Introduction
It is well known that subsequent to the infection of
hepatitis B virus (HBV) into the target cells, the interactions of
the virus genome and proteins with the genes and proteins in the
target cells are significant in determining HBV replication, immune
evasion and chronic infection (1).
In previous years, it has been revealed that a complex
trans-regulatory mechanism is involved in the interaction of HBV
with the target cells, and the HBV proteins are trans-regulatory in
the gene expression of these cells (2). The critical antigen components of HBV
associated with trans-regulatory function have been identified in
target cells and the specific mechanisms have been clarified, which
are of great significance in confirming the pathogenic mechanisms
of HBV and identifying effective prevention and treatment methods.
HBV DNA polymerase transactivated protein 1 (HBVDNAPTP1) is a
protein that is worth studying. Thus, suppression subtractive
hybridization technology (GenBank accession no. AY450389). has been
used to study the trans-regulatory target genes of the HBV DNA
polymerase, which was verified by dot blot hybridization. The HepG2
hepatoblastoma cell line was screened to obtain a novel gene
(GenBank accession no. AY450389), which was located on the long arm
of chromosome 9, region 2, band 2, sub-band 31 (9q22.31). With the
use of the Unigene database (http://www.ncbi.nlm.nih.gov/unigene) for an expression
analysis of tissue distribution, this gene was observed to be
expressed in a variety of tissues, excluding the pituitary gland,
tonsil, tongue, thymus, trachea and umbilical cord. A preliminary
study clarified that HBVDNAPTP1 is localized in the cytoplasm
(3). The yeast two-hybrid system
is currently one of the most effective ways to study the function
and proteins of novel genes. Thus, the present study mainly focused
on the screening of HBVDNAPTP1 interacting proteins in order to
reveal their biological functions.
Materials and methods
Vector, strain and cell lines
The pGEM-T vector was purchased from Promega
Corporation (Madison, WI, USA). The yeast cell expression vector,
pGBKT7, and the AH109 haploid yeast cell line were obtained from
the Infectious Disease Research Institute of Beijing Ditan
Hospital, Capital Medical University (Beijing, China). The
mammalian cell expression vectors, pCMV-Myc and pCMV-HA, were
purchased from Clontech Laboratories, Inc. (Mountain View, CA,
USA). E. coli DH5α, HepG2 hepatoblastoma and HEK293 human
embryonic kidney cell lines were preserved in our laboratory
(Dalian, China).
Reagents
TRIzol reagent and Lipofectamine PLUS were purchased
from Gibco-BRL (Carlsbad, CA, USA). The DNA Glass-Milk Rapid
Purification kit was purchased from BioDev (Milano, Italy). DNA
marker, T4 ligase, Takara Ex Taq kit and restriction endonuclease
enzymes EcoRI, BamHI, BgIII and SalI
were purchased from Takara Biotechnology Inc. (Dalian, China).
Yeast extract, tryptone and yeast nitrogen base were obtained from
Oxoid (Hampshire, UK). Matchmaker GAL4 Two-hybrid system 3,
pre-transformed Matchmaker libraries, yeast YPD medium, deficient
amino acid mixtures and disulfuric acid adenosine were all
purchased from Clontech Laboratories, Inc. The glutathione-S
transferase (GST) gene fusion system was obtained from Amersham
plc, (Piscataway, NJ, USA). c-myc Tag monoclonal antibody, goat
anti-mouse horseradish peroxidase (HRP)-IgG and the concentrated
3,3′-diaminobenzidine kit were all purchased from Zhongshan Golden
Bridge Biotechnology Co., Ltd. (Beijing, China). Human influenza
hemagglutinin (HA)-Tag monoclonal antibody, mouse IgG, Protein A+G
Sepharose 4B and the hypersensitive enhanced chemiluminescence kit
were purchased from the Beyotime Institute of Biotechnology,
(Haimen, China). Primer synthesis and DNA sequencing were performed
by Takara Biotechnology Inc.
Amplification of the HBVDNAPTP1 gene by
quantitative polymerase chain reaction (qPCR)
Specific primers with EcoRI/BamHI
sites were designed according to the HBVDNAPTP1 gene sequence from
the National Center for Biotechnology Information: Primer 1:
5′-GAATTCATGATGTTTGTGCTGCTAAAC-3′ and primer 2:
5′-GGATCCATAAGTCCTCTCTAAAATTGCG-3′. The total RNA from the HepG2
cells was extracted and transcribed into cDNA using oligo dT. The
PCR amplification of the HBVDNAPTP1 gene was performed using the
following conditions: 95°C for 5 min, followed by 30 cycles at 94°C
for 30 sec, 55°C for 45 sec and 72°C for 1 min, and then 72°C for
10 min.
Construction of yeast cell bait
expression vector, pGBKT7-HBVDNAPTP1
The purified HBVDNAPTP1 PCR fragment was ligated
into a pGEM-T vector and transformed into DH5α competent cells.
Positive colonies from X-gal/isopropyl β-D-1-thiogalactopyranoside
selection were used for plasmid isolation. Following sequencing,
the pGEM-T-HBVDNAPTP1 plasmid was digested by
EcoRI/BamHI, and the purified fragment was ligated
into linearized pGBKT7 by EcoRI/BamHI and then
transformed into DH5α competent cells. The recombinant plasmid,
pGBKT7-HBVDNAPTP1, was extracted by an alkaline lysis method and
identified by enzyme digestion.
Transformation of AH109 yeast cells with
pGBKT7-HBVDNAPTP1
The AH109 yeast haploid cells were transformed with
recombinant pGBKT7-HBVDNAPTP1 or pGBKT7 empty vector by the LiAc
method, and spread onto synthetic dropout medium (SD)/-Trp filter
plates. Positive yeast colonies were inoculated on synthetic
defined/-Trp medium and agitated overnight at 30°C. The yeast
protein was extracted using the urea/SDS method. In total, 50 μl of
the extracted yeast protein was subjected to SDS-PAGE, transferred
to a membrane and blocked with 5% skimmed milk overnight at 4°C.
The membrane was incubated with primary antibody (1:100; c-myc Tag
monoclonal antibody) and secondary antibody (1:3,000; HRP-goat
anti-mouse IgG). The expression of the bait protein, HBVDNAPTP1,
was detected by 3,3′-diaminobenzidine chromogenic methods.
Yeast two-hybrid screening
The positive yeast colonies were screened according
to the manufacturer’s instructions for the yeast two-hybrid system
(Clontech Laboratories, Inc., Mountain View, CA, USA). The yeast
plasmid was extracted and transformed into DH5α competent cells.
The plasmid was extracted by the alkaline lysis method and
identified by enzyme digestion. The positive clones (500–2,000 bp)
obtained from enzyme digestion were verified further by sequencing.
A homology analysis of the sequencing results was conducted by a
basic local alignment search tool (BLAST) comparison (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Verification of candidate library
vector
The extract containing the library vector of E.
coli DH5α and the AH109 yeast cells containing the
pGBKT7-HBVDNAPTP1 bait vector were incubated in inverted quadruple
dropout medium (QDO; SD/-Ade/-His/-Leu/-Trp) plates at 30°C for 7
days. The yeast colonies were selected again and inoculated into
QDO/X-α-gal plates. pGBKT7-HBVDNAPTP1 with candidate library
vectors was individually co-transformed into AH109 cells. The
cotransformed library and the empty vector, pGBKT7, served as
negative controls.
Interaction between HBVDNAPTP1 and the
paired immunoglobulin-like type 2 receptor α (PILRA)
intracellular domain by immunoprecipitation
The eukaryotic expression plasmids
pCMV-Myc-HBVDNAPTP1 and pCMV-HA-PILRA intracellular domains [the
cytoplasmic domain of PILRA, (PILRACD)] were constructed and
co-transfected into HEK293 cells in 6-well plates. The cells were
lysed by NP-40 lysis buffer. In total, 2 μg HA-tag monoclonal
antibody or mouse IgG was added to the cell lysates and incubated
at 4°C overnight. The cell lysates were incubated with 30 μl
protein A+G Sepharose 4B beads (Beyotime Institute of
Biotechnology) overnight. The beads were collected by
centrifugation at 3,000 rpm for 3 min at 4°C, washed three times
with lysis buffer and then boiled with 1X SDS-PAGE protein loading
buffer for 5 min. In total, 20 μl of the supernatant was separated
by SDS-PAGE and transferred to a PVDF membrane. Following blocking
with 5% skimmed milk at 4°C, the membrane was incubated with
primary antibody (1:200) and secondary antibody (1:5,000; HRP-goat
anti-mouse IgG), and then developed using the chemiluminescence
method. The co-transfection of HEK293 cells with
pCMV-Myc-HBVDNAPTP1 and pCMV-HA was used as a negative control.
Interaction between HBVDNAPTP1 and the
PILRA intracellular domain by GST pull-down
The prokaryotic cell expression plasmid,
pGEX-4T-1-PILRACD, was constructed and transformed into BL21 (DE3)
competent cells. The positive colony was inoculated overnight and
scaled-up in a 1:20 dilution until the bacterial concentration
reached A600=0.6. A final concentration of 0.5 mmol/l
isopropyl β-D-1-thiogalactopyranoside was added and cultured at
30°C for 4 h. The bacterial cells were harvested, washed and
suspended in 1X phosphate-buffered saline. Following
ultrasonication (Sonics & Materials, Inc., Newtown, CT, USA),
the cell supernatant was collected subsequent to centrifugation. In
total, 400 μl Glutathione Sepharose 4B (Beyotime Institute of
Biotechnology) was added to the bacterial lysis supernatant and
incubated at 4°C for 2 h. The mixture was centrifuged at 1,500 × g
(Beckman Coulter, Inc., Brea, CA, USA) at 4°C for 3 min to remove
the supernatant, and the beads were washed with lysis buffer. The
BL21 (DE3) competent cells transformed with pGEX-4T-1 were used as
a negative control. The GST pull-down method was performed in the
same way as aforementioned, but protein A+G Sepharose 4B
cross-linked with HA-Tag monoclonal antibody was replaced by
Glutathione Sepharose 4B cross-linked with the GST-PILRACD fusion
protein.
Results
Construction of yeast cell bait
expression plasmid pGBKT7-HBVDNAPTP1
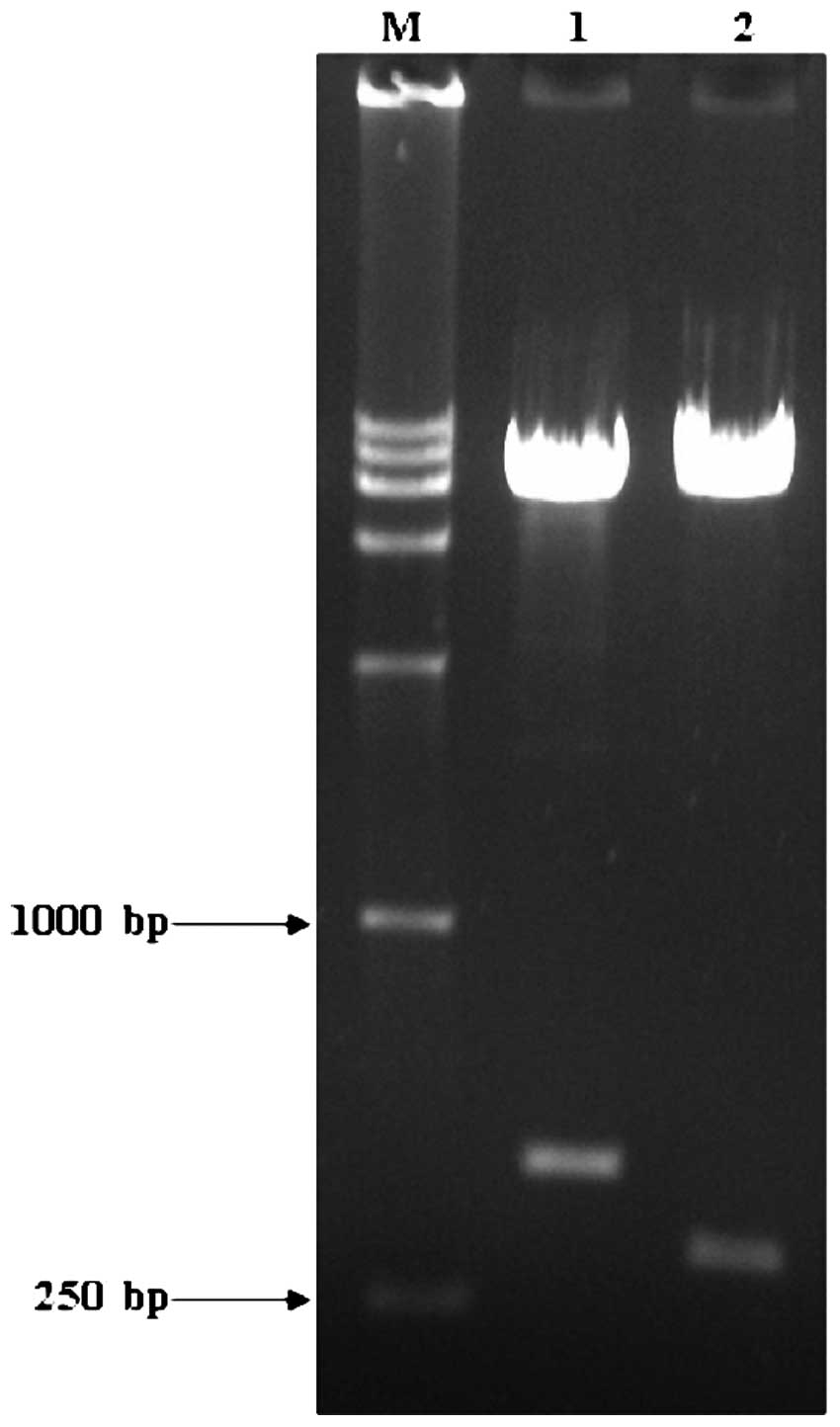
Using cDNA from the HepG2 cells as the template, the
444-bp HBVDNAPTP1 gene was amplified and ligated into the yeast
cell expression vector, pGBKT7, by TA cloning and then identified
by EcoRI/BamHI and Bg1II/SalI
digestions. Gene fragments of 444 and 319 bp were obtained
(Fig. 1). This indicated that the
HBVDNAPTP1 gene was accurately ligated into the pGBKT7 vector.
Expression of HBVDNAPTP1 in yeast
cells
As the pGBKT7 vector has a C-myc tag sequence, the
expression of the fusion proteins was examined by detecting the
C-myc tag. Subsequent to the yeast cells being transformed by
pGBKT7-HBVDNAPTP1, one clear band was observed and its molecular
weight was consistent with the expected size of HBVDNAPTP1
(Fig. 2), indicating that
HBVDNAPTP1 was successfully expressed in the yeast cells.
Results of yeast two-hybrid screen and
verification
The transformed yeast culture, AH109, and the
library cultures were combined together to screen for genes that
interact with HBVDNAPTP1 in the leukocyte library. In total, 15
positive colonies were selected (data not shown). Since the pACT2
library plasmid contained two BglII restriction sites, the
BglII enzyme was used to release the leukocyte cDNA library
fragment, which was 400–2,000 bp in size. The plasmids from
positive colonies were cut by Bgl II and sent for
sequencing. Using BLAST for the sequence comparison, four genes
were obtained, including PILRA, zymogen granule protein 16,
carboxylic acid lipase 1 and β transduction element protein 2, and
their homology to the target genes was recorded as 98, 100, 99 and
98%, respectively.
Interaction between HBVDNAPTP1 and the
PILRA intracellular domain by co-immunoprecipitation
The full length of the PILRA protein is 303 aa and
is composed of five domains; a signal peptide and the antibody
variable, hinge, transmembrane and intracellular domains, of which
the intracellular domain consists of 85 aa. Since the subcellular
location of HBVDNAPTP1 is in the cytoplasm, it was speculated that
it interacts with PILRA via the intracellular domain. The results
of the present study demonstrated that the HA-PILRACD fusion
protein could bind with the Myc-HBVDNAPTP1 fusion protein (Fig. 3). This result indicated that the
intracellular domain of PILRACD is capable of specifically binding
to HBVDNAPTP1 in vivo.
Interaction of HBVDNAPTP1 and the
PILRA intracellular domain by GST pull-down
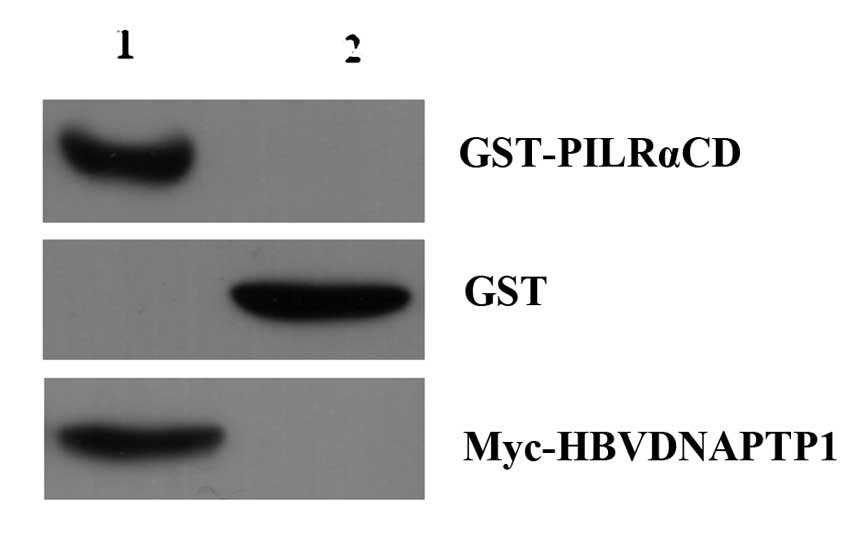
To verify the interaction of HBVDNAPTP1 and PILRA,
GST-PILRACD fusion proteins and GST-tagged proteins were incubated
with Glutathione Sepharose 4B, then incubated with the cell lysates
of pCMV-Myc-HBVDNAPTP1 transfected cells. The GST pull-down
experiment demonstrated that the GST-PILRACD fusion protein could
bind with Myc-HBVDNAPTP1, but that the GST-tagged proteins were not
capable of binding (Fig. 4). Thus,
the GST pull-down experiment further validated the interaction of
HBVDNAPTP1 and the PILRA intracellular domain.
Discussion
Chronic infection with HBV remains a worldwide
health problem. The liver is the main target organ of HBV
infection, but a number of extrahepatic tissues and organs,
including the heart, spleen, lung and kidney, can also be infected
by HBV (4–6). In previous years, various forms of
HBV DNA, replicative RNA intermediates and antigen components have
been detected in the peripheral blood mononuclear cells (PBMCs) of
chronic HBV-infected individuals (7). The major clinical effect of the HBV
infection of PBMCs is to cause host immune dysfunction, which leads
to chronic HBV infection and causes latent chronic infection and
mother-to-child transmission. Reinfection may occur following liver
transplantation or using drugs to clear viruses from the serum
(8,9). Since HBV DNA can be replicated and
transcribed by integration into the PBMC genome, the PBMCs have
become significant replication sites and propagation vectors for
HBV (10). PBMCs are mainly
composed of lymphocytes, monocytes, granulocytes and other
immunocompetent cells, which are supposed to be crucial in the
body’s immune response against HBV infection. However, HBV
infection with PBMCs leads to cell dysfunction and a decline in the
number of PBMCs, which indicates that HBV DNA replication and
transcription in PBMCs cause cell apoptosis and inhibition of cell
proliferation. Due to the inadequate HBV-specific cellular immune
response, the removal of the virus is difficult (10,11).
Compared with the PBMCs of healthy volunteers, those
of patients with chronic hepatitis B have upregulated expression of
inducing apoptosis death receptor, FasL, and downregulated
expression of resisting apoptosis decoy receptor, RAIL-R3 (12). Due to its antiviral and
immunomodulatory effects, interferon (IFN)-α 2a is the drug
treatment of choice for chronic hepatitis; however, IFN-α 2a cannot
induce PBMC apoptosis and there is no significant change in the
level of FasL expression in patients prior to and following
treatment (12–14). There is a T-helper (Th)1/Th2
imbalance in patients with chronic hepatitis B. In patients with
high levels of HBV DNA, the levels of interleukin (IL)-4 and IL-10
(Th2-type cytokines) in the peripheral blood are significantly
higher compared with those in healthy volunteers, but the levels of
IFN-γ and IL-12 (Th1-type cytokines) are significantly lower. Th2
cytokines, particularly IL-4, can promote PBMC apoptosis in
patients with chronic hepatitis B (14–15).
In the present study, a yeast two-hybrid system was
used to screen a human leukocyte cDNA library, which was validated
by intracellular co-immunoprecipitation. To the best of our
knowledge, this study is the first to report that HBVDNAPTP1
interacts with the PILRA intracellular domain. PILRA is a
transmembrane receptor with the effect of inhibitory regulation,
which is not expressed in lymphocytes, but mostly in monocytes
(16). The intracellular domain
contains two immune-receptor tyrosine-based inhibition motifs
(ITIMs). As PILRA binds with its ligand (primary cytokines), it is
triggered to form homodimers and cause self-phosphorylation and
activation of the non-receptor tyrosine kinase, Janus-activated
kinase (JAK), which is followed by catalyzing the phosphorylation
of tyrosine residues in ITIMs. At the same time, potential docking
sites containing two Src homology 2 (SH2) domains of SH2-containing
phosphatase 1 (SHP-1) tyrosine phosphatase are formed (16,17).
Once SHP-1 is docked at this docking site, the dephosphorylation of
JAK and its downstream signaling molecules, signal transducers and
activators of transcription (STAT), can be catalyzed, and thereby
terminate the cell proliferation signal or induce an apoptosis
signal (18,19). It is of note to mention that in
normal PBMCs, PILRA binds to cytokines, but does not terminate cell
proliferation or trigger apoptosis. This means that PILRA requires
a positive regulatory role to activate downstream signaling
pathways. Due to the interaction of HBVDNAPTP1 and PILRA, it is
easy to speculate that HBVDNAPTP1 is likely to bind with PILRA to
mediate the negative regulation of the JAK/STAT signaling pathway.
In the case of no ligand binding to PILRA, the downstream JAK/STAT
signaling pathway cannot be activated. However, if a ligand binds
to PILRA, the interaction of PILRA and HBVDNAPTP1 exhibits a
positive role in the regulation, which can coordinate the
activation of the downstream JAK/STAT signaling pathway, and thus
activate the monocyte apoptosis signal. The low expression of
HBVDNAPTP1 under normal conditions cannot effectively exhibit a
positive regulatory role and cannot coordinate the activation of
the JAK/STAT signaling pathway and induce apoptosis.
HBVDNAPTP1 interacting with the cytoplasmic domain
of PILRA was examined successfully by GST pull-down in vitro
and co-immunoprecipitation assay in vivo, respectively.
HBVDNAPTP1 may be involved in the negative regulation of
PILRA-mediated JAK/STAT signaling pathway.
Acknowledgements
The authors would like to thank the technical staff
of the Institute of Infectious Diseases, Beijing Ditan Hospital,
Beijing, China, for their excellent technical assistance.
References
|
1
|
Eyre NS, Phillips RJ, Bowden S, Yip E,
Dewar B, Locarnini SA and Beard MR: Hepatitis B virus and hepatitis
C virus interaction in Huh-7 cells. J Hepatol. 51:446–457. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cheng J: Trans-regulation mechanism in the
pathogenesis of viral hepatitis. Shi Jie Wei Chang Bing Xue Za Zhi.
11:888–896. 2003.(In Chinese).
|
|
3
|
Lun YZ, Lei S, Cheng J, Wang YJ, Wang Q,
Shen LT, Jiang HH and Zhang Y: Subcellular localization and
structure prediction of HBVDNAPTP1 transactivated by hepatitis B
virus DNA polymerase. Jie Fang Jun Yi Xue Za Zhi. 34:280–282.
2009.(In Chinese).
|
|
4
|
Chotiyaputta W, Pelletier SJ, Fontana RJ
and Lok AS: Long-term efficacy of nucleoside monotherapy in
preventing HBV infection in HBsAg-negative recipients of
anti-HBc-positive donor livers. Hepatol Int. 4:707–715. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lai M and Liaw YF: Chronic hepatitis B:
past, present, and future. Clin Liver Dis. 14:531–546. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pontisso P, Vidalino L, Quarta S and Gatta
A: Biological and clinical implications of HBV infection in
peripheral blood mononuclear cells. Autoimmun Rev. 8:13–17. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu L, Zhang HY, Yueng YH, Cheung KF, Luk
JM, Wang FS and Lau GK: Intracellular levels of hepatitis B virus
DNA and pregenomic RNA in peripheral blood mononuclear cells of
chronically infected patients. J Viral Hepat. 16:104–112. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Coppola N, Pisapia R, Tonziello G, Martini
S, Imparato M, Piai G, Stanzione M, Sagnelli C, Filippini P,
Piccinino F and Sagnelli E: Virological pattern in plasma,
peripheral blood mononuclear cells and liver tissue and clinical
outcome in chronic hepatitis B and C virus coinfection. Antivir
Ther. 13:307–318. 2008.
|
|
9
|
Gatta A, Giannini C, Lampertico P,
Pontisso P, Quarta S, Zignego AL, Atzeni F and Sarzi-Puttini P:
Hepatotropic viruses: new insights in pathogenesis and treatment.
Clin Exp Rheumatol. 26(1 Suppl 48): S33–S38. 2008.PubMed/NCBI
|
|
10
|
Hollinger FB and Sood G: Occult hepatitis
B virus infection: a covert operation. J Viral Hepat. 17:1–15.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bai GQ, Li SH, Yue YF and Shi L: The study
on role of peripheral blood mononuclear cell in HBV intrauterine
infection. Arch Gynecol Obstet. 283:317–321. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hou W, Liu KZ, Li MW and Wo JE: Effect of
IFNalpha-2a on Fas expression and apoptosis rate of peripheral
blood cytotoxic T cells in patients with hepatitis B. Hepatobiliary
Pancreat Dis Int. 4:403–405. 2005.PubMed/NCBI
|
|
13
|
Karan MA, Oztürk S, Yenerel M, Erten N,
Cefle K, Palanduz S and Tascioglu C: The in vitro effect of
interferon-alpha 2a on CD95 expression of T cells in hepatitis B.
Hepatogastroenterology. 50:2031–2034. 2003.PubMed/NCBI
|
|
14
|
Xing TJ, Zhang L, Luo KX, Hou JL, Zhang MX
and Feng XR: Effect of cytokines on lymphocyte apoptosis in the
patients with chronic hepatitis B in vitro. Med J Chin PLA.
25:16–18. 2000.(In Chinese).
|
|
15
|
Park Y, Park Y, Han KH and Kim HS: Serum
cytokine levels in patients with chronic hepatitis B according to
lamivudine therapy. J Clin Lab Anal. 25:414–421. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fournier N, Chalus L, Durand I, Garcia E,
Pin JJ, Churakova T, Patel S, Zlot C, Gorman D, Zurawski S, Abrams
J, Bates EE and Garrone P: FDF03, a novel inhibitory receptor of
the immunoglobulin superfamily, is expressed by human dendritic and
myeloid cells. J Immunol. 165:1197–1209. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mousseau DD, Banville D, L’Abbé D,
Bouchard P and Shen SH: PILRalpha, a novel immunoreceptor
tyrosine-based inhibitory motif-bearing protein, recruits SHP-1
upon tyrosine phosphorylation and is paired with the truncated
counterpart PILRbeta. J Biol Chem. 275:4467–4474. 2000. View Article : Google Scholar
|
|
18
|
Saha B, Jyothi Prasanna S, Chandrasekar B
and Nandi D: Gene modulation and immunoregulatory roles of
interferon gamma. Cytokine. 50:1–14. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xiao W, Kashiwakura J, Hong H, Yasudo H,
Ando T, Maeda-Yamamoto M, Wu D, Kawakami Y and Kawakami T:
Phospholipase C-β3 regulates FcɛRI-mediated mast cell activation by
recruiting the protein phosphatase SHP-1. Immunity. 34:893–904.
2011.
|