Introduction
Renal cell carcinoma (RCC) accounts for 2–3% of all
adult cancers, is more common in males than females (2:1) and most
commonly occurs in patients aged 50–70 years (1). The incidence of RCC has increased and
RCC is the most lethal of all urological malignancies (2). The development of macroscopic
metastases is the predominant cause of tumor-associated mortality
(3,4). For RCC metastatic disease affects
one-third of patients at diagnosis and an additional one-third
develop metastatic disease following radical nephrectomy (5). Patients who present with metastatic
disease have a median survival time of 7–11 months and a 5-year
mortality rate of >90% (6).
Additionally, clear-cell RCC (ccRCC) is the most common type
(70–80%) of RCC metastatic disease (7). Therefore, molecular markers are
urgently required to predict individual metastatic risk and
prognosis in ccRCC patients. Such markers represent the
prerequisite for optimal follow-up and treatment of patients
following surgery.
Glycoprotein non-metastatic melanoma protein B
(Gpnmb) is a transmembrane glycoprotein that is expressed in
various types of cancer. This protein was first cloned from a
melanoma cell line and is expressed at a high level in a number of
melanoma cell lines (8). The
gpnmb gene is located on human chromosome 7q15 and encodes a
type I transmembrane protein that is expressed in a wide variety of
human tissues and cells, including the embryonic nervous system,
developing nephrons, the germinal cells of hair follicles,
osteoblasts, osteoclasts, myocytes, retinal pigment epithelium,
renal tubules, macrophages and dendritic cells (9–11).
The Gpnmb protein consists of at least four domains: An N-terminal
domain with a signal peptide, a polycystic kidney disease domain, a
transmembrane domain and an ARG-GLY-ASP domain. The Arg-Gly-Asp
(RGD) cell attachment domain is for integrin-mediated cell
attachment and migration. Further analysis has suggested that Gpnmb
is highly glycosylated and has two isoforms: A secreted type and a
transmembrane type. Moreover, the expression of Gpnmb is directly
regulated by microphthalmia-associated transcription factor
(12). Using immunohistochemistry,
Hong et al (8) were the
first to report Gpnmb expression in several cases of RCC. Since
Gpnmb promotes the migration, invasion and metastasis of tumor
cells, this protein represents a potential immunomarker for
RCC.
Previous studies have revealed that the differential
expression of proteomic markers in primary tumors and metastatic
ccRCC tissues is important in drug resistance (1,5,13–15).
Furthermore, poor prognoses and therapeutic outcomes may be
attributed to a high level of these molecules, which are associated
with metastasis. To the best of our knowledge, no studies have
evaluated the difference in Gpnmb expression between primary and
metastatic ccRCCs. In the present study, ccRCC specimens were
selected according to a strict protocol to detect differences in
Gpnmb expression levels between primary and matched metastatic
ccRCC samples. In addition, the prognostic significance and
therapeutic potential of these differential expression levels were
analyzed.
Materials and methods
Patient and tumor characteristics
Two tissue microarrays were constructed. The first
array consisted of primary ccRCCs and their matched metastases
(n=12), including seven ccRCCs with bone metastases, one with lung
metastases, two with adrenal metastases and two with lymph node
metastases near the inferior vena and/or renal hilus. The second
array consisted of a subset that included only primary ccRCC
samples (n=43). Twelve ccRCC patients from the archives of the
Department of Pathology at the People’s Hospital of Peking
University (Beijing, China) were initially recruited for the
present study according to a protocol that was approved by the
Institutional Review Board. The clinicopathological characteristics
of the patients are summarized in Table I. The second array was designed by
Shanghai Outdo Biotech Co., Ltd. (Shanghai, China) and contained
material from 43 primary ccRCC patients. Clinical follow-up data
were available for 43 patients who had undergone nephrectomy. The
follow-up duration was calculated as the interval in months between
initial presentation and either the final visit or RCC-related
mortality. The length of the follow-up time ranged from 1–74
months. At the end of follow-up, 14 patients had succumbed to
disease progression.
 | Table IPatient characteristics of the 12
paired renal cell carcinoma cases. |
Table I
Patient characteristics of the 12
paired renal cell carcinoma cases.
| Characteristic | Variable |
|---|
| Case number | 12 |
| Age (years), median
(range) | 53.5 (21–74) |
| Gender,
male/female | 9/3 |
| Pathological
findings |
| Clear cell type,
n | 12 |
| Fuhrman grade, n |
| ≤II | 7 |
| >II | 5 |
Immunohistochemical analysis and
evaluation
Immunohistochemistry was performed using the
Envision™ ABC kit (Dako, Capinteria, CA, USA) according to the
manufacturer’s instructions. After washing in phosphate-buffered
saline, the tissue microarray and slides were incubated in 0.3%
hydrogen peroxide for 10 min to quench endogenous peroxidase
activity. The tissues were then pre-incubated with goat serum and
incubated overnight at 4°C in a humidified chamber with a primary
antibody against Gpnmb (monoclonal mouse; 1:100; MAB25501, R&D
Systems, Minneapolis, MN, USA). The primary antibody was detected
using the Envision™ ABC kit. The color was developed with
diaminobenzidine. Following counterstaining with hematoxylin, the
sections were dehydrated and mounted. Gpnmb expression was scored
as positive only upon observation of membranous staining and was
measured using the H-score [intensity (1, 2, 3 or 4) plus the
distribution (%)]. The expression was divided into three groups
according to positive distribution of the tumor cell membranes: 1,
0–5%; 2, 6–50%; and 3, >50%. To assess intratumor heterogeneity,
three distinct microscopic fields (magnification, ×200) per
specimen were used to evaluate the expression of Gpnmb with a Leica
DM400B (Leica Microsystems, Wetzlar, Germany), using a DM2500 image
analysis system (exposure value, 16.5 ms; saturation, 1.75; Gamma,
0.95; Gain, 1.0). Subsequently, the scores were averaged to obtain
a single concatenated score for each tissue for Gpnmb expression.
Gpnmb expression was assessed by two independent pathologists who
were blinded to the clinicopathological data. The data are
expressed as the mean value of the triplicate experiments.
Selection of cutoff score for positive
Gpnmb expression
The selection of clinically important cutoff scores
for Gpnmb expression was determined by receiver operating
characteristic (ROC) curve analysis in the 43 primary ccRCCs
(16). To determine the Gpnmb
score, the sensitivity and specificity for each outcome in the
study were plotted, thus generating various ROC curves. The score
was selected as the cutoff value closest to the points of maximum
sensitivity and maximum specificity. The tumors designated as
having ‘low expression’ of Gpnmb were those with scores below or
equal to the cutoff value, whereas tumors with ‘high expression’
were those with scores above the cutoff value. To facilitate ROC
curve analysis, the cases were dichotomized as follows: death vs.
others (censored, alive).
Follow-up
All patients provided follow-up records for more
than four years. Following completion of therapy, the patients were
observed at 3-month intervals during the first three years and at
6-month intervals thereafter. Overall survival (OS) was defined as
the time from diagnosis to the date at which the patient succumbed
to the disease or the latest date of census if the patient was
still alive.
Statistical analysis
The mean scores and the differences between primary
and metastatic tumors were analyzed using paired t-tests. For
survival analysis, the optimal cutoff point for Gpnmb expression
was obtained by ROC analysis of the primary ccRCC set (n=43). For
validation, the correlation between Gpnmb expression, which was
classified using a ROC analysis-generated cutoff point, and OS was
evaluated in this set. The χ2 test or Fisher’s exact
test was employed to evaluate the correlation between Gpnmb
expression and clinicopathological variables. The multivariate Cox
proportional-hazards model was utilized to estimate the hazard
ratios for patient outcome. The correlations between Gpnmb
expression and OS were determined by Kaplan-Meier analysis.
Log-rank tests were performed to assess differences in survival
probabilities among patient subsets. All P-values were two-sided,
and P<0.05 was considered to indicate a statistically
significant difference. Statistical analysis was performed using
SPSS 17.0 (SPSS, Inc., Chicago, IL, USA).
Results
Gpnmb expression in the two ccRCC
sets
The clinical features of 12 pairs of primary ccRCCs
and matched metastatic tissues from patients, including age,
gender, histologic differentiation and Fuhrman grade are summarized
in Table I. Gpnmb expression was
examined in 12 pairs of primary ccRCCs and matched metastatic
tissues by immunohistochemistry and Gpnmb was found to be strongly
expressed in the metastatic lesions, with significant upregulation
in metastatic RCCs compared with matched primary tissue (Fig. 1, P=0.036). To further assess
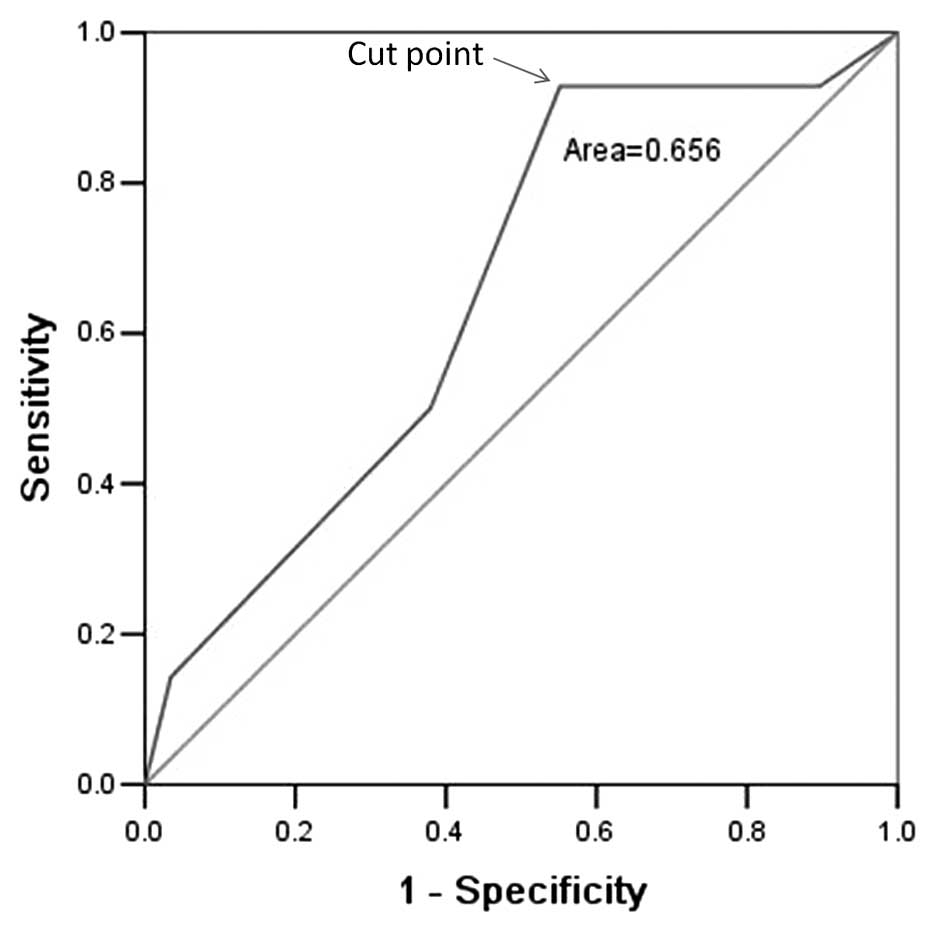
survival, ROC curve analysis was employed to determine the cutoff
score for Gpnmb expression. The Gpnmb cutoff score for OS in 43
primary ccRCCs was 5 (P<0.05, Fig.
2; the cutoff value closest to the points of maximum
sensitivity, 0.929, and maximum specificity, 0.448). Therefore, a
Gpnmb expression score of 5 (>5 vs. ≤5) was selected as the
uniform cutoff point for survival analysis in the tested set.
Correlation between Gpnmb expression and
ccRCC patient clinicopathologic features
In the cohort of 43 primary ccRCCs, high Gpnmb
expression was observed in 29/43 (67.44%) of the ccRCC samples. A
correlation analysis revealed that there was no significant
correlation identified between Gpnmb expression and
clinicopathological features, including patient gender, age,
clinical stage and Fuhrman grade (P>0.05, Table II).
 | Table IIAssociation between Gpnmb expression
and patient characteristics in renal cell carcinoma. |
Table II
Association between Gpnmb expression
and patient characteristics in renal cell carcinoma.
| Variable | n | Positive n (%) | P-valuea |
|---|
| Gender |
| Male | 25 | 16 (64.0) | 0.570 |
| Female | 18 | 13 (72.0) | |
| Age (years)b |
| ≤57 | 23 | 15 (65.2) | 0.739 |
| >57 | 20 | 14 (70.0) | |
| Clinical stage |
| 1 | 19 | 11 (57.9) | 0.235 |
| 2,3 | 24 | 18 (75.0) | |
| Fuhrman grade |
| ≤II | 20 | 11 (55.0) | 0.104 |
| >II | 23 | 18 (78.3) | |
Gpnmb expression and survival analysis:
Univariate survival analysis
Kaplan-Meier analysis demonstrated that elevated
Gpnmb expression predicted inferior OS in the 43 primary ccRCCs
(P=0.02, Fig. 3).
Independent prognostic factors for
ccRCC
As the variables that were observed to have a
prognostic influence on ccRCC patients in the univariate analyses
may be correlated, the expression of Gpnmb, clinical stage and
Fuhrman grade, which were significant in the univariate analyses in
the two cohorts, were further evaluated in a multivariate analysis.
Gpnmb expression in ccRCC tissues was found to represent an
independent prognostic factor for poor OS in the 43 primary ccRCCs
(Table III).
 | Table IIIResults of the multivariate Cox
proportional-hazards analysis. |
Table III
Results of the multivariate Cox
proportional-hazards analysis.
| Variable | Hazard ratio | P-value |
|---|
| Age ≤57.00 years (vs.
>57 years) | 1.037 | 0.950 |
| Male gender (vs.
female) | 0.654 | 0.446 |
| Clinical stage III
(vs. II+I) | 1.315 | 0.663 |
| Fuhrman grade ≤II
(vs. >II) | 3.301 | 0.070 |
| Gpnmb-positive (vs.
negative) | 7.719 | 0.049 |
Discussion
Previous studies have identified gene expression
changes resulting in aggressive behavior or metastatic potential in
RCC (17,18). In addition, a bone-seeking clone of
metastatic ccRCC was developed (19), which demonstrated that the
metastasis of ccRCC cells may be the result of a more invasive
subclone derived from primary ccRCC cells. A study by Thompson
et al (20) showed that
high B7-H1 expression is associated with poor prognosis in primary
and metastatic RCC; although only one patient was represented in
each cohort, metastatic specimens demonstrated higher B7-H1
expression than primary specimens (54.3% vs. 44.4%, respectively).
Mutant p53 expression was also significantly higher in metastatic
tumors compared with primary tumors (51.8% vs. 22.8%) in a study by
Zigeuner et al (21), and
p53 overexpression (risk ratio, 2.9; 95% confidence interval,
1.3–6.7; P=0.01) was revealed to be an independent prognostic
factor for metastasis-free survival, although the specimens were
not matched. In a study of mammalian target of rapamycin- and
hypoxia-induced pathway members, which included 135 primary RCCs
and 41 unrelated metastases, differential global patterns of
expression were measured (22).
The levels of p-AKT, p-S6, 4EBP1 and c-myc were higher in
metastatic lesions compared with primary tissues.
Differential protein expression between primary
ccRCC and subsequent metastases may reveal molecules responsible
for metastasis. This may aid in the development of strategies to
overcome drug resistance and novel therapies (1,5,13,14).
A larger study of 168 metastatic RCC patients who received targeted
therapy in situ for their primary tumors reported negligible
decreases in the size of the primary tumors (23). Karashima et al (15) reported that the expression levels
of a number of angiogenesis-related genes in metastases were
relatively higher than in primary tumors. In addition, an in
vitro proliferation assay demonstrated the relatively increased
resistance of metastatic ccRCC cells to targeted therapy compared
with matched primary tumor cells. Clinical observations of
discordance in the responses of primary and metastatic tumors also
suggest possible differences among stages of tumor biology. One
potential weakness of previous studies involves the use of
non-matched primary and metastatic tumor tissue. For example,
metastatic ccRCCs may have different tumor grades and stages in the
primary setting when compared with primary ccRCCs, which may have
influenced the distribution of the biomarker levels evaluated. In
the present study, immunostaining was performed on a tissue set
from 12 patients with primary ccRCCs and paired metastases, and the
result demonstrated that the expression of Gpnmb was upregulated in
metastatic ccRCC compared with primary carcinoma tissue
(P=0.036).
Metastasis is a predominant cause of mortality in
clear-cell carcinoma. However, one significant problem in the
clinical management of patients presenting with localized ccRCC is
the inability to determine tumor aggressiveness and accurately
predict which patients are at greater risk of experiencing distant
metastases following surgery. A ‘metastasis signature’ derived from
primary RCCs with different prognoses may be used to classify
tumors with and without metastases at the time of surgery (1). The purpose of the present study was
to identify molecules associated with a ‘metastasis signature’
indicative of poor prognosis. Gpnmb is a transmembrane glycoprotein
that is expressed in various types of cancer and promotes the
migration, invasion and metastasis of tumor cells. This protein is
also highly expressed in several aggressive types of cancer,
including melanoma (a cancer with high propensity for metastasizing
to bone), glioma and breast cancer (24–27).
Finally, the ectopic expression of Gpnmb is sufficient to enhance
invasive phenotypes in vitro and metastatic capabilities
in vivo (28). Hong et
al (8) previously demonstrated
that Gpnmb is important in RCC. The present study assessed
differential Gpnmb expression between primary ccRCCs and paired
metastases, with the hypothesis that metastasized ccRCC with a high
expression of Gpnmb may be the result of a more invasive subclone
derived from the primary ccRCC and may be responsible for poor
prognosis. Furthermore, the present study assessed the differential
prognosis of another 43 primary ccRCCs following radical
nephrectomy, which revealed that the high expression of Gpnmb was
positively associated with poor survival (P=0.02). The present
study also identified that Gpnmb served as an independent
prognostic biomarker for OS in ccRCC (P=0.049).
In conclusion, these findings provide evidence that
the elevated expression of Gpnmb in ccRCC may promote a malignant
phenotype with a poor prognosis. To develop an objective Gpnmb
cutoff point for the survival analysis, a ROC curve analysis was
performed to generate a cutoff score for the 43 ccRCCs. Gpnmb is
expressed at the surface of cancer cells (24) but is predominantly expressed
intracellularly in normal cells, such as macrophages and
melanocytes (29,30). This expression pattern renders
Gpnmb particularly attractive for antibody-based therapies since,
as a target, the protein is more readily accessible in cancer cells
than normal cells, thereby reducing potential complications due to
bystander effects. A monoclonal antibody-drug conjugate,
CR011-vcMMAE, is currently under development for the treatment of
Gpnmb-expressing cancers. The results of the present study identify
a molecule that is responsible for metastasis and highlight the
requirement for further pathway analysis of metastatic ccRCC to
overcome drug resistance and develop novel therapeutic
strategies.
References
|
1
|
Tan X, Zhai Y, Chang W, et al: Global
analysis of metastasis-associated gene expression in primary
cultures from clinical specimens of clear-cell renal-cell
carcinoma. Int J Cancer. 123:1080–1088. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fang Y, Wei J, Cao J, et al: Protein
expression of ZEB2 in renal cell carcinoma and its prognostic
significance in patient survival. PLoS One. 8:e625582013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Minn AJ, Kang Y, Serganova I, et al:
Distinct organ-specific metastatic potential of individual breast
cancer cells and primary tumors. J Clin Invest. 115:44–55. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nguyen DX and Massagué J: Genetic
determinants of cancer metastasis. Nat Rev Genet. 8:341–352. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Laird A, O’Mahony FC, Nanda J, et al:
Differential expression of prognostic proteomic markers in primary
tumour, venous tumour thrombus and metastatic renal cell cancer
tissue and correlation with patient outcome. PLoS One.
8:e604832013. View Article : Google Scholar
|
|
6
|
Abou Youssiff T, Fahmy MA, Koumakpayi IH,
et al: The mammalian target of rapamycin pathway is widely
activated without PTEN deletion in renal cell carcinoma metastases.
Cancer. 117:290–300. 2011.PubMed/NCBI
|
|
7
|
Reuter VE: The pathology of renal
epithelial neoplasms. Semin Oncol. 33:534–543. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hong SB, Oh H, Valera VA, Baba M, Schmidt
LS and Linehan WM: Inactivation of the FLCN tumor suppressor gene
induces TFE3 transcriptional activity by increasing its nuclear
localization. PLoS One. 5:e157932010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hoashi T, Sato S, Yamaguchi Y, Passeron T,
Tamaki K and Hearing VJ: Glycoprotein nonmetastatic melanoma
protein b, a melanocytic cell marker, is a melanosome-specific and
proteolytically released protein. FASEB J. 24:1616–1629. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Abdelmagid SM, Barbe MF, Hadjiargyrou M,
et al: Temporal and spatial expression of osteoactivin during
fracture repair. J Cell Biochem. 111:295–309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Safadi FF, Xu J, Smock SL, Rico MC, Owen
TA and Popoff SN: Cloning and characterization of osteoactivin, a
novel cDNA expressed in osteoblasts. J Cell Biochem. 84:12–26.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Loftus SK, Antonellis A, Matera I, et al:
Gpnmb is a melanoblast-expressed, MITF-dependent gene. Pigment Cell
Melanoma Res. 22:99–110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tan X, He S, Han Y, et al: Establishment
and characterization of clear cell renal cell carcinoma cell lines
with different metastatic potential from Chinese patients. Cancer
Cell Int. 13:202013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ohno Y, Izumi M, Tachibana M, et al:
Characterization and gene expression analysis of novel matched
primary and metastatic renal cell carcinoma cell lines. Oncol Rep.
20:501–509. 2008.PubMed/NCBI
|
|
15
|
Karashima T, Fukuhara H, Tamura K, et al:
Expression of angiogenesis-related gene profiles and development of
resistance to tyrosine-kinase inhibitor in advanced renal cell
carcinoma: characterization of sorafenib-resistant cells derived
from a cutaneous metastasis. Int J Urol. 20:923–930. 2013.
View Article : Google Scholar
|
|
16
|
Saw RP, Morgan M, Koorey D, et al: p53,
deleted in colorectal cancer gene, and thymidylate synthase as
predictors of histopathologic response and survival in low, locally
advanced rectal cancer treated with preoperative adjuvant therapy.
Dis Colon Rectum. 46:192–202. 2003. View Article : Google Scholar
|
|
17
|
Jones J, Otu H, Spentzos D, et al: Gene
signatures of progression and metastasis in renal cell cancer. Clin
Cancer Res. 11:5730–5739. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kosari F, Parker AS, Kube DM, et al: Clear
cell renal cell carcinoma: gene expression analyses identify a
potential signature for tumor aggressiveness. Clin Cancer Res.
11:5128–5139. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J, Chen A, Yang C, Zeng H, Qi J and
Guo FJ: A bone-seeking clone exhibits different biological
properties from the ACHN parental human renal cell carcinoma in
vivo and in vitro. Oncol Rep. 27:1104–1110. 2012.PubMed/NCBI
|
|
20
|
Thompson RH, Gillett MD, Cheville JC, et
al: Costimulatory molecule B7-H1 in primary and metastatic clear
cell renal cell carcinoma. Cancer. 104:2084–2091. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zigeuner R, Ratschek M, Rehak P, Schips L
and Langner C: Value of p53 as a prognostic marker in histologic
subtypes of renal cell carcinoma: a systematic analysis of primary
and metastatic tumor tissue. Urology. 63:651–655. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schultz L, Chaux A, Albadine R, et al:
Immunoexpression status and prognostic value of mTOR and
hypoxia-induced pathway members in primary and metastatic clear
cell renal cell carcinomas. Am J Surg Pathol. 35:1549–1556. 2011.
View Article : Google Scholar
|
|
23
|
Abel EJ, Culp SH, Tannir NM, et al:
Primary tumor response to targeted agents in patients with
metastatic renal cell carcinoma. Eur Urol. 59:10–15. 2011.
View Article : Google Scholar
|
|
24
|
Tse KF, Jeffers M, Pollack VA, et al:
CR011, a fully human monoclonal antibody-auristatin E conjugate,
for the treatment of melanoma. Clin Cancer Res. 12:1373–1382. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rich JN, Shi Q, Hjelmeland M, et al:
Bone-related genes expressed in advanced malignancies induce
invasion and metastasis in a genetically defined human cancer
model. J Biol Chem. 278:15951–15957. 2003. View Article : Google Scholar
|
|
26
|
Kuan CT, Wakiya K, Dowell JM, et al:
Glycoprotein nonmetastatic melanoma protein B, a potential
molecular therapeutic target in patients with glioblastoma
multiforme. Clin Cancer Res. 12:1970–1982. 2006. View Article : Google Scholar
|
|
27
|
Rose AA, Pepin F, Russo C, Abou Khalil JE,
Hallett M and Siegel PM: Osteoactivin promotes breast cancer
metastasis to bone. Mol Cancer Res. 5:1001–1014. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Onaga M, Ido A, Hasuike S, et al:
Osteoactivin expressed during cirrhosis development in rats fed a
choline-deficient, L-amino acid-defined diet, accelerates motility
of hepatoma cells. J Hepatol. 39:779–785. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ripoll VM, Irvine KM, Ravasi T, Sweet MJ
and Hume DA: Gpnmb is induced in macrophages by IFN-γ and
lipopolysaccharide and acts as a feedback regulator of
proinflammatory responses. J Immunol. 178:6557–6566.
2007.PubMed/NCBI
|
|
30
|
Tomihari M, Hwang SH, Chung JS, Cruz PD Jr
and Ariizumi K: Gpnmb is a melanosome-associated glycoprotein that
contributes to melanocyte/keratinocyte adhesion in a RGD-dependent
fashion. Exp Dermatol. 18:586–595. 2009. View Article : Google Scholar : PubMed/NCBI
|