbaI of pcDNA3-ZHX1-N, yielding the recombinant
eukaryotic expression plasmid, pcDNA3-ZHX1.
The construction strategy of recombinant eukaryotic
expression plasmid pcDNA3-ZHX1 is presented in a schematic
representation (Fig. 1B).
Cell culture and transient
transfection
SMMC-7721 cells were cultured in RPMI-1640 medium
supplemented with 10% FCS, in an incubator at 37ºC, with 5%
CO2 and saturated humidity. The day prior to
transfection, 2.5×105 cells were inoculated in a 24-well
plate. When cells reached 70–80% confluency, transfections were
performed with pcDNA3 and pcDNA3-ZHX1, respectively, following the
Lipofectamine 2000 manufacturer’s instructions. Cells were
collected 48 h following transfection.
Reverse transcription-PCR (RT-PCR) and
qPCR for ZHX1 mRNA detection
RT-PCR was used to analyze the ZHX1 mRNA expression
in SMMC-7721 cells. Briefly, total RNA was extracted from
transfected SMMC-7721 cells using TRIzol reagent, according to the
manufacturer’s instructions. cDNA was subsequently synthesized from
the total RNA with the ReverTra Ace qPCR RT kit (Toyobo). Primers,
designed using the software Primer Premier v5.0 (Premier Biosoft,
Palo Alto, CA, USA), were as follows: Sense:
5′-GAAATCAAACCAGACCGTGAAGA-3′ and antisense:
5′-ATGCAGCATTGTAGGTGGGAA-3′ for ZHX1; and sense:
5′-AGTTGCGTTACACCCTTTC-3′ and antisense: 5′-CCTTCACCGTTCCAGTTT-3′
for β-actin. PCR amplifications were performed for 26 (β-actin ) or
30 (ZHX1) cycles of denaturation at 94ºC for 35 sec, annealing at
58ºC for 35 sec and extension at 72ºC for 35 sec, followed by a
final 10-min extension at 72ºC. PCR products were separated on 1.5%
agarose gel by electrophoresis and visualized by ethidium bromide
staining.
To quantify the ZHX1 mRNA expression in HCC
patients, qPCR was performed. Total RNA of liver tissue samples
from 12 HCC patients was prepared using TRIzol reagent and
converted to cDNA with the ReverTra Ace qPCR RT kit. qPCR was
performed in a mixture of 20 μl containing10 μl SYBR-Green PCR
Master mix, 1.5–3 μl cDNA and 0.4 μl of each paired primer (10 μM).
qPCR was performed on the LightCycler 2.0 Instrument (Roche
Diagnostics GmbH, Mannheim, Germany). Parameters were as follows: A
cycle of 95ºC for 30 sec and 40 consecutive cycles of 5 sec at
95ºC, 5 sec at 58ºC (for ZHX1 and β-actin) and 30 sec at 72ºC. The
expression levels of ZHX1 gene in HCC patients were normalized to
that of β-actin.
Western blot analysis for ZHX1 protein
detection
Transfected SMMC-7721 cells were collected and lysed
in cell lysis buffer. The protein concentration was measured with
the bicinchoninic acid assay. Following boiling for 10 min, cell
lyses with an equal quantity of protein were resolved by sodium
dodecyl sulfate-polyacrylamide gel electrophoresis and
electroblotted onto polyvinylidene fluoride membranes. ZHX1 was
immunoblotted with the rabbit anti-ZHX1 antibody as follows:
Membranes were blocked with phosphate-buffered saline buffer
containing 3% non-fat milk overnight, followed by incubation with
polyclonal rabbit anti-ZHX1 antibody (1:1500) at room temperature
for 1 h and subsequently incubated with the secondary horseradish
peroxidase-Immunoglobulin G antibody (1:10,000) at room temperature
for 1 h. After three washes, the immunoreactive proteins were
visualized by enhanced chemiluminescence.
Cell growth curve assay
To evaluate the effect of ZHX1 on SMMC-7721 cell
proliferation, a CCK-8 assay was employed. SMMC-7721 cells were
seeded into 96-well plates with 6,000 cells/well and transfected
with pcDNA3-ZHX1 (experimental group) or pcDNA3 (control group).
The cell number was quantified daily by CCK-8 according to the
manufacturer’s instructions. Cell absorbance was read using a Model
680 microplate reader (Bio-Rad Laboratories, Hercules, CA, USA)
with a 450 nm filter and expressed as optical density values.
Statistical analysis
GraphPad Prism (GraphPad Software, San Diego, CA,
USA) was used for data analysis. Two-way analysis of variance was
employed to analyze statistical differences between different
groups of SMMC-7721 cells at various time points. Wilcoxon matched
pairs test was applied to evaluate the differences between
cancerous and adjacent cirrhotic liver tissues from patients.
P<0.05 or P<0.001 was considered to indicate a statistically
significant difference.
Results
Cloning of hZHX1 CDS and identification
of recombinant plasmids
Two specific PCR amplifications were performed, one
with P1F/P1R and another with P2F/P2R. The PCR products were
subsequently analyzed by agarose gel electrophoresis. A band
between 750 and 1 kb (ZHX1-N) and another at ~2 kb (ZHX1-C) were
separated (Fig. 2A) and were
consistent with the expected molecular mass. ZHX-N and ZHX-C were
respectively cloned into pEASY-T1 vectors, which were identified by
restriction enzymes and DNA sequencing. The identified ZHX1-N and
ZHX1-C fragments were subcloned into a pcDNA3 vector to produce
pcDNA3-ZHX1. To confirm the construction of pcDNA3-ZHX1,
HindIII and BamHI/XbaI restriction enzymes and
agarose gel electrophoresis was used. Two DNA bands with 6.2 and
1.9 kb were obtained with HindIII digestion and two DNA
bands of 5.4 and 2.7 kb were obtained with BamHI and
XbaI digestion (Fig. 2B).
DNA sequencing further confirmed the successful construction of
pcDNA3-ZHX1 (data not shown).
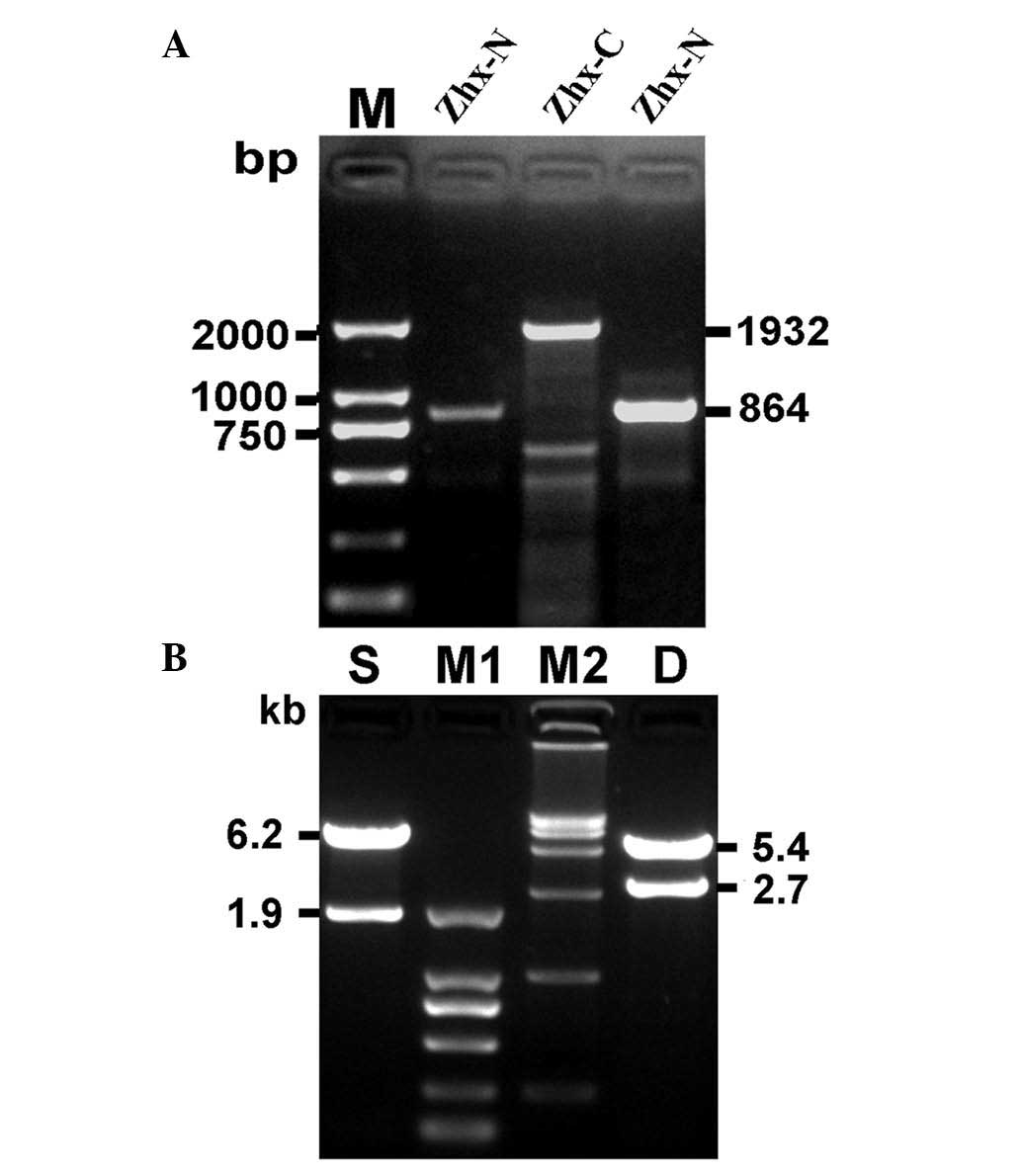 | Figure 2Cloning of hZHX1 coding sequence and
identification of recombinant plasmid pcDNA3-ZHX1. (A)
Electrophoresis of the target genetic fragments of hZHX1. (B)
Identification of the recombinant plasmid pcDNA3-ZHX1 by
single/double restriction endonuclease analysis. M, DNA marker
DL2,000; ZHX-N, a target fragment of 864 bp; ZHX-C, a target
fragment of 1932 bp; S, single restriction endonuclease analysis
with HindIII; M1, DNA marker DL2,000; M2, DNA marker
DL15,000; D, double restriction endonuclease analysis with
BamHI and XbaI. hZHX1, human zinc-fingers and
homeoboxes 1; ZHX-N, N-terminal ZHX; ZHX-C, C-terminal ZHX. |
Expression of the recombinant plasmid
pcDNA3-ZHX1 in HCC cells
Forty-eight hours after transfection, SMMC-7721
cells were collected for RT-PCR and western blot analysis. As shown
in Fig. 3A and B, SMMC-7721 cells
transfected with pcDAN3-ZHX1 showed significantly increased levels
of ZHX1 mRNA and protein expression, while cells without
transfection or transfected with empty plasmid pcDNA3 showed
low-level expression of endogenous ZHX1, indicating that a
functional eukaryotic expression plasmid pcDNA3-ZHX1 was
obtained.
Identification of biological activity by
cell growth curve assay
To investigate the involvement of ZHX1 in HCC, a
CCK-8 assay was employed to evaluate cell proliferation. As shown
in Fig. 4, SMMC-7721 cells
transfected with pcDNA3-ZHX1 exhibited a decreased proliferation
rate compared with cells transfected with pcDNA3, indicating that
the overexpression of ZHX1 may inhibit the proliferation of
SMMC-7721 cells.
Reduced ZHX1 expression among cancer
tissue from HCC patients
qPCR was used to evaluate the differential
expression of ZHX1 in adjacent cirrhotic and cancer liver tissue
from the same HCC patients. Following normalizing against the
house-keeping gene, 7/12 cases showed a 2–5-fold reduction of ZHX1
mRNA compared with the adjacent cirrhotic liver tissues (Fig. 5). A significant difference was
confirmed by Wilcoxon matched pairs test. These results
demonstrated that reduced ZHX1 expression was widespread among
cancer tissues from HCC patients and indicated that ZHX1 may be
responsible for hepatocarcinogenesis.
Discussion
The ZHX1 gene was originally cloned by
immunoscreening with a monoclonal B92 antibody (12). Similar to its family members, ZHX1
is expressed in various tissues, is localized in the cell nucleus
and appears to act as a transcriptional repressor (12,14,15).
It was previously reported that ZHX1, together with ZHX2 and ZHX3,
were involved in regulating gene expression in podocyte disease
(21,22). However, a number of studies
indicated that ZHX1 may be involved in tumorigenesis, including in
hepatocarcinogenesis (11,13,14,18–20).
In the current study, the expression vector of hZHX1 was
successfully prepared. Moreover, the present results demonstrated
the expression pattern and involvement of ZHX1 in HCC.
To determine the involvement of ZHX1 in HCC, a
vector containing the full length of hZHX1 CDS was required.
However, due to the size of hZHX1 CDS, more than one amplification
was necessary. A two-step strategy was employed to obtain the full
length hZHX1 gene. Using NEBCutter analysis, an EcoRI site,
one of the multiple cloning sites of pCDNA3, was identified in the
gene sequence of hZHX1. Therefore, two pairs of primers were
designed to obtain two partial genetic fragments of hZHX1; ZHX1-N
and ZHX1-C, which had an overlapping sequence including the
EcoRI site. The two partial genetic fragments were subcloned
into the EcoRI site of pcDNA3, and pcDNA3-ZHX1 was
successfully constructed and expressed in SMMC-7721 cells.
With the successful construction of pcDNA3-ZHX1, the
potential involvement of ZHX1 in HCC was evaluated for the first
time. Results of the CCK-8 assay showed that overexpression of ZHX1
may inhibit the proliferation of SMMC-7721 cells. This is
consistent with the detected ZHX1 expression pattern in HCC.
Compared with the adjacent cirrhotic tissues, ZHX1 expression was
significantly lower in cancer tissues from patients with HCC
(Fig. 5).
As a member of the ZHX family, ZHX1 contains two
zinc finger and five homeobox domains, indicating the potential of
ZHX1 interaction with DNA and proteins. Proteins known to interact
with ZHX1 include ZHX2 (13),
NF-YA (14) and DNMT 3B (18). ZHX2 prevents AFP expression but
also inhibits the proliferation of HCC cells (10,11).
NF-YA is a subunit of NF-Y, which is an important transcription
factor capable of binding to the CCAAT box to trigger transcription
of numerous eukaryotic genes and is involved in tumorigenesis
(19). DMNT3B is important in the
development of tumorigenesis (23), including hepatocarcinogenesis
(20,24). The interaction of ZHX1 with these
tumor-associated proteins indicates that ZHX1 may be involved in
hepatocarcinogenesis. However, the molecular mechanism underlying
the involvement of ZHX1 in the pathology of hepatocarcinogenesis
remains unclear, thus further studies are required.
In conclusion, the recombinant eukaryotic expression
plasmid, pcDNA3-ZHX1, was successfully constructed. It contained
the full length sequence of the hZHX1 gene and demonstrated that
overexpression of ZHX1 may inhibit the proliferation of SMMC-7721
cells. This study provides preliminary data for further studies
regarding the function and molecular mechanism of the ZHX1 gene in
HCC.
Acknowledgements
This study was supported in part by grants from the
National Science Foundation of China (grant no. 30972753), the
Program for NCET-10-0524 and Shandong Provincial Nature Science
Foundation for Distinguished Young Scholars (grant no. JQ200907).
The authors would like to thank the staff at the Institute of
Immunology, Shandong University School of Medicine, who contributed
to this study.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Bosch FX, Ribes J, Díaz M and Cléries R:
Primary liver cancer: worldwide incidence and trends.
Gastroenterology. 127(5 Suppl 1): S5–S16. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zender L, Villanueva A, Tovar V, Sia D,
Chiang DY and Llovet JM: Cancer gene discovery in hepatocellular
carcinoma. J Hepatol. 52:921–929. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mínguez B and Lachenmayer A: Diagnostic
and prognostic molecular markers in hepatocellular carcinoma. Dis
Markers. 31:181–190. 2011.
|
|
5
|
Sakamoto M, Effendi K and Masugi Y:
Molecular diagnosis of multistage hepatocarcinogenesis. Jpn J Clin
Oncol. 40:891–896. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gomaa AI, Khan SA, Leen EL, Waked I and
Taylor-Robinson SD: Diagnosis of hepatocellular carcinoma. World J
Gastroenterol. 15:1301–1314. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Forner A, Reig M and Bruix J:
Alpha-fetoprotein for hepatocellular carcinoma diagnosis: the
demise of a brilliant star. Gastroenterology. 137:26–29. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Perincheri S, Dingle RW, Peterson ML and
Spear BT: Hereditary persistence of alpha-fetoprotein and H19
expression in liver of BALB/cJ mice is due to a retrovirus
insertion in the Zhx2 gene. Proc Natl Acad Sci USA. 102:396–401.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Morford LA, Davis C, Jin L, Dobierzewska
A, Peterson ML and Spear BT: The oncofetal gene glypican 3 is
regulated in the postnatal liver by zinc fingers and homeoboxes 2
and in the regenerating liver by alpha-fetoprotein regulator 2.
Hepatology. 46:1541–1547. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shen H, Luan F, Liu H, Gao L, Liang X,
Zhang L, Sun W and Ma C: ZHX2 is a repressor of alpha-fetoprotein
expression in human hepatoma cell lines. J Cell Mol Med.
12:2772–2780. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yue X, Zhang Z, Liang X, et al: Zinc
fingers and homeoboxes 2 inhibits hepatocellular carcinoma cell
proliferation and represses expression of Cyclins A and E.
Gastroenterology. 142:1559–1570. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barthelemy I, Carramolino L, Gutiérrez J,
Barbero JL, Márquez G and Zaballos A: zhx-1: a novel mouse
homeodomain protein containing two zinc-fingers and five
homeodomains. Biochem Biophys Res Commun. 224:870–876. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kawata H, Yamada K, Shou Z, Mizutani T,
Yazawa T, Yoshino M, Sekiguchi T, Kajitani T and Miyamoto K:
Zinc-fingers and homeoboxes (ZHX) 2, a novel member of the ZHX
family, functions as a transcriptional repressor. Biochem J.
373:747–757. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamada K, Printz RL, Osawa H and Granner
DK: Human ZHX1: cloning, chromosomal location, and interaction with
transcription factor NF-Y. Biochem Biophys Res Commun. 261:614–621.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamada K, Kawata H, Matsuura K, Shou Z,
Hirano S, Mizutani T, Yazawa T, Yoshino M, Sekiguchi T, Kajitani T
and Miyamoto K: Functional analysis and the molecular dissection of
zinc-fingers and homeoboxes 1 (ZHX1). Biochem Biophys Res Commun.
297:368–374. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yamada K, Kawata H, Shou Z, Hirano S,
Mizutani T, Yazawa T, Sekiguchi T, Yoshino M, Kajitani T and
Miyamoto K: Analysis of zinc-fingers and homeoboxes
(ZHX)1-interacting proteins: molecular cloning and characterization
of a member of the ZHX family, ZHX3. Biochem J. 373:167–178. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hirano S, Yamada K, Kawata H, et al: Rat
zinc-fingers and homeoboxes 1 (ZHX1), a nuclear
factor-YA-interacting nuclear protein, forms a homodimer. Gene.
290:107–114. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim SH, Park J, Choi MC, Kim HP, Park JH,
Jung Y, Lee JH, Oh DY, Im SA, Bang YJ and Kim TY: Zinc-fingers and
homeoboxes 1 (ZHX1) binds DNA methyltransferase (DNMT) 3B to
enhance DNMT3B-mediated transcriptional repression. Biochem Biophys
Res Commun. 355:318–323. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dolfini D, Gatta R and Mantovani R: NF-Y
and the transcriptional activation of CCAAT promoters. Crit Rev
Biochem Mol Biol. 47:29–49. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fan H, Chen L, Zhang F, et al: MTSS1, a
novel target of DNA methyltransferase 3B, functions as a tumor
suppressor in hepatocellular carcinoma. Oncogene. 31:2298–2308.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu G, Clement LC, Kanwar YS, Avila-Casado
C and Chugh SS: ZHX proteins regulate podocyte gene expression
during the development of nephrotic syndrome. J Biol Chem.
281:39681–39692. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Clement LC, Liu G, Perez-Torres I, Kanwar
YS, Avila-Casado C and Chugh SS: Early changes in gene expression
that influence the course of primary glomerular disease. Kidney
Int. 72:337–347. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Baylin SB, Esteller M, Rountree MR,
Bachman KE, Schuebel K and Herman JG: Aberrant patterns of DNA
methylation, chromatin formation and gene expression in cancer. Hum
Mol Genet. 10:687–692. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Datta J, Kutay H, Nasser MW, et al:
Methylation mediated silencing of MicroRNA-1 gene and its role in
hepatocellular carcinogenesis. Cancer Res. 68:5049–5058. 2008.
View Article : Google Scholar : PubMed/NCBI
|