Introduction
Colorectal carcinoma (CRC) is one of the most common
types of malignant gastrointestinal tumors. It is the third most
common type of cancer in the United States (1) and affects ~1 million people each
year, with a 5-year survival rate of 62%. Although surgery,
radiotherapy and chemotherapy have been widely used in the
treatment of CRC, they are limited in preventing the progression of
the disease and reducing mortality rates. Advances in human genome
studies have revealed that gene alterations are key in the
oncogenesis and progression of CRC (2,3).
Cip1 interacting zinc finger protein 1 (CIZ1) is a
nuclear protein and is encoded by 17 exons that map to chromosome
9q34 (4). It was initially
isolated in 1999 (5) and is
reported to be important in mammalian DNA replication and
transcription (6). CIZ1 was
previously shown to be expressed in several cancer cell lines,
including HeLa cells and ZR-75 human breast ductal carcinoma cells,
and was suggested to be involved in the pathogenesis of lung
cancer, lymphoma, pancreatic cancer and a number of
estrogen-related tumors (7,8). In
addition, CIZ1 was observed to bind to the N-terminal region of
p21Cip1/Waf1 through the first zinc finger motif and to
subsequently result in translocation from the nucleus to the
cytoplasm. p21Cip1/Waf1 is a downstream target of the
P53 tumor suppressor gene (9) and
has been suggested to be one of the primary causes of CRC
progression (9,10). However, the involvement of CIZ1 in
human CRC has not previously been addressed.
In the current study, CIZ1 expression in several
human CRC cell lines was examined and the effect of CIZ1 on cell
proliferation, cell cycle and apoptosis of one of these lines, RKO,
was investigated.
Materials and methods
Cell culture
Five CRC cell lines, RKO, DLD-1, HCT116, SW620 and
LOVO, were obtained from American Type Cell Culture (ATCC;
Manassas, VA, USA) and cultured in ATCC-recommended media in a
humidified incubator at 5% CO2 and 37°C. For lentiviral
transfection, RKO cells were resuspended in 0.25% trypsin and
plated in six-well plates (2,000 cells/well). For colony formation,
RKO cells were allowed to proliferate for 14 days. Following
washing with phosphate-buffered saline (PBS), cells were treated
with Giemsa for 20 min and washed three times with
ddH2O. Images were captured with a fluorescence
microscope (Micropublisher 3.3RTV; Olympus Corporation, Shibuya,
Japan) and the colonies were counted.
Transfection
Small interfering RNA (siRNA) sequences targeting
human CIZ1 (GenBank accession no. GCG-291708) were designed
by Genechem Co., Ltd. (Shanghai, China) using the following
template: GCACTTAGTGCTGCAACAGAA. Following confirmation by
sequencing, the sequences were cloned into a pGCSIL-GFP vector
(GeneChem, Shanghai, China) to generate CIZ1-siRNA lentiviral
vectors, which were then used to infect RKO cells. The CIZ1
lentiviral vector-transfected and non-transfected cells were
included as a control. CIZ1 expression was measured by green
fluorescent protein (GFP) expression using fluorescence microscopy
(Micropublisher 3.3RTV)three days following infection; and the
knockdown efficiency was measured by quantitative PCR (qPCR) and
western blot analysis 5 days following infection.
qPCR
Total RNA was isolated by the TRIzol method
(Invitrogen Life Technologies, Carlsbad, CA, USA). First-strand
cDNA was generated by two-step qPCR using the following primers:
Forward: 5′-GCCAAACAATCC TTGCGAC-3′ and reverse: 5′-CAACCCACAGCGTCC
ACT-3′ for CIZI; and forward: 5′-TGACTTCAACAGCG ACACCCA-3′ and
reverse: 5′-CACCCTGTTGCTGTAGC CAAA-3′ for GAPDH. qPCR
comprised of an initial denaturation step at 95°C for 15 sec, 45
cycles at 95°C for 5 sec and 60°C for 30 sec. Results are presented
as Ct values, defined as the threshold PCR cycle number at which an
amplified product was first detected. The average Ct was calculated
for CIZ1 and GAPDH and ΔCt was determined
as the mean of the triplicate Ct values for CIZ1 minus the
mean of the triplicate Ct values for GAPDH. The
2−(ΔΔCt) method was used to analyze the relative changes
in gene expression. All samples were examined at least three
times.
Western blot analysis
Cells were harvested in RIPA lysis buffer that was
supplemented with protease and phosphatase inhibitor cocktails
(Shanghai Chemical Reagent Co., Ltd., Shanghai, China). Proteins
were separated by sodium dodecyl sulfate-polyacrylamide gel
electrophoresis, transferred onto polyvinylidene fluoride membranes
and incubated with the following antibodies: Anti-CIZ1 (1:3,000;
Sigma-Aldrich St. Louis, MO, USA) and anti-GAPDH (1:5,000;
Santa-Cruz Biotechnology, Inc., Santa Cruz, CA, USA). Secondary
antibodies conjugated to horseradish peroxidase and enhanced
chemiluminescence western blotting reagents were used for
detection.
Cellomics method
Cellomics Whole Cell Stains (Thermo Scientific,
Waltham, MA, USA) provide excellent staining for high content
screening (HCS) assays and fluorescence microscopy (Micropublisher
3.3RTV). Cell growth was measured via multiparametric HCS. Briefly,
RKO cells at 10 days after infection with either CIZ1-siRNA
lentivirus and NC lentivirus were seeded at 2,000 cells per well in
96-well plates, then incubated at 37°C with 5% CO2 for
five days. Plates were processed with the ArrayScan HCS software
(Thermo Scientific) and kept at +4°C for up to 24 h before each
day’s analysis. The system is a computerized, automated
fluorescence-imaging microscope that automatically identifies
stained cells and reports the intensity and distribution of
fluorescence in each individual cell. Images were acquired for each
fluorescence channel, using suitable filters and a 20× objective.
In each well, at least 800 cells were analyzed. Images and data
were stored in a Microsoft SQL database (Microsoft, Remond, WA,
USA) for easy retrieval.
Flow cytometry
Cells were collected when the cell density had
reached 80%. For cell cycle detection, cells were washed twice with
ice-cold PBS, fixed with 70% ice-cold ethanol and stained with
propidium iodide. For cell apoptosis analysis, cells were stained
with Annexin V-APC at room temperature in the dark for 10–15 min
and detected by flow cytometry (FACSCalibur; Becton Dickinson,
Franklin Lakes, NJ, USA). Cells were analyzed using flow cytometry.
All experiments were repeated at least three times.
Statistical analysis
Values are expressed as the mean ± SEM of these
observations. Statistical analysis was performed using SPSS
software (Release 13.0, SPSS Inc., Chicago, IL, USA). The
difference between the two groups was analyzed by Student’s t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
CIZ1 was highly expressed in RKO CRC
cells and was successfully knocked down by CIZ1-siRNA
The expression of CIZ1 was clearly detected in all
five CRC cell lines examined (RKO, DLD-1, HCT116, SW620 and LOVO
cells; Fig. 1A). Among these, the
expression level of CIZ1 in RKO cells was significantly higher
compared with the other cell lines; thus, RKO cells were used for
subsequent experiments. To investigate the effect of CIZ1 on RKO
cells, a CIZ1-siRNA-GFP lentivirus was generated and RKO cells were
transfected to knockdown CIZ1 gene expression. To verify the
knockdown efficiency of CIZ1, we detected its mRNA expression in
RKO cells by qPCR. CIZ1 expression was observed to be
significantly reduced compared with the negative control that was
transfected with CIZ1 alone (P<0.05; Fig. 1B). This result was further
confirmed by the detection of CIZ1 protein expression by western
blot analysis (Fig. 1C),
demonstrating that the CIZ1 gene was efficiently knocked
down by CIZ1-siRNA. CIZ1 expression was also visualized by GFP
expression under fluorescent microscopy three days following
infection (Fig. 1D), >80% of
RKO cells expressed GFP. Under light microscopy (XSP-8CA; Shanghai
Optical Instrument Factory, Shanghai, China), no significant
difference was identified between the negative control and the
non-transfected groups, indicating that the lentivirus itself and
the transfection process had no effect on cell proliferation
(Fig. 1D).
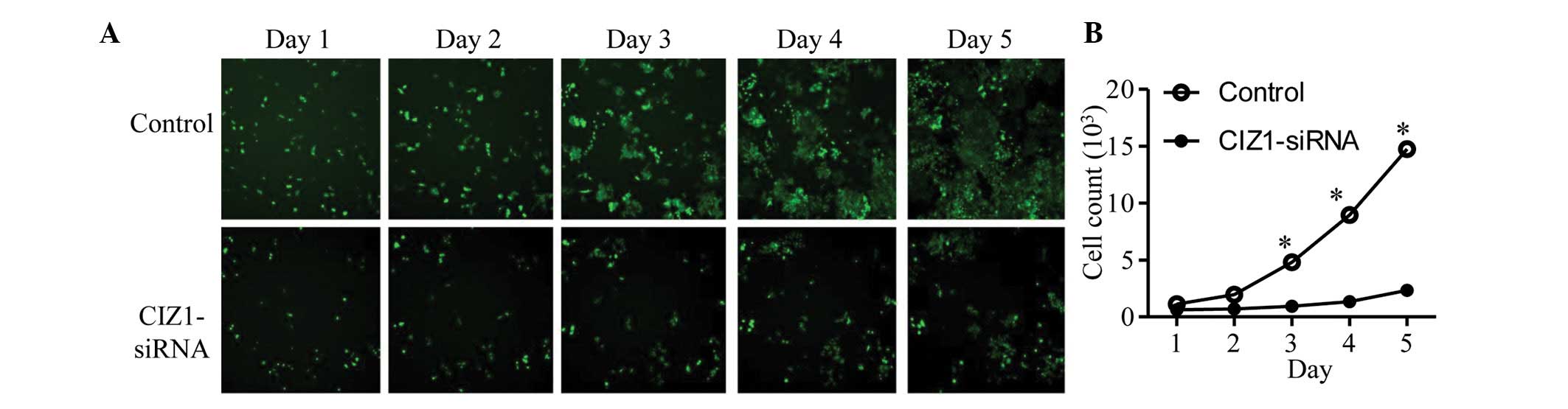
Knockdown of CIZ1 inhibits RKO cell
proliferation, induces cell cycle arrest and increases
apoptosis
The effect of CIZ1 expression on RKO cell
proliferation was examined. Cells transfected with CIZ1-siRNA
lentivirus and lentivirus alone were resuspended in 0.25% trypsin
and replicates were plated in five wells of six-well plates to
equalize the results. Cells were counted once daily for five days
using the Cellomics method (Fig.
2A). Cells from the two groups divided slowly for the first two
days. However, on day three, a significantly increased number of
cells were counted in the control group compared with the
CIZ1-siRNA group and the effect was more pronounced on days four
and five (Fig. 2B; P<0.05),
indicating that knockdown of CIZ1 may significantly inhibit the
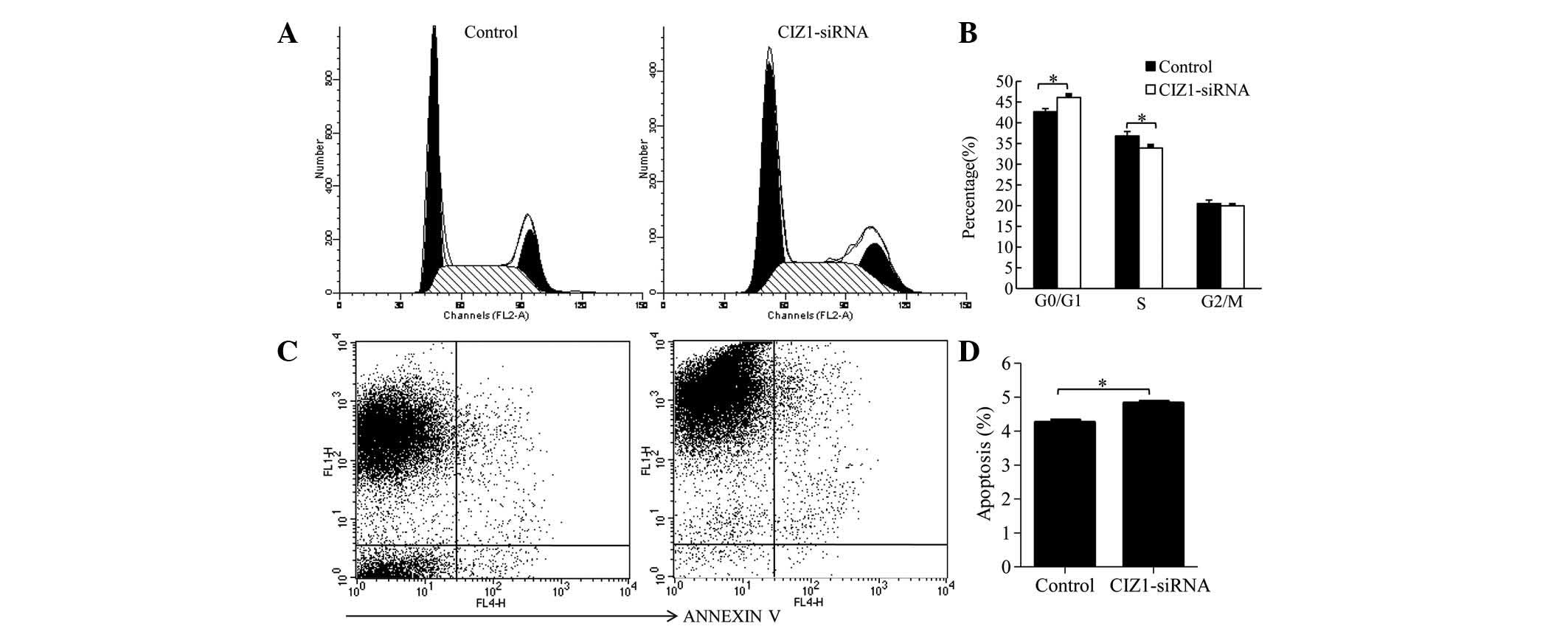
proliferation of RKO cells. In addition, the effect of CIZ1 on cell
cycle distribution was investigated using flow cytometry. The
percentage of G0/G1 stage cells was increased in the CIZ1-siRNA
group (46.11±0.64) compared with the control group (42.66±0.75;
P=0.003), while the percentage of cells in the S phase was
decreased (33.91±0.58) compared with the control group (36.83±1.05)
(P=0.02; Fig. 3A and B). These
data suggested that CIZ1 blocked the progression of the cell cycle.
Furthermore, the effect of CIZ1 on RKO cell apoptosis was examined.
Cell were stained with Annexin V and detected by flow cytometry.
Knockdown of the CIZ1 gene markedly increased the percentage
of apoptotic cells (Fig. 3C and
D).
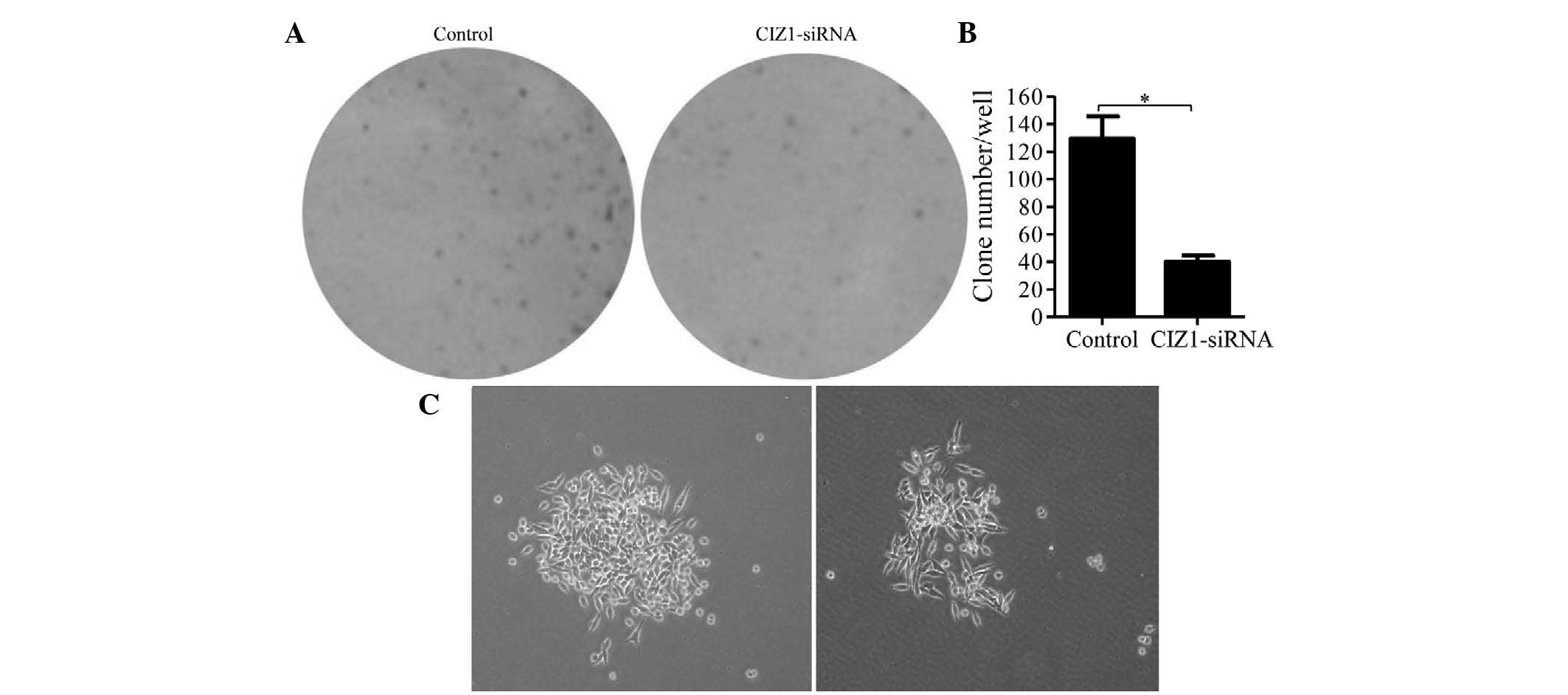
Knockdown of CIZ1 repressed RKO cell
colony formation
To investigate the effect of CIZ1 on colony
formation, RKO cells were allowed to proliferate for 14 days to
form colonies in six-well plates. Colonies were stained with
Giemsa, counted and analyzed. CIZ1-siRNA lentiviral infection
markedly reduced the colonies of RKO cells by 69.0%, compared with
the control lentivirus-infected groups (P<0.01; Fig. 4A and B). Moreover, the colonies of
the CIZ1-siRNA-infected group were also smaller and more dispersed
compared with the control group (Fig.
4C).
Discussion
In this study, the expression levels of CIZ1
mRNA in human CRC cell lines and their function in a human CRC cell
line, RKO, were investigated. The results demonstrated that
knockdown of CIZ1 using CIZ1-siRNA inhibited RKO cell
proliferation, induced cell cycle arrest, increased apoptosis and
eventually repressed RKO cell colony formation.
CIZ1, as a DNA replication regulator, was initially
characterized to interact with p21Cip1/Waf1(5). p21Cip1/Waf1 is induced by
the P53 tumor suppressor protein and is a cell cycle regulator
(12). Previous studies showed
that p21Cip1/Waf1 inhibits cell cycle progression by
binding to G1 cyclin/cyclin-dependent kinase (CDK) complexes and
proliferating cell nuclear antigen through its N- and C-terminal
domains, respectively (5,13). Therefore, p21Cip1/Waf1
has been suggested to be critical in inhibiting progression of
cancer cells, including fibroblasts, HaCaT and breast cancer cells
(14,15). CIZ1 was observed to bind to
p21Cip1/Waf1, competitively inhibiting the interaction
of p21Cip1/Waf1 with CDK and subsequently regulating
cytoplasmic distribution (5).
Therefore, it is reasonable to assume that CIZ1 regulates tumor
cell progression by affecting the cell cycle. The current study
demonstrated that knockdown of CIZ1 gene inhibits RKO cell
proliferation and promotes cell cycle arrest.
Further studies are required to determine the
mechanism by which CIZ1-siRNA inhibits RKO cell proliferation as
CIZ1 is also a DNA replication factor as well as an inhibitor of
p21Cip1/Waf1. Normally, CIZ1 is attached to the nuclear
matrix and resides within foci that partially colocalize at sites
of DNA replication (16). CIZ1
promotes the initiation of mammalian DNA replication by
coordinating the sequential functions of cyclin E- and A-dependent
protein kinases (17,18). It is also indirectly involved in
DNA replication by modulating the expression of genes, including
cyclin D, that affect cell proliferation (6). Therefore, it is also possible that
CIZ1 regulates RKO cell proliferation, cell cycle and apoptosis by
affecting DNA replication, which is a p21Cip1/Waf1
independent pathway.
The present study showed that knockdown of
CIZ1 expression negatively controlled the progression of RKO
CRC cells in vitro. These results indicate that CIZ1-siRNA
may be a promising therapeutic strategy for CRC.
Acknowledgements
This study was funded by a grant from the Shandong
Provincial Natural Science Foundation (grant no. ZR2010HQ055).
References
|
1
|
Kaur M, Mandair R, Agarwal R and Agarwal
C: Grape seed extract induces cell cycle arrest and apoptosis in
human colon carcinoma cells. Nutr Cancer. 60(Suppl 1): 2–11. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tomlinson IP, Webb E, Carvajal-Carmona L,
et al: A genome-wide association study identifies colorectal cancer
susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet.
40:623–630. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tenesa A, Farrington SM, Prendergast JG,
et al: Genome-wide association scan identifies a colorectal cancer
susceptibility locus on 11q23 and replicates risk loci at 8q24 and
18q21. Nat Genet. 40:631–637. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rahman FA, Aziz N and Coverley D:
Differential detection of alternatively spliced variants of Ciz1 in
normal and cancer cells using a custom exon-junction microarray.
BMC Cancer. 10:4822010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mitsui K, Matsumoto A, Ohtsuka S, Ohtsubo
M and Yoshimura A: Cloning and characterization of a novel
p21(Cip1/Waf1)-interacting zinc finger protein, ciz1. Biochem
Biophys Res Commun. 264:457–464. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
den Hollander P, Rayala SK, Coverley D and
Kumar R: Ciz1, a novel DNA-binding coactivator of the estrogen
receptor alpha, confers hypersensitivity to estrogen action. Cancer
Res. 66:11021–11029. 2006.PubMed/NCBI
|
|
7
|
Warder DE and Keherly MJ: Ciz1, Cip1
interacting zinc finger protein 1 binds the consensus DNA sequence
ARYSR(0–2)YYAC. J Biomed Sci. 10:406–417. 2003.PubMed/NCBI
|
|
8
|
Rahman F, Ainscough JF, Copeland N and
Coverley D: Cancer-associated missplicing of exon 4 influences the
subnuclear distribution of the DNA replication factor CIZ1. Hum
Mutat. 28:993–1004. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stein GH, Drullinger LF, Soulard A and
Dulić V: Differential roles for cyclin-dependent kinase inhibitors
p21 and p16 in the mechanisms of senescence and differentiation in
human fibroblasts. Mol Cell Biol. 19:2109–2117. 1999.PubMed/NCBI
|
|
10
|
Kinzler KW and Vogelstein B: Lessons from
hereditary colorectal cancer. Cell. 87:159–170. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Samowitz WS, Curtin K, Ma KN, Edwards S,
Schaffer D, Leppert MF and Slattery ML: Prognostic significance of
p53 mutations in colon cancer at the population level. Int J
Cancer. 99:597–602. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Poon RY and Hunter T: Expression of a
novel form of p21Cip1/Waf1 in UV-irradiated and transformed cells.
Oncogene. 16:1333–1343. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lim DY, Tyner AL, Park JB, Lee JY, Choi YH
and Park JH: Inhibition of colon cancer cell proliferation by the
dietary compound conjugated linoleic acid is mediated by the CDK
inhibitor p21CIP1/WAF1. J Cell Physiol. 205:107–113. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wei M, Liu B, Gu Q, Su L, Yu Y and Zhu Z:
Stat6 cooperates with Sp1 in controlling breast cancer cell
proliferation by modulating the expression of p21(Cip1/WAF1) and
p27 (Kip1). Cell Oncol (Dordr). 36:79–93. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zamkova M, Khromova N, Kopnin BP and
Kopnin P: Ras-induced ROS upregulation affecting cell proliferation
is connected with cell type-specific alterations of
HSF1/SESN3/p21Cip1/WAF1 pathways. Cell Cycle. 12:826–836. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ainscough JF, Rahman FA, Sercombe H, Sedo
A, Gerlach B and Coverley D: C-terminal domains deliver the DNA
replication factor Ciz1 to the nuclear matrix. J Cell Sci.
120:115–124. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Coverley D, Marr J and Ainscough J: Ciz1
promotes mammalian DNA replication. J Cell Sci. 118:101–112. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Copeland NA, Sercombe HE, Ainscough JF and
Coverley D: Ciz1 cooperates with cyclin-A-CDK2 to activate
mammalian DNA replication in vitro. J Cell Sci. 123:1108–1115.
2010. View Article : Google Scholar : PubMed/NCBI
|