Introduction
Breast cancer is one of the most common types of
cancer in females in developed and developing countries (1). The large number of novel breast
cancer cases arising annually and the high mortality rate of breast
cancer (2,3) encourage researchers to investigate
the correlation between the potential environmental and genetic
factors, and the risk of developing breast cancer. A number of
genetic factors are assumed to correlate with the modification of
the risk of breast cancer according to several of the most recently
published studies (4–8). Adipose metabolism-related genetic
variations may also modify the risk of breast cancer (9).
The peroxisome proliferator-activated receptors
(PPARs) are a cluster of nuclear transcription factors, which are
members of the nuclear hormone receptor super-family, and function
in cellular differentiation and the regulation of carbohydrate and
lipid metabolism (10).
Polymorphisms in these receptors are assumed to affect the
pathology of cancers and other diseases.
PPARs are classified into three predominant
sub-types: PPAR-α, -β and -γ (11). PPAR-γ, also termed PPARG, is
located on chromosome 3p25 in humans and dimerizes with the
retinoid X receptor (RXR) to regulate target genes involved in
adipocyte differentiation and insulin sensitization (12,13).
PPAR-γ is also assumed to be correlated with malignant breast
cancer epithelial cells (12).
PPAR-γ2 is a sub-type of PPAR that is only expressed in adipose
tissue (14). The
Pro12Ala single nucleotide (rs1801282) polymorphism is a
C/G mutation that may be associated with the modifications of the
risk of a number of diseases (15–18).
Numerous studies have also been conducted to
estimate the association between the Pro12Ala
(rs1801282) polymorphism in the PPAR-γ2 gene and the risk of breast
cancer, however the results have not always been consistent
(19–22).
In the present study a meta-analysis on the eligible
case-control studies was undertaken in order to analyze the
association between PPAR-γ2 Pro12Ala polymorphisms and
breast cancer susceptibility.
Materials and methods
Search strategy
Multi-databases, including PubMed (http://www.ncbi.nlm.nih.gov/pubmed/), EMBASE
(http://www.embase.com/home) and the
COCHRANE Library (http://www.thecochranelibrary.com/view/0/index.html)
in English and VIP (http://lib.cqvip.com/), CNKI (http://www.cnki.net/) and Sinomed (http://www.sinomed.ac.cn/) in Chinese, were used to
search the potential related published papers (all papers were
published before November 11, 2012). The following keywords and
subject terms were used: ‘PPARγ2’, ‘PPARG’ or
‘proliferator-activated receptor gamma2’ and ‘breast cancer’. In
addition, the search terms ‘PPARG’, ‘breast cancer’ and ‘genetic
association’ were used in the HuGE Navigator. All the search terms
were restricted to studies in humans. The references of the studies
obtained were also searched in PubMed.
Inclusion criteria
Studies included in this meta-analysis were defined
as: a) Case-control studies (including nested case-control studies;
b) non-family based studies; and c) those evaluating the
correlation between the PPAR-γ2 Pro12Ala (rs1801282)
polymorphism and the risk of breast cancer.
Exclusion criteria
The articles that were case reports, system reviews,
editorials, clinical guidelines and information articles for
patients were all excluded. A study was also rejected if it did not
provide information concerning PPAR-γ2 Pro12Ala
polymorphisms.
Data extraction
Two investigators (QX Mao and HL Guo) searched and
screened the potential associated articles for inclusion and
appraisal. If there were any discrepancies, a discussion would be
conducted in which other reviewers (LG Gao and HW Wang) would also
be involved until an agreement was reached. The data abstracted
from each publication consisted of first author, year of
publication, country, ethnicity, study design, sample size,
resources of controls and the PPAR-γ2 Pro12Ala
polymorphism information. The study quality was quantified by the
Newcastle-Ottawa-Scale (NOS) for case-control studies (23).
Statistical analysis
An unadjusted odds ratio (OR) and the corresponding
95% confidence interval (CI) of every eligible study was initially
calculated. The Z-test was used to examine the pooled OR. The
Q-statistic and I2 statistical tests were used to
measure the heterogeneity among the eligible studies. Fixed-effects
models using Mantel-Haenszel methods and random-effects models were
used in the meta-analysis. The Hardy-Weinberg (H-W) equilibrium was
examined by a Pearson χ2 test for the controls in every
individual study. Potential publication bias was assessed by a
Funnel plot and Egger’s linear regression.
All analyses were performed by the Stata software,
version 8.0 (Stata Corp LP, College Station, TX, USA). The tests
were two-sided and P<0.05 was used to indicate a statistically
significant difference.
Results
Study characteristics and meta-analysis
database
According to the search terms from the databases of
the HuGE Navigator, PubMed, EMBASE and the COCHRANE Library when
using the English language, fourteen potential correlated studies
were collected. No correlated study published in Chinese was
identified. Among these thirteen articles, one was excluded due to
the family-based design (24). Six
of the articles did not analyze the correlation between the
Pro12Ala (rs1801282) polymorphism and the risk of breast
cancer (9,11,25–28).
One cohort study that investigated the correlation among benign
breast cancer patients was also deleted (29). Two studies were based on the same
population, and the former of them was excluded (30). Another study that lacked the full
text was also excluded (31).
Therefore, four individual studies remained for further analysis
(19–22). A total of 2,279 cases and 2,360
controls available from the included reports for the PPAR-γ2
Pro12Ala polymorphism information were obtained. Breast
cancer was confirmed by clinical examinations and from clinical
records.
A dataset based on the extracted information from
each included report was established (Table I). A quality assessment for the
eligible studies according to the NOS is shown in Table II.
 | Table ICharacteristics of studies included in
this analysis. |
Table I
Characteristics of studies included in
this analysis.
| ID | First author | Year | Country | Ethnicity | Source of
controls | Genotyping
method | Sample size
case/control | Polymorphism
distribution of PPARγ Pro12Ala case/control | Allele distribution
of PPARγ Pro12Ala case/control |
|---|
|
|
|---|
| CC | CG | GG | C | G |
|---|
| 1 | Kim KZ | 2012 | Korea | Asian | PB | Sequence | 400/452 | 366/406 | 33/40 | - | - | |
| 2 | Petersen RK | 2012 | Denmark | Not mentioned | PB | TaqMan | 798/798 | 616/569 | 167/209 | 15/20 | 182/229 | 197/240 |
| 3 | Justenhoven C | 2008 | Germany | European | PB | TaqMan | 593/622 | 452/462 | 135/145 | 6/15 | 1039/1069 | 147/175 |
| 4 | Wang Y | 2007 | USA | Mixed | PB | TaqMan | 488/488 | 376/375 | 87/98 | 15/5 | 839/848 | 117/108 |
 | Table IIQuality assessment for the eligible
studies according to the NOS. |
Table II
Quality assessment for the eligible
studies according to the NOS.
| ID | First author | Selection, n
stars | Comparability, n
stars | Exposure, n
stars |
|---|
| 1 | Kim KZ | 4 | 2 | 1 |
| 2 | Petersen RK | 4 | 2 | 1 |
| 3 | Justenhoven C | 4 | 2 | 1 |
| 4 | Wang Y | 4 | 2 | 1 |
Quantitative synthesis
The average proportions of the frequencies of the G
allele and the CG genotype from three eligible populations were
12.3 and 20.5%, respectively, in the patient cases and 13.7 and
23.2%, respectively, in the controls. The corresponding proportion
of the CC genotype from four eligible populations was 80.5% in the
patient cases and 78.1% in the controls. The genotype distributions
of the G allele in the controls from every eligible study
population satisfied the H-W equilibrium (all P>0.05).
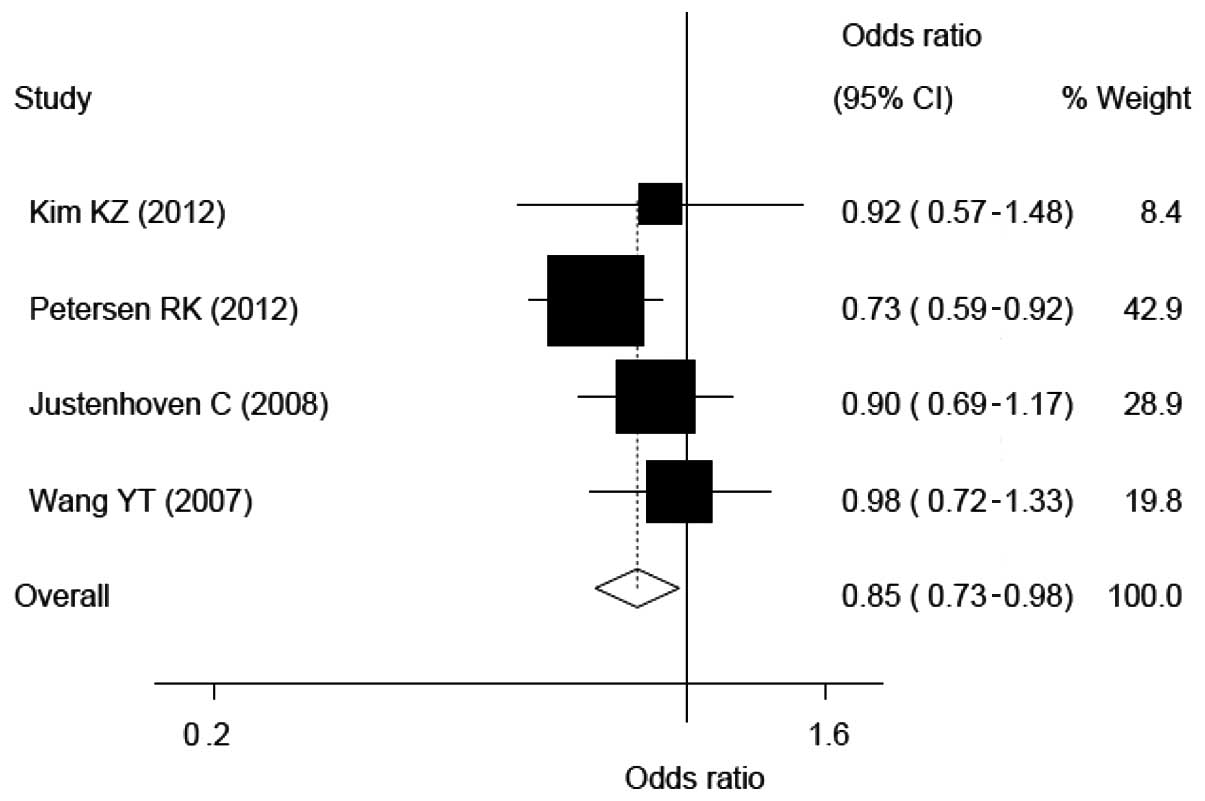
Compared with the CC genotype, the CG genotype and
CG+GG mixed genotypes carriers had a lower risk of breast cancer
according to the three and four eligible populations, respectively.
The ORs, CIs and heterogeneity values for CG and CG+GG on the risk
of breast cancer were 0.84, 0.72–0.98 and 0.347 and 0.85, 0.73–0.98
and 0.441, respectively (see Fig.
2 and 4).
As the GG genotype did not modify the risk of breast
cancer statistically (OR, 0.92; 95% CI, 0.32–2.61; heterogeneity,
0.015; Fig. 3) compared with the C
allele carriers, those with the G allele did not have a
statistically significant effect on the risk of breast cancer
either. The corresponding OR, 95% CI and heterogeneity values were
0.98, 0.84–1.13 and 0.397, respectively (Fig. 5).
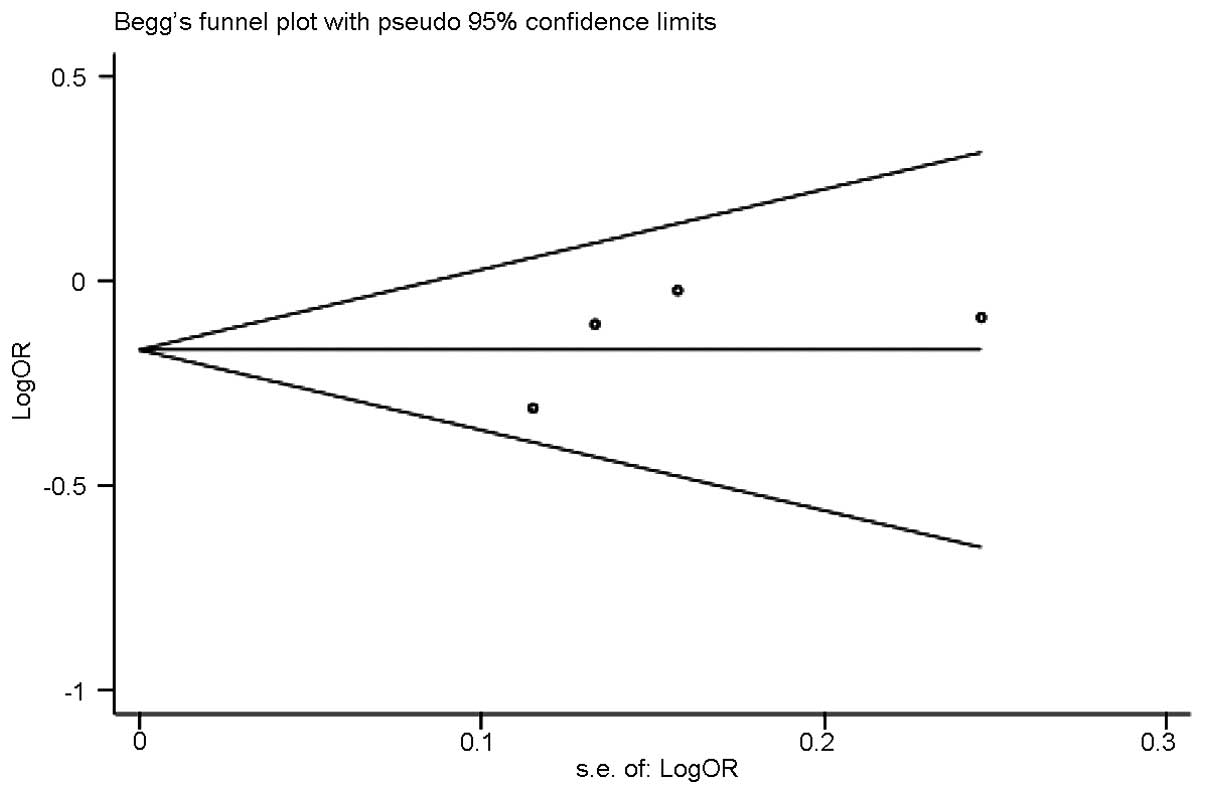
Publication bias
Funnel plots and Egger’s tests were conducted to
examine the publication bias (Fig.
6). No publication bias was identified (P=0.410).
Discussion
A total of 2,279 cases and 2,360 controls from four
eligible individual studies were included in the present study in
order to investigate the association between the PPAR-γ2
Pro12Ala (rs1801282) polymorphism and the risk of breast
cancer.
According to the results of the present study, the
Pro12Ala polymorphism was demonstrated to be correlated
with modifying the risk of breast cancer. The CG heterozygote and
the CG+GG genotype carriers exhibited lower breast cancer incident
risks in comparison with the GG genotype carriers. The
corresponding ORs and 95% CIs were 0.84 and 0.72–0.98,
respectively, for the CG carriers and 0.85 and 0.73–0.98,
respectively, for the CG+GG carriers. Although, no statistical
association between the Pro12Ala polymorphism and the
breast cancer incident risk was demonstrated, when the comparisons
were conducted between the GG and CC homozygotes or between the G
and C alleles, there remained a potential effect from the GG
homozygote or the G allele on the risk of breast cancer. The
corresponding ORs and 95% CIs were 0.92 and 0.32–2.61,
respectively, for the GG versus the CC homozygotes and 0.98 and
0.84–1.13, respectively, for the G versus C alleles.
The results of the present study were supported by
certain previous studies. In a case-control study in Denmark
conducted by Vogel et al(30), compared with the CC homozygote, the
CG heterozygote and the CG+GG mixed genotype groups had a decreased
risk of breast cancer. In addition, no statistically significant
effect was observed from the GG homozygote on the breast cancer
incident risk compared with the CC homozygote. Even in the
multivariate adjusted model, such results did not change markedly.
The corresponding multivariate-adjusted ORs and 95% CIs were 0.66
and 0.45–0.96, respectively, for CG versus. CC, 0.67 and 0.46–0.97,
respectively, for CG+GG versus CC and 0.81 and 0.29–2.29,
respectively, for GG versus CC, respectively. The results of this
study were also partially supported by German (19) and American (29) studies. In the German study (688
cases and 724 population-based controls), neither the CG
heterozygote nor the GG homozygote modified the risk of breast
cancer significantly. The corresponding ORs and 95% CIs were 0.96
and 0.74–1.27, respectively, for CG versus CC and 0.41 and
0.16–1.08, respectively, for GG versus CC (19). In the American study, a total of
994 post-menopausal females with benign breast disease were
included in the cohort study, among which, 61 participants
developed breast cancer after 14 years of follow-up. All the breast
cancer patients were regarded as the cases and the others were
analyzed as the controls. No statistically significant correlation
was revealed between the Pro12Ala polymorphism and the
breast cancer risk among the post-menopausal females with benign
breast cancer. The corresponding ORs and 95% CIs were 0.53 and
0.24–1.19, respectively, for CG vs. CC, 0.79 and 0.10–6.03,
respectively, for GG vs. CC and 0.55 and 0.26–1.19, respectively,
for CG+GG vs. CC (29).
Contrary results were identified in the study
conducted by Wang et al(20). In the nested case-control study,
which included 488 cases and 488 controls, compared with the CC
homozygote, the GG homozygote increased the risk of breast cancer
(OR, 2.91; 95% CI, 1.05–8.04). At the same time, the CG
heterozygote did not modify the risk of breast cancer significantly
(OR, 0.88; 95% CI, 0.63–1.24).
The majority of the results, including the present
meta-analysis, did not reveal that the GG homozygote modified the
risk of breast cancer. However, the CG heterozygote and the CG+GG
mixed genotype group modified the risk of breast cancer in certain
studies (21). The majority of the
results indicated the potential protective effect from the G allele
on the risk of breast cancer. The lower frequency of the G allele
in the study population included in the analyses may be a possible
reason that a statistically significant correlation between the G
allele/GG homozygote and the risk of breast cancer could not be
demonstrated. Further studies based on a larger population are
required to be undertaken in order to investigate such an
association.
Several limitations of the present meta-analysis
should be considered when interpreting the results. Due to the
lower between-study heterogeneity and the limited number of studies
involved in this meta-analysis, a sensitivity analysis was not
conducted. In addition, a stratified analysis was not performed as
the number of eligible published studies was insufficient for such
a comprehensive analysis. Moreover, the language limitation may
mean that information published in other languages may have been
missed. Furthermore, no original data of the individual studies was
obtained so only the summarized data about the potential
confounding variables could be collected, and only unadjusted
estimates were performed in the meta-analysis. However, the
meta-analysis also had several advantages. All the cases and
controls were pooled from different studies, which significantly
increased the statistical power of the analysis. Furthermore, the
quality of the eligible studies included in the current
meta-analysis was satisfactory, as they met the inclusion criterion
and received a high quality score according to the NOS. All the
study populations were also in H-W equilibrium.
In conclusion, this meta-analysis indicated that the
G allele modestly modified the risk of breast cancer. However, due
to insufficient comparative published studies involved, a
systematic analysis of the correlation between the G allele and the
risk of breast cancer could not be confirmed, but the study may
have developed our understanding of the effect of the G allele on
breast cancer. Further evidence from epidemiological studies is
required in order to provide a clearer characterization of the
involvement of the G allele and its genotypes in the genetic
susceptibility to developing breast cancer.
Acknowledgements
This study was supported by the National Nature
Science Foundation for Young Scientists of China (grant no.
81102142) and the National Research Institute for Family Planning
(grant no. 2010GJSSJKA10).
References
|
1
|
World Health Organization. Breast cancer:
prevention and control. www.who.int/cancer/detection/breastcancer/en/https://www.who.int/cancer/detection/breastcancer/en/.
Accessed Jan 6, 2013
|
|
2
|
World health statistics 2008: Part 1: Ten
highlights in health statistics. http://www.who.int/whosis/whostat/EN_WHS08_Part1.pdf.
Accessed Jan 6, 2013
|
|
3
|
The global burden of disease: 2004 update.
www.who.int/healthinfo/global_burden_disease/2004_report_update/en/https://www.who.int/healthinfo/global_burden_disease/2004_report_update/en/.
Accessed Jan 6, 2013
|
|
4
|
Guo H, Ming J, Liu C, et al: A common
polymorphism near the ESR1 gene is associated with risk of breast
cancer: evidence from a case-control study and a meta-analysis.
PLoS One. 7:e524452012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li LW and Xu L: Menopausal status modifies
breast cancer risk associated with ESR1 PvuII and
XbaI polymorphisms in Asian women: a HuGE review and
meta-analysis. Asian Pac J Cancer Prev. 13:5105–5111. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu L and Chen J: Association of MTHFR
Ala222Val (rs1801133) polymorphism and breast cancer
susceptibility: An update meta-analysis based on 51 research
studies. Diagn Pathol. 7:1712012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wei G, Wang Y, Zhang P, Lu J and Mao JH:
Evaluating the prognostic significance of FBXW7 expression level in
human breast cancer by a meta-analysis of transcriptional profiles.
J Cancer Sci Ther. 4:299–305. 2012.PubMed/NCBI
|
|
8
|
Mao Q, Gao L, Wang H, Wang Q and Zhang T:
The alcohol dehydrogenase 1C(rs698) genotype and breast cancer: A
meta-analysis. Asia Pac J Public Health. May 31–2012.(Epub ahead of
print).
|
|
9
|
Wu MH, Chu CH, Chou YC, Chou WY, Yang T,
Hsu GC, Yu CP, Yu JC and Sun CA: Joint effect of peroxisome
proliferator-activated receptor γ genetic polymorphisms and
estrogen-related risk factors on breast cancer risk: results from a
case-control study in Taiwan. Breast Cancer Res Treat. 127:777–784.
2011.
|
|
10
|
Cho MC, Lee K, Paik SG and Yoon DY:
Peroxisome proliferators-activated receptor (PPAR) modulators and
metabolic disorders. PPAR Res. 2008:6791372008.PubMed/NCBI
|
|
11
|
Memisoglu A, Hankinson SE, Manson JE,
Colditz GA and Hunter DJ: Lack of association of the codon 12
polymorphism of the peroxisome proliferator-activated receptor
gamma gene with breast cancer and body mass. Pharmacogenetics.
12:597–603. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dallongeville J, Iribarren C, Ferrières J,
et al: Peroxisome proliferator-activated receptor gamma
polymorphisms and coronary heart disease. PPAR Res.
2009:5437462009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He W: PPARγ2 polymorphism and human
health. PPAR Res. 2009:8495382009.
|
|
14
|
Spiegelman BM: PPAR-gamma: adipogenic
regulator and thiazolidinedione receptor. Diabetes. 47:507–514.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ho JS, Germer S, Tam CH, et al:
Association of the PPARG Pro12Ala polymorphism with type 2 diabetes
and incident coronary heart disease in a Hong Kong Chinese
population. Diabetes Res Clin Pract. 97:483–491. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Prakash J, Srivastava N, Awasthi S, et al:
Association of PPAR-γ gene polymorphisms with obesity and
obesity-associated phenotypes in North Indian population. Am J Hum
Biol. 24:454–459. 2012.
|
|
17
|
Alsaleh A, Frost GS, Griffin BA, et al;
RISCK Study Investigators. PPARγ2 gene Pro12Ala and PPARα gene
Leu162Val single nucleotide polymorphisms interact with dietary
intake of fat in determination of plasma lipid concentrations. J
Nutrigenet Nutrigenomics. 4:354–366. 2011.
|
|
18
|
Poliska S, Penyige A, Lakatos PL, et al;
Hungarian IBD Study Group. Association of peroxisome
proliferator-activated receptor gamma polymorphisms with
inflammatory bowel disease in a Hungarian cohort. Inflamm Bowel
Dis. 18:472–479. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Justenhoven C, Hamann U, Schubert F, et
al: Breast cancer: a candidate gene approach across the estrogen
metabolic pathway. Breast Cancer Res Treat. 108:137–149. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, McCullough ML, Stevens VL, et al:
Nested case-control study of energy regulation candidate gene
single nucleotide polymorphisms and breast cancer. Anticancer Res.
27:589–593. 2007.PubMed/NCBI
|
|
21
|
Petersen RK, Larsen SB, Jensen DM, et al:
PPARgamma-PGC-1alpha activity is determinant of alcohol related
breast cancer. Cancer Lett. 315:59–68. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim KZ, Shin A, Lee YS, Kim SY, Kim Y and
Lee ES: Polymorphisms in adiposity-related genes are associated
with age at menarche and menopause in breast cancer patients and
healthy women. Hum Reprod. 27:2193–2200. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wells GA, Shea B, O’Connell D, et al: The
Newcastle-Ottawa Scale (NOS) for assessing the quality of
nonrandomised studies in meta-analyses. Available from: URL:
www.ohri.ca/programs/clinical_epidemiology/oxford.asphttps://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
Accessed Nov 26, 2011
|
|
24
|
Wirtenberger M, Tchatchou S, Hemminki K,
et al: Associations of genetic variants in the estrogen receptor
coactivators PPARGC1A, PPARGC1B and EP300 with familial breast
cancer. Carcinogenesis. 27:2201–2208. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Li Y, Wedrén S, et al: Genetic
variation of ESR1 and its co-activator PPARGC1B is synergistic in
augmenting the risk of estrogen receptor-positive breast cancer.
Breast Cancer Res. 13:R102011. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Paynter RA, Hankinson SE, Colditz GA,
Hunter DJ and De Vivo I: No evidence of a role for PPARgamma
Pro12Ala polymorphism in endometrial cancer susceptibility.
Pharmacogenetics. 14:851–856. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Posch MG, Zang C, Mueller W, Lass U, von
Deimling A and Elstner E: Somatic mutations in peroxisome
proliferator-activated receptor-gamma are rare events in human
cancer cells. Med Sci Monit. 10:BR250–BR254. 2004.PubMed/NCBI
|
|
28
|
Ondrey F: Peroxisome
proliferator-activated receptor gamma pathway targeting in
carcinogenesis: implications for chemoprevention. Clin Cancer Res.
15:2–8. 2009. View Article : Google Scholar
|
|
29
|
Gallicchio L, McSorley MA, Newschaffer CJ,
et al: Body mass, polymorphisms in obesity-related genes, and the
risk of developing breast cancer among women with benign breast
disease. Cancer Detect Prev. 31:95–101. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vogel U, Christensen J, Nexø BA, Wallin H,
Friis S and Tjønneland A: Peroxisome profilerator-activated
[corrected] receptor-gamma2 [corrected] Pro12Ala, interaction with
alcohol intake and NSAID use, in relation to risk of breast cancer
in a prospective study of Danes. Carcinogenesis. 28:427–434.
2007.
|
|
31
|
Lee E, Hsu C, Van den Berg D, et al:
Genetic variation in peroxisome proliferator-activated receptor
gamma, soy, and mammographic density in Singapore Chinese women.
Cancer Epidemiol Biomarkers Prev. 21:635–644. 2012. View Article : Google Scholar : PubMed/NCBI
|