Introduction
Bone formation is a complex process that involves
the recruitment of progenitor cells, proliferation and
differentiation of these cells into osteoblasts, and the resultant
secretion of abundant bone extracellular matrix proteins that
coordinates the mineralization process (1,2).
Differentiation into cells of the osteoblast lineage is dependent
upon multiple extracellular cues received by the cell, and involves
complex pathways regulated at the transcriptional and
posttranscriptional levels. However, the regulation of these
pathways is not fully understood. Bone-anabolic agents, such as
bone morphogenetic protein (BMP)-2, parathyroid hormone and
canonical Wnts promote osteoblast differentiation by stimulating
distinct second messenger pathways (3). The canonical Wnt signaling pathway is
crucial for the regulation of bone formation and osteoblastic
differentiation in humans (4,5). In
addition, we have previously demonstrated that canonical Wnt
signaling induces the expression of osteoprotegerin (6).
MicroRNAs (miRNAs) are small non-coding RNAs that
are ~22 nucleotides in length. One strand of this short-lived
duplex is degraded by an unknown nuclease, while the other strand
is selected and incorporated into the effector complex termed the
RNA-induced silencing complex (RISC). RISC interacts with the mRNA
target in a sequence-specific manner and regulates specific target
genes by repressing their translation and/or promoting the
degradation of their transcribed mRNAs by binding to their
3′-untranslated regions (7–9).
More than 1,000 miRNAs have been discovered in mammals, a number of
which are expressed in a tissue-specific manner (10). An accumulating number of studies
have suggested that the regulation of cell differentiation by
miRNAs is a notable component of the regulatory machinery. In a
previous study, striated-muscle-specific miR-206 was shown to be
downregulated by BMP-2 at the post-transcriptional level in C2C12
cells (11). In addition, several
studies have demonstrated that certain miRNAs act as important
orchestrators of osteoblast-specific genes that are required for
osteoblast differentiation, acting through their regulation of
diverse signaling molecules and pathways (12–19).
In the present study, microRNA profiling studies
were conducted during the induction of osteoblast differentiation
by canonical Wnt signaling. miR-34b-5p and miR-34c were identified
to be upregulated by the activation of canonical Wnt signaling in
C2C12 cells. Thus, the present study also investigated the
involvement of miR-34b/c throughout osteoblast differentiation.
During the maturation of preosteoblasts, such as MC3T3-E1 cells,
these miRNAs are upregulated at the mineralization stage. These
results indicated that miR-34b/c regulated osteoblast
differentiation, controlling the expression of osteocalcin and
alkaline phosphatase (ALP).
Materials and methods
Cell cultures
The C2C12 mouse myoblast cell line and the MC3T3-E1
mouse calvarial osteoblast cell line were obtained as described
previously (20). The Kusa-O and
ST-2 mouse mesenchymal cell lines were obtained from the RIKEN Cell
Bank (Tsukuba, Japan). Wnt3a-C2C12 cells were established as
described previously (20). Cells
were cultured in α-minimum essential medium (MEM) supplemented with
10% fetal bovine serum (SAFC Biosciences, Inc., Lenexa, KS, USA) at
37°C in a 5% CO2 humidified atmosphere. For osteoblastic
differentiation, the cells were cultured in α-MEM containing 5 mM
β-glycerophosphate, 100 μg/ml ascorbic acid and 10−8 M
dexamethasone (Sigma-Aldrich, St. Louis, MO, USA) for 1–3 weeks.
The culture medium was replaced every 3 days.
miRNA microarray and data analysis
miRNA was extracted from the cells using the mirVana
miRNA isolation kit (Ambion, Austin, TX, USA). Labeled miRNAs were
hybridized onto 3D-Gene miRNA oligo chips containing >700
antisense probes (Toray Industries Inc., Tokyo, Japan). The
microarray images obtained were analyzed using Genepix Pro™ 4.0
software (Molecular Devices, Sunnyvale, CA, USA). Differences in
the total fluorescence intensity between arrays were adjusted by
global normalization. The mean values for duplicate microarrays
were calculated and used for comparison between the groups. When
the difference in relative miRNA expression between the two groups
was >2.0-fold, this was defined as a change in the
expression.
Transfection assay
The antisense inhibitors were designed to bind to
specific endogenous miRNAs and inhibit their activities when
introduced into cells. The following 5′- and 3′-phosphorylated RNAs
with full 2′-O-methyl modifications were chemically
synthesized by Nippon Bio Services Co. Ltd. (Asaka, Japan):
5′-ACAAUCAGCUAAUUACACUGCCU-3′ for antimiR-34b5p;
5′-GCAAUCAGCUAACUACACUGCCU-3′ for antimiR-34c; and
5′-UCAGCAGCACAGUCAAUACUGG-3′ for antimiR-16. The antimiRNAs were
transfected into C2C12 cells using lipofectamine 2000 cationic
liposomes (Invitrogen Life Technologies, Carlsbad, CA, USA)
according to the manufacturer's instructions and were then cultured
for 2 days. AntimiR-16 served as a control for nonspecific
effects.
Quantitation of miRNA expression
To analyze the miRNA level, RNA was extracted from
the cells using a mirVana miRNA isolation kit (Ambion) and cDNA was
synthesized with specific miRNA primers from the TaqMan
MicroRNA assay (Applied Biosystems, Carlsbad, CA, USA). The
resulting cDNA was amplified by PCR using the TaqMan
MicroRNA assay system with the StepOne® Real Time PCR
system (Applied Biosystems) (21).
The relative expression level of miRNA was quantified using the
comparative CT method with sno234 RNA as the
endogenous control.
Quantitation of mRNA expression by
quantitative polymerase chain reaction (qPCR)
To analyze the mRNA expression level, qPCR was
performed using assay-on-demand TaqMan probes (Applied
Biosystems) with the StepOne® real time PCR system
(Applied Biosystems). The relative gene expression level was
quantified using the comparative CT method with
glyceraldehyde 3-phosphate dehydrogenase as the endogenous
control.
ALP activity and ALP staining
Cells were washed twice with phosphate-buffered
saline, 200 μl lysis buffer was added to the cell layer and the
cells were incubated on ice for 5 min. The cell lysate was then
sonicated for 1 min (Virtis Virsonic; Virtis Co., Gardiner, NY,
USA) and centrifuged at 1,000 × g and 4°C for 10 min (MX-300; Tomy,
Tokyo, Japan). ALP activity was assayed by a spectrophotometric
method using a LabAssay™ ALP kit (Wako Pure Chemical Industries
Ltd., Osaka, Japan). The absorbance of each well at 405 nm was
measured with a microplate reader (iMark; Bio-Rad, Richmond, CA,
USA).
Statistical analysis
All experiments were repeated three to four times
and representative results are shown. The data are presented as the
mean ± SD, and were analyzed by Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of miRNAs in Wnt3a-C2C12
cells
C2C12 is a multipotent cell line that provides a
well-characterized model system and has been demonstrated to
differentiate into myotubes and osteoblasts. To investigate the
involvement of canonical Wnt signaling in osteoblastic
differentiation, we previously generated a C2C12 cell line stably
expressing Wnt3a (20). C2C12
cells were transfected with the Wnt3a expression plasmid to
activate canonical Wnt signaling and were then selected using G418
to establish a stable cell line, termed Wnt3a-C2C12 (20). To confirm the activation of
canonical Wnt signaling in these cells, the reporter plasmid that
carries six tandem repeats of the Lef1/Tcf binding site was
transfected with Topflash, which demonstrated that the promoter
activity of Topflash is enhanced in Wnt3a-C2C12 cells. Previously,
ALP activity was observed, which indicated that canonical Wnt
signaling is active in these cells (20). Therefore, in the present study,
total RNA was extracted from Wnt3a-C2C12 and C2C12 cells, and the
expression of miRNAs was analyzed using an established microarray
platform to determine the potential involvement of miRNAs in
canonical Wnt signaling-mediated osteoblastic differentiation. From
the miRNAs that were expressed at significant levels and showed
changes in expression in response to Wnt signaling, five miRNAs
were identified, which were upregulated in Wnt3a-C2C12 cells
compared with C2C12 cells. Two of the miRNAs were downregulated by
Wnt3a stimulation, while in the upregulated group, miR-34b-5p,
miR-34b-3p and miR-34c were among the most markedly changed. These
changes in expression of miR-34b-5p and miR-34c in Wnt3a-C2C12 and
C2C12 cells were confirmed by qPCR. As expected, expression of
miR-34b-5p and miR-34c was upregulated in Wnt3a-C2C12 cells
compared with that in the vehicle-transfected C2C12 cells (Fig. 1). No regulation of non-specific
miR-16 expression was observed in the Wnt3a-C2C12 cells. However,
miR-206 was highly expressed in the C2C12 cells, whereas its
expression was lower in the Wnt3a-C2C12 cells (Fig. 1). These results indicated that the
canonical Wnt signaling pathway may be involved in regulating the
expression of miR-34b-5p and miR-34c.
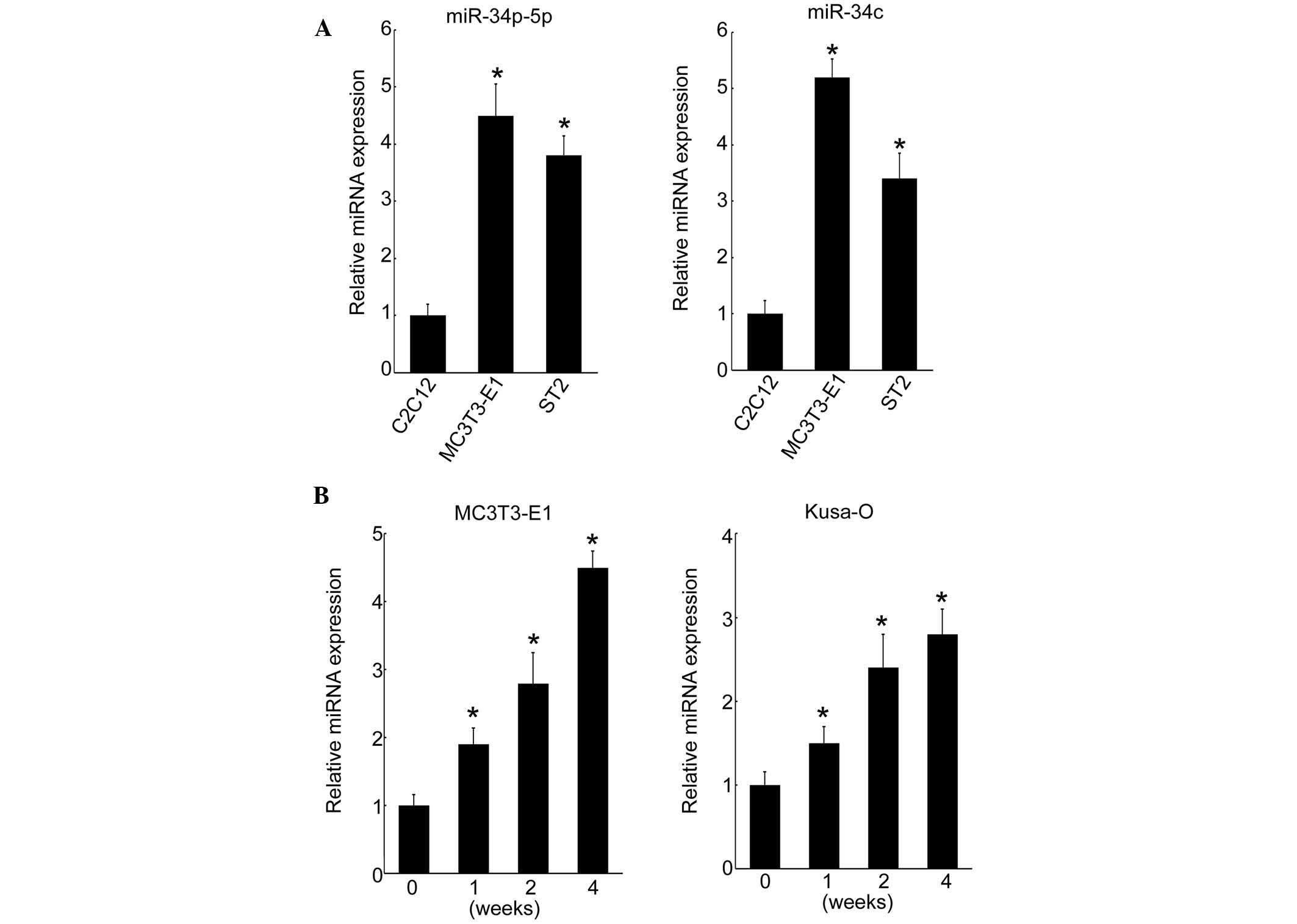
Cell type-specific expression of
miR-34b-5p and miR-34c
The cell type-specific expression of miR-34b-5p and
miR-34c was investigated in a myoblastic cell line and two
osteoblastic cell lines. In MC3T3-E1 and ST-2 osteoblastic cells,
miR-34b-5p and miR-34c were highly expressed compared with that in
myoblastic C2C12 cells (Fig. 2A).
By contrast, miR-16 expression levels were not altered (data not
shown). MC3T3-E1 cells derived from fetal mouse calvaria undergo
maturation when cultured in osteogenic medium. In the present
study, it was demonstrated that the expression of miR-34b-5p and
miR-34c was induced by canonical Wnt signaling, which suggests that
these miRNAs may act as stimulators of osteoblastic
differentiation. To understand the involvement of miR-34b/c
expression in osteoblastic differentiation, the effect of cell
culture on the level of miRNA expression over time was
investigated. Expression of miR-34c increased over the culture
period from day 0 and reached maximum levels at four weeks in
MC3T3-E1 cells (Fig. 2B). This
pattern of expression of miRNA was confirmed in the Kusa-O mouse
mesenchymal cell line, with an increase up to four weeks when
proliferation ceased in the multilayered cell nodules (Fig. 2B). These results indicated that
miR-34b/c was induced during osteoblastic differentiation in
vitro.
Expression of osteoblastic genes
following transfection with anti-miRNA
To further analyze the function of miRNAs in
osteoblastic differentiation, anti-miRNA was transfected and the
expression of osteoblastic genes was measured. Following the
transfection of MC3T3-E1 cells with anti-miR34c, the expression
levels of miR-34b-5p and miR-34c significantly diminished to
undetectable levels (Fig. 3A). In
these cells, the expression of specific genes that characterize
osteoblastic differentiation (such as osteocalcin) increased,
whereas the expression levels of ALP mRNA were reduced by miR34
inhibition (Fig. 3B). In addition,
ALP activity declined following knockdown of miR-34 in MC3T3-E1
cells (Fig. 3C), supporting the
hypothesis that miR-34 regulates osteoblastic gene expression.
Discussion
To date, several studies have demonstrated a
regulatory role of miRNAs in osteoblast differentiation (12–19).
Certain miRNAs have also been shown to act as negative regulators
of osteoblast differentiation, such as miR-26a and miR-125b. In the
present study a novel miRNA that regulates canonical Wnt signaling
was investigated. Canonical Wnt signaling induces miR-34b/c
expression.
Three miR-34s have been identified, miR-34b and
miR-34c are encoded by a gene located on mouse chromosome 9, and
miR-34a is encoded by a gene located on mouse chromosome 4.
miR-34b-5p and miR-34b-3p are transcribed in tandem, in the same
primary miRNA (22). In general,
miRNAs are transcribed by RNA polymerase II or III, and
transcription is regulated by the interaction of trans-acting
factors with the promoter region (23). According to the current model of
canonical Wnt action, glycogen synthase kinase (GSK)-3β
phosphorylates β-catenin and thereby induces rapid degradation of
β-catenin in cells that lack Wnt signaling. Wnt inhibits GSK-3β,
thus stabilizing β-catenin, which interacts with several molecules
in the cytosol, including lymphoid enhancer factor 1/T cell factor
(Lef1/Tcf). A complex involving the transcription factor Lef1/Tcf
and β-catenin regulates the expression of several target genes via
Lef1/Tcf binding sites within gene promoter regions (24). The results suggested that the
miR-34b/c gene promoter region may contain an Lef1/Tcf binding site
that responds to canonical Wnt signaling.
A previous study indicated that human miR-29a
expression is induced by canonical Wnt signaling and that miR-29a
subsequently downregulates the Wnt signaling antagonists Dkk1,
Kremen2 and sFRP2, potentiating Wnt signaling (13). These two actions promote a gene
expression program that is required for osteoblast differentiation.
In the present study, miR-34b/c and miR-29a expression increased in
Wnt3a-C2C12 cells, as shown by microarray analysis (data not
shown). However the correlation between miR-34b/c and miR-29a is
not clear when canonical Wnt signaling is activated in osteoblasts.
An advantage of using multiple miRNAs may be the provision of a
variety of options for biological regulation in cells.
It has been demonstrated that p53 transactivates
miR-34, which then suppresses Lef1/Tcf complexes by targeting the
canonical Wnt pathway (25).
Furthermore, Wei et al(26)
demonstrated that miR34b/c affects skeletogenesis during embryonic
development and influences bone mass accrual following birth. It
was suggetsed that two molecular mechanisms may be involved.
Initially, miR-34b/c inhibits the osteoblast proliferation by
suppressing CyclinD1, CDK4 and CDK6 accumulation, and miR34b/c then
inhibits terminal differentiation of osteoblasts through the
inhibition of SATB2, a nuclear matrix protein (26). Similar to the results of the
present study, Wei et al demonstrated that osteocalcin
expression was increased in miR-34−/− osteoblasts. In our study,
inhibition of miR34b/c downregulated the expression of ALP, which
is a marker of osteoblastic differentiation. The exact mechanisms
by which miR-34b/c regulates gene expression or its function during
osteoblastic differentiation remains to be elucidated. miRNAs
provide a mechanism for fine-tuning intricate cellular processes.
In combination with a variety of signaling molecules and
transcription factors, miRNAs may aid in controlling the complex
program of osteoblastic differentiation.
The present study supports the hypothesis that
canonical Wnt signaling regulates miR-34b/c expression and in turn
regulates gene expression by altering cellular function and/or
signaling activities, thus controlling osteoblastic differentiation
in bone tissue. This regulatory circuit provides additional insight
into the manner in which canonical Wnt signaling modulates the
expression of miRNAs during osteoblast differentiation and may be
of value in the future, aiding in the development of novel
therapies to improve bone mass in patients with bone disorders.
Acknowledgements
This study was supported in part by the Japan
Ministry of Education, Culture, Sports, Science and Technology
Grants-in-aid (grant no. 22390346) (MT).
References
|
1
|
Young MF: Bone matrix proteins: more than
markers. Calcif Tissue Int. 72:2–4. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ortega N, Behonick DJ and Werb Z: Matrix
remodeling during endochondral ossification. Trends Cell Biol.
14:86–93. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Canalis E: Update in new anabolic
therapies for osteoporosis. J Clin Endocrinol Metab. 95:1496–1504.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krishnan V, Bryant H and Macdougald OA:
Regulation of bone mass by Wnt signaling. J Clin Invest.
116:1202–1209. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tamura M, Nemoto E, Sato MM, Nakashima A
and Shimauchi H: Role of the Wnt signaling pathway in bone and
tooth. Front Biosci (Elite Ed). 2:1405–1413. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sato M, Nakashima A, Nashimoto M, Yawaka Y
and Tamura M: Bone morphogenetic protein-2 enhances
Wnt/beta-catenin signaling-induced osteoprotegerin expression.
Genes Cells. 14:141–153. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Filipowicz W, Bhattacharyya S and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ding XC, Weiler J and Grosshans H:
Regulating the regulators: mechanisms controlling the maturation of
microRNAs. Trends Biotechnol. 27:27–36. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sayed D and Abdellatif M: MicroRNAs in
development and disease. Physiol Rev. 91:827–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Williams AE: Functional aspects of animal
microRNAs. Cell Mol Life Sci. 65:545–562. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sato M, Nashimoto M, Katagiri T, Yawaka Y
and Tamura M: Bone morphogenetic protein-2 down-regulates miR-206
expression by blocking its maturation process. Biochem Biophys Res
Commun. 383:125–129. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kapinas K, Kessler CB and Delany AM:
miR-29 suppression of osteonectin in osteoblasts: regulation during
differentiation and by canonical Wnt signaling. J Cell Biochem.
108:216–224. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kapinas K, Kessler C, Ricks T, Gronowicz G
and Delany AM: miR-29 modulates Wnt signaling in human osteoblasts
through a positive feedback loop. J Biol Chem. 285:25221–25231.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schoolmeesters A, Eklund T, Leake D, et
al: Functional profiling reveals critical role for miRNA in
differentiation of human mesenchymal stem cells. PLoS One.
4:e56052009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Itoh T, Nozawa Y and Akao Y: MicroRNA-141
and −200a are involved in bone morphogenetic protein-2-induced
mouse pre-osteoblast differentiation by targeting distal-less
homeobox 5. J Biol Chem. 284:19272–19279. 2009.
|
|
16
|
Hu R, Liu W, Li H, et al: A
Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse
osteoblast differentiation. J Biol Chem. 286:12328–12339. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hassan MQ, Gordon JA, Beloti MM, et al: A
network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster
regulates the osteoblast differentiation program. Proc Natl Acad
Sci USA. 107:19879–19884. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Eskildsen T, Taipaleenmäki H, Stenvang J,
et al: MicroRNA-138 regulates osteogenic differentiation of human
stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci USA.
108:6139–6144. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Xie RL, Croce CM, et al: A
program of microRNAs controls osteogenic lineage progression by
targeting transcription factor Runx2. Proc Natl Acad Sci USA.
108:9863–9868. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nakashima A, Katagiri T and Tamura M:
Cross-talk between Wnt and bone morphogenetic protein 2 (BMP-2)
signaling in differentiation pathway of C2C12 myoblasts. J Biol
Chem. 280:37660–37668. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Uyama M, Sato MM, Kawanami M and Tamura M:
Regulation of osteoblastic differentiation by the proteasome
inhibitor bortezomib. Genes Cells. 17:548–558. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Landgraf P, Rusu M, Sheridan R, et al: A
mammalian microRNA expression atlas based on small RNA library
sequencing. Cell. 29:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee Y, Kim M, Han J, et al: MicroRNA genes
are transcribed by RNA polymerase II. EMBO J. 23:4051–4060. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yochum G, McWeeney S, Rajaraman V, Cleland
R, Peters S and Goodman RH: Serial analysis of chromatin occupancy
identifies beta-catenin target genes in colorectal carcinoma cells.
Proc Natl Acad Sci USA. 104:3324–3329. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim NH, Kim HS, Kim NG, et al: p53 and
microRNA-34 are suppressors of canonical Wnt signaling. Sci Signal.
4:ra712011.PubMed/NCBI
|
|
26
|
Wei J, Shi Y, Zheng L, et al: miR-34s
inhibit osteoblast proliferation and differentiation in the mouse
by targeting SATB2. J Cell Biol. 197:509–521. 2012. View Article : Google Scholar : PubMed/NCBI
|