Introduction
The microRNA-34 (miR-34) family comprises three
homologous molecules, miR-34a, miR-34b and miR-34c. Previous
studies have demonstrated that p53 regulates the transcription of
miR-34a, and that miR-34a is involved in the p53 pathway (1–4).
Members of the miR-34 family have been widely suggested to act as
important tumor suppressors and mediators of p53 effects (5–9).
Reduced miR-34a expression is also observed in leukemia-related
thrombocythemia, polycythemia, and primary myelofibrosis. He et al
(10) showed that cyclin E2
(CCNE2), cyclin-dependent kinase 4 (CDK4) and the
hepatocyte growth factor receptor (MET) genes are targets of
miR-34a. Navarro et al (11)
reported that miR-34a directly regulates the expression of
MYB, facilitating megakaryocyte differentiation, and the
expression of CDK4 and CDK6, to inhibit the G1/S
cell cycle transition. Yamakuchi et al (5) showed that miR-34a-mediated repression
of the expression of the silent information regulator 1 gene
(SIRT1) leads to an increase in acetylated p53 and
expression of p21 and PUMA, transcriptional targets
of p53 that regulate the cell cycle and apoptosis, respectively.
The levels of miR-34s are reported to be undetectable or reduced in
various types of cancer, including nasopharyngeal carcinoma,
gastric, pancreatic and prostate cancer, retinoblastoma and
leukemia (12–17).
The human promyelocytic cell line HL60, derived from
an acute promyelocytic leukemia patient, provides a continuous
source of human cells for studying the molecular events of myeloid
differentiation and the effects of physiologic, pharmacologic, and
virologic elements in this process (18). The expression of CDK4,
MYB and SIRT1 in peripheral blood samples collected
from children with or without acute myelogenous leukemia was
examined, as well as the correlation between the expression of
miRNA-34a and that of CDK4, MYB and SIRT1.
Then, the interfering miRNA-34a was used to downregulate the
expression of CDK4, MYB and SIRT1 in the HL-60
cell line, in order to further investigate its role in regulation
of these genes in AL.
Materials and methods
Ethical considerations
The study was approved by the Ethics Committee of
the Capital Institute of Pediatrics-Affiliated Children’s Hospital
in May 2011, which approved the blood sample extraction and data
collection procedures. All the experiments were undertaken
following the provisions of the Declaration of Helsinki. Written
informed consent was obtained from the children’s parents.
Subjects
A total of 29 pediatric patients (12 male and 17
female) with acute leukemia (AL) were recruited from the Hematology
Department of the Capital Institute of Pediatrics-Affiliated
Children’s Hospital and the Department of Pediatrics in the General
Hospital of the Chinese PLA between February and November, 2011.
These AL patients had a median age of 5.1 years (range, 1–13.5
years), and were diagnosed with acute leukemia based on the
morphologic, immunologic and cytogenetic (MIC) system, where the
morphologic test was conducted on bone marrow cells. Patients with
the AML-M3 subtype of AL were not included in this study. All the
patients were diagnosed within 7 days, with 25 diagnosed with acute
lymphoblastic leukemia [7 high risk (HR), 7 intermediate risk (IR)
and 15 standard risk (SR) cases] and the remaining 4 patients
diagnosed with acute myeloid leukemia. Twenty-one age-matched
healthy children were recruited as controls. Clinical information
on all the participants, including age, gender, peripheral white
blood cell (WBC) count, MIC classification and response to
prednisone was recorded (Table I),
and all the patients were divided into subgroups based on the above
parameters to analyze the associations between these factors and
CDK4, MYB and SIRT1 expression.
 | Table IAnalysis of mRNA levels of
CDK4 and MYB in peripheral blood of pediatric
patients with acute leukemia (AL). |
Table I
Analysis of mRNA levels of
CDK4 and MYB in peripheral blood of pediatric
patients with acute leukemia (AL).
| Variables | | No. (%) | MYBa | P | CDK4a | P |
|---|
| Age | <3 years | 8 (27.59) | 1.26±0.37 | P<0.05 | 1.11±0.45 | P>0.05 |
| >3 years | 21 (72.42) | 2.69±0.26 | | 1.78±0.33 | |
| Gender | Male | 12 (41.38) | 3.40±0.21 | P>0.05 | 1.62±0.41 | P>0.05 |
| Female | 17 (58.62) | 0.01±0.32 | | 1.60±0.31 | |
| WBCs | ≤50 | 22 (41.38) | 5.64±0.38 | P<0.01 | 1.61±0.28 | P>0.05 |
| >50 | 7 (24.14) | 0.50±0.29 | | 1.44±0.58 | |
| AL Type | ALL | 25 (86.07) | 3.64±0.15 | P>0.05 | 1.78±0.31 | P>0.05 |
| ANLL | 4 (13.80) | 1.24±0.31 | | 0.91±0.39 | |
| Risk | HR | 7 (24.14) | 1.35±0.30 | P>0.05 | 1.46±0.40 | P>0.05 |
| IR | 7 (24.13) | 2.29±0.41 | | 1.76±0.23 | |
| SR | 15 (52.72) | 2.91±0.30 | | 1.39±0.41 | |
| Prednisone
responseb | Sensitive | 18 (69.23) | 2.10±0.31 | P>0.05 | 1.20±0.28 | P>0.05 |
| Insensitive | 8 (30.78) | 4.20±0.29 | | 2.35±0.30 | |
|
Immunophenotypec | B | 19 (76.00) | 3.45±0.34 | P>0.05 | 1.46±0.36 | P>0.05 |
| T | 6 (24.00) | 10.03±0.20 | P>0.05 | 2.15±0.43 | |
| Gene fusion
testd | Positive | 6 (22.22) | 1.39±0.36 | | 1.37±1.37 | P>0.05 |
| Negative | 11 (40.74) | 5.22±0.26 | | 1.73±0.43 | |
Blood samples and RNA extraction
A total of 3 ml of peripheral blood was collected
from the participants of each group. Heparin (10 IU/ml) was used
for anticoagulation, and mononuclear cells were isolated by
lymphocyte separation solution (Huamei Biotechnology Co., Beijing,
China). Total RNA was isolated using the TRIzol reagent (Invitrogen
Life Technologies, Carlsbad, CA, USA) following the manufacturer’s
protocol.
miRNA-34a expression analysis
Isolation of peripheral blood mononuclear cells
(PBMCs) from blood of AL patients was achieved by centrifugation in
a Ficoll gradient as previously described (19). After washing with
phosphate-buffered saline (PBS), cells were resuspended in
RPMI-1640 medium to a final concentration of 1×107
cells/ml. Total RNA samples extracted from PBMCs were reverse
transcribed and quantitative PCR (qPCR) was performed on the
resulting cDNA to determine the level of miR-34a using a ReverTra
Ace® qPCR RT Kit (Toyobo Co., Ltd., Osaka, Japan) and
ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA,
USA). The expression of the small RNA RNU6B was used as internal
reference for normalization. miR-34a Primers were synthesized as
follows: forward, CTTGAACTCCTGGGGCCTGAAG; reverse,
GCCAAAGAAACACTCACAGCT. Amplifi cation consisted of 2 min at 95°C,
followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec.
Melting curve analysis was performed at the end of the PCR cycles
in order to validate the specificity of the expected PCR product.
All samples were run in duplicate, including blank controls without
cDNA. The cycle threshold (Ct) was defined as the number of cycles
required for the fluorescent signal to cross the threshold in qPCR.
The formula 2ΔCt was used to calculate the levels of
miRNAs in the serum, where ΔCt=mean (Ct of internal references) −
Ct of target miRNA.
Cell culture and in vitro
transfection
Human HL-60 cells were purchased from the American
Type Culture Collection (ATCC; Manassas, VA, USA). Cells were
maintained in RPMI-1640 medium (Neuronbc, Beijing, China)
supplemented with 10% fetal bovine serum (FBS) from HyClone
Laboratories, Inc. (Logan, UT, USA) and were kept under 5%
CO2 at 37°C. Three groups were defined for the in
vitro experiments: i) test group, in which cells were
transfected with a sequence specific to miRNA-34a (pre-miR34a); ii)
negative control (NC) group, transfected with scrambled miRNA; iii)
blank control (BC) group, where cells were not transfected.
Transient transfection of cells was performed on
six-well plates, using the following sequences (Invitrogen Life
Technologies, Grand Island, NY, USA): pre-miRNA34a, 5′-UGG CAG UGU
CUU AGC UGG UUG U-3′; scrambled miRNA, 5′-UCA CAA CCU CCU AGA AAG
AGU AGA-3′. The Lipofectamine® RNAiMAX transfection
reagent (Invitrogen Life Technologies) was used according to the
manufacturer’s instructions. A total of 5 μl Lipofectamine was used
for transfection of 30 nM miRNA (pre-miRNA34a or scrambled) per
well.
Target gene expression analysis
After 48 h of transfection, total RNA was extracted,
and first strand (cDNA) was synthesized using the ReverTra
Ace® reverse transcription kit (Toyobo Co., Ltd., Osaka,
Japan). The CDK4, MYB and SIRT1 genes were
amplified using the ABI 7500 real-time PCR system, with the
glyceraldehyde 3-phosphate dehydrogenase gene (GAPDH)
serving as an internal control. The amplification was performed as
follows: one cycle of pre-denaturation at 95°C for 5 min, followed
by 40 cycles of denaturation at 95°C for 15 sec and extension at
60°C for 1 min. Sequences of the upstream and downstream primers
used in these reactions were: CDK4, 5′-GCC CAA TCA GGT CAA
AGA TT-3′ and 5′-ACA TGT GGA GTG TTG GCT GT-3′; MYB, 5′-TGT
TCC ATT CTG TTC CAC CA-3′ and 5′-GAC TAT GAT GGG CTG CTT CC-3′;
SIRT1, 5′-TGA CAG AGA GAT GGC TGG AA-3′ and 5′-CCA GAT CCT
CAA GCG ATG TT-3′; GAPDH, 5′-CGA GAT CCC TCC AAA ATC AA-3′
and 5′-TTC ACA CCC ATG ACG AAC AT-3′.
MTT assay
Cells were cultured in 96-well plates. Following
transfection, 20 μl of 5 mg/ml MTT (Sigma-Aldrich, St. Louis, MO,
USA) was added to each well. After incubation at 37°C for 4 h, 50
μl of the supernatant was carefully removed and 150 μl dimethyl
sulfoxide (DMSO; Sigma-Aldrich) was added. Samples were agitated
for 10 min, and absorbance was measured at 550 nm on a microplate
reader (SPECTRA Fluor, Tecan, Männedorf, Switzerland). Three
replicated wells were measured per group. The BC group contained no
cells.
Cell viability test
The trypan blue assay was used to assess the
viability of HL-60 cells. Briefly, the HL-60 cell suspension was
mixed with 0.4% trypan blue solution (9:1) (Huamei Biotechnology
Co., Wuhan, China). Dead cells were stained blue and living cells
remained unstained. Unstained and stained cells were counted within
3 min. HL-60 cell viability = living cells/(living cells + dead
cells) ×100%.
Western blot analysis
Cells were washed twice with ice-cold PBS and then
treated with lysis buffer for 30 min at 4°C. The supernatants were
centrifuged at 12,000 × g for 30 min at 4°C. Protein samples were
electrophoresed on 7.5% SDS-PAGE gels and transferred to
polyvinylidene fluoride (PVDF) membranes. Non-specific reactivity
was blocked by incubating with 5% non-fat dry milk in a mixture of
Tris-buffered saline and Tween 20 (TBS/T) for 1 h at room
temperature. The membrane was then treated with primary antibodies
targeting CDK4, MYB and SIRT1 proteins. Each sample was also
treated with anti-β-actin antibody (Sigma-Aldrich) as a
control.
Statistical analysis
Data are expressed as mean ± standard deviation (SD)
and were analyzed by Student’s t-tests. Pearson’s correlation
analysis was used to determine the relationship between expression
of miRNA-34a and that of CDK4, MYB and SIRT1.
P<0.05 was considered to indicate statistically significant
differences. The analyses were performed with the SAS 9.1
statistical software (SAS Institute, Cary, NC, USA).
Results
In vivo expression of miR-34a, CDK4, MYB
and SIRT1
The expression of miRNA-34a in acute leukemia
children patients was significantly lower compared to healthy
controls (P<0.01). The miR-34a levels were highly variable among
patients, ranging from undetectable to 35% of average control. The
average relative expression of miR-34a in AL patients was only
14.93% of the healthy controls (0.66 of 2−ΔΔct vs. 3.11
of 2−ΔΔct in controls).
The expression levels of CDK4 and MYB
in the peripheral blood of AL children were significantly higher
compared to those of healthy children (P<0.01), while the
expression of SIRT1 showed no significant difference between
the two groups (Fig. 1). Sub-group
analysis for AL children showed that MYB expression was
significantly higher in patients >3 years of age compared to
patients <3 years of age, and a similar result was obtained when
comparing patients with WBC counts above and below
5×1010/l (Table I).
There was no significant difference in the expression of
CDK4 and MYB between subgroups when they were
classified by age (for CDK4), gender, AL subtype,
immunophenotype, WBC count in peripheral blood at diagnosis (for
CDK4), risk factors, appearance of fusion genes, and
response to prednisone therapy.
Correlation analysis
Pearson’s correlation analysis demonstrated that the
expression of miRNA-34a was negatively correlated with the mRNA
level of CDK4 and MYB (P<0.01), but not with that
of SIRT1 (P>0.05) (Table
II).
 | Table IICorrelation analysis between miR-34a
and CDK4, MYB, SIRT1 expression in AL
patients. |
Table II
Correlation analysis between miR-34a
and CDK4, MYB, SIRT1 expression in AL
patients.
| Parameters | CDK4a | MYBa |
SIRT1a |
|---|
| R2 | 0.933 | 0.887 | 0.305 |
| P | 0.000 | 0.000 | 0.108 |
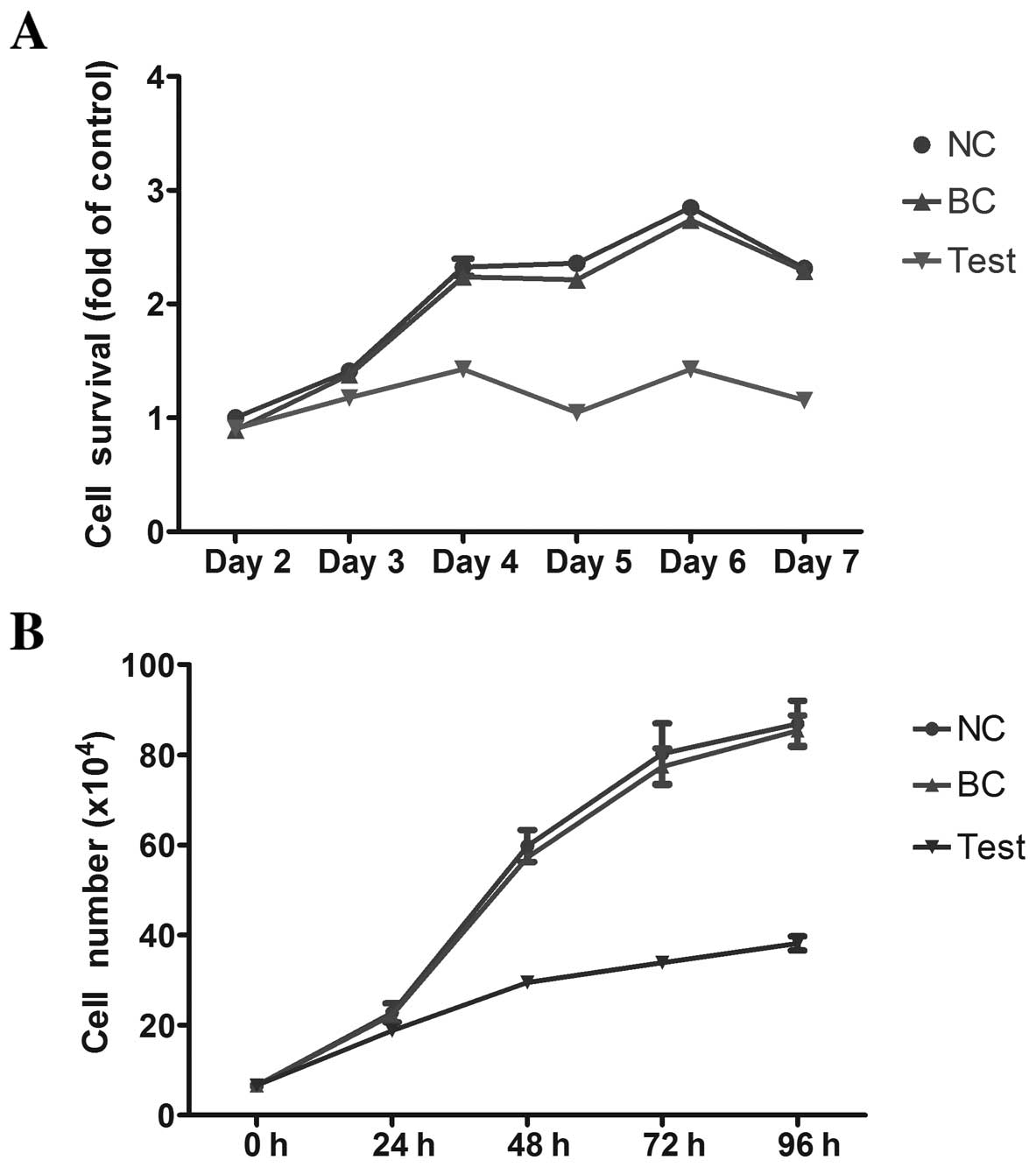
Viability of HL-60 cells following
miRNA-34a transfection
Fig. 2A shows the
viability of HL-60 cells as assessed by the MTT assay. The BC group
had the highest viability in all tested time-points, while the test
group had the lowest viability, suggesting that exogenous miRNA-34a
can inhibit the viability of HL-60 cells. A similar result was
obtained from trypan blue staining assays, which demonstrated that
miRNA-34a effectively inhibits the proliferation of HL-60 cells
(Fig. 2B).
Expression of CDK4, MYB and SIRT1 in
HL-60 cells following transfection
We measured the mRNA expression of CDK4,
MYB and SIRT1 48 h after transfection of HL-60 cells
with miRNA. The levels of these genes were significantly lower in
the miR-34a-transfected (test) group compared to the other two
groups (P<0.05), while there was no difference in their levels
when BC and NC groups were compared (Fig. 3). Compared to the NC group
(transfected with scrambled miRNA), the expression of CDK4,
MYB, and SIRT1 was decreased by 43.3, 53.2 and 33.5%,
respectively. Western blot analysis also demonstrated that
exogenous miRNA-34a can reduce the protein level of CDK4, MYB and
SIRT1 proteins (Fig. 4).
Discussion
Tumorigenesis is a complex biological process that
involves gene mutations and aberrant expression. Characterizing
differences in the expression of miRNA target genes in cancer
patients is of high value in cancer research. Rücker et al
(20) reported the aberrant
expression of miR-34a in acute myeloid leukemia patients with
complex karyotype (CK-AML). The authors showed that in the clinic,
low miR-34a expression and tumor protein p53 (TP53) alterations
were associated with resistance to chemotherapy and inferior
outcome of the treatment. Notably, in TP53-unaltered CK-AML
patients, high miR-34a expression predicted inferior overall
survival (OS), whereas in TP53-biallelic-altered CK-AML patients,
high miR-34a expression predicted better OS. Thus, targeted
treatment based on the expression status of cells for miR-34a is
necessary.
Numerous miRNAs have been directly associated with
the development of human tumors such as lung, breast, liver and
colon cancer, as well as leukemia (21). Some of these miRNAs act as tumor
suppressors. For example, He et al (7) reported in 2007 that miRNA-34
molecules (miR-34a, miR-34b and miR-34c) are direct targets of the
tumor suppressor protein p53, and suggested a new mechanism
explaining the p53-mediated inhibition of mammalian cell growth:
the evolutionally conserved miR-34s may respond to DNA damage and
tumor formation via a p53-dependent pathway. The authors presented
evidence showed that alternatively spliced miR-34 may exert the
same biological effects as p53, such as the induction of cell cycle
arrest and apoptosis, probably via the downregulation of mitogenic
and anti-apoptotic genes.
Pediatric leukemia is unique for its clinical and
biological properties: response to treatment, cure rate, and
immunophenotype. The development of leukemia relates not only to
chromosome abnormalities and gene mutations, but is also closely
associated with epigenetic characteristics and miRNA expression
profiles. Previous studies showing that miRNAs can modulate the
expression of target genes at the post-transcriptional level
(22,23) and that miRNA expression is also
subject to epigenetic regulation (24,25)
contributed to the clinical application of miRNAs.
A number of genes, including CDK4, CDK6,
MYB, SIRT1, and cell cycle-associated protein 2, has
been shown to be targeted by miR-34 (11,26,27).
The current study showed that the expression of CDK4 and
MYB in peripheral blood cells is significantly higher in
children with acute leukemia compared to healthy ones. Stratified
(sub-group) analyses showed that the expression of MYB in
children with acute leukemia <3 years old is significantly
higher compared to that observed in children >3 years, and it
was also significantly higher in children with peripheral WBC
counts >5×1010/l at diagnose compared to those with
WBC counts ≤5×1010/l, who may have lower miRNA-34a
expression. These findings strongly suggest that the expression
level of CDK4 and MYB may be associated with the
development of leukemia. Aberrant expression of CDK4 was
previously shown to associate with differentiation of liposarcoma
and enhanced proliferation of laryngeal cancer cells (28,29).
In addition, Weber et al (30)
showed that alternative splicing of the c-MYB mRNA resulting
in enhanced MYB expression occurred in hematologic
malignancies and caused excessive proliferation and differentiation
of the malignant cells.
Yamakuchi et al (5)
found that there is a 3′-UTR binding site in the SIRT1 gene
sequence for miR-34a, and confirmed that SIRT1 is a target
of miR-34a by luciferase reporter gene assays, and by showing that
miR-34a causes SIRT1 mRNA degradation. In our study, there
was no significant difference in SIRT1 expression between
leukemic and healthy controls, but the relatively small number of
studied patients may have affected this result. It is therefore
worth extending the present study in a larger population.
In conclusion, the development of leukemia involves
the activation of multiple genes and related signaling pathways.
Our study has demonstrated that miRNA-34a inhibits HL-60 cell
proliferation by downregulating the expression of CDK4,
MYB and SIRT1. Thus, miR-34 may target these genes
and play an important role in leukemic cell proliferation.
Intervention with endogenous miR-34a may be a suitable treatment
strategy.
Acknowledgements
This study was supported by the China National grant
863, permit no. 2012AA020804.
References
|
1
|
Yamakuchi M and Lowenstein CJ: MiR-34,
SIRT1 and p53: the feedback loop. Cell Cycle. 8:712–715. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rokhlin OW, Scheinker VS, Taghiyev AF,
Bumcrot D, Glover RA and Cohen MB: MicroRNA-34 mediates
AR-dependent p53-induced apoptosis in prostate cancer. Cancer Biol
Ther. 7:1288–1296. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Paris R, Henry RE, Stephens SJ, McBryde M
and Espinosa JM: Multiple p53-independent gene silencing mechanisms
define the cellular response to p53 activation. Cell Cycle.
7:2427–2433. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mert U, Ozgür E, Tiryakioglu D, Dalay N
and Gezer U: Induction of p53-inducible microRNA miR-34 by gamma
radiation and bleomycin are different. Front Genet. 3:2202012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yamakuchi M, Ferlito M and Lowenstein CJ:
miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci
USA. 105:13421–13426. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tazawa H, Tsuchiya N, Izumiya M and
Nakagama H: Tumor-suppressive miR-34a induces senescence-like
growth arrest through modulation of the E2F pathway in human colon
cancer cells. Proc Natl Acad Sci USA. 104:15472–15477. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He X, He L and Hannon GJ: The guardian’s
little helper: microRNAs in the p53 tumor suppressor network.
Cancer Res. 67:11099–11101. 2007.
|
|
8
|
Tarasov V, Jung P, Verdoodt B, et al:
Differential regulation of microRNAs by p53 revealed by massively
parallel sequencing: miR-34a is a p53 target that induces apoptosis
and G1-arrest. Cell Cycle. 6:1586–1593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kumamoto K, Spillare EA, Fujita K, et al:
Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2
and up-regulate mir-34a, mir-34b, and mir-34c expression, and
induce senescence. Cancer Res. 68:3193–3203. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He L, He X, Lim LP, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Navarro F, Gutman D, Meire E, et al:
miR-34a contributes to megakaryocytic differentiation of K562 cells
independently of p53. Blood. 114:2181–2192. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li L, Wu J, Sima X, et al: Interactions of
miR-34b/c and TP-53 polymorphisms on the risk of nasopharyngeal
carcinoma. Tumour Biol. 34:1919–1923. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ji Q, Hao X, Meng Y, et al: Restoration of
tumor suppressor miR-34 inhibits human p53-mutant gastric cancer
tumorspheres. BMC Cancer. 8:2662008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ji Q, Hao X, Zhang M, et al: MicroRNA
miR-34 inhibits human pancreatic cancer tumor-initiating cells.
PLoS One. 4:e68162009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pang Y, Young CY and Yuan H: MicroRNAs and
prostate cancer. Acta Biochim Biophys Sin (Shanghai). 42:363–369.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dalgard CL, Gonzalez M, deNiro JE and
O’Brien JM: Differential microRNA-34a expression and tumor
suppressor function in retinoblastoma cells. Invest Ophthalmol Vis
Sci. 50:4542–4551. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zenz T, Mohr J, Eldering E, et al: miR-34a
as part of the resistance network in chronic lymphocytic leukemia.
Blood. 113:3801–3808. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang J, Liu L, Xie L, Xiang G and Zhou Y:
Induction of differentiation-specific miRNAs in TPA-induced myeloid
leukemia cells through MEK/ERK activation. Int J Mol Med. 31:59–66.
2013.PubMed/NCBI
|
|
19
|
Vissers MC, Jester SA and Fantone JC:
Rapid purification of human peripheral blood monocytes by
centrifugation through Ficoll-Hypaque and Sepracell-MN. J Immunol
Methods. 110:203–207. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rücker FG, Russ AC, Cocciardi S, et al:
Altered miRNA and gene expression in acute myeloid leukemia with
complex karyotype identify networks of prognostic relevance.
Leukemia. 27:353–361. 2013.PubMed/NCBI
|
|
21
|
Visone R and Croce CM: MiRNAs and cancer.
Am J Pathol. 174:1131–1138. 2009. View Article : Google Scholar
|
|
22
|
Kelly L, Bryan K, Kim SY, et al:
Post-transcriptional dysregulation by miRNAs is implicated in the
pathogenesis of gastrointestinal stromal tumor [GIST]. PLoS One.
8:e641022013.PubMed/NCBI
|
|
23
|
Tuccoli A, Poliseno L and Rainaldi G:
miRNAs regulate miRNAs: coordinated transcriptional and
post-transcriptional regulation. Cell Cycle. 5:2473–2476. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Isken F, Steffen B, Merk S, et al:
Identification of acute myeloid leukaemia associated microRNA
expression patterns. Br J Haematol. 140:153–161. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Garzon R, Heaphy CE, Havelange V, et al:
MicroRNA 29b functions in acute myeloid leukemia. Blood.
114:5331–5341. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun F, Fu H, Liu Q, et al: Downregulation
of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett.
582:1564–1568. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tivnan A, Tracey L, Buckley PG, Alcock LC,
Davidoff AM and Stallings RL: MicroRNA-34a is a potent tumor
suppressor molecule in vivo in neuroblastoma. BMC Cancer.
11:332011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pilotti S, Della Torre G, Mezzelani A, et
al: The expression of MDM2/CDK4 gene product in the differential
diagnosis of well differentiated liposarcoma and large deep-seated
lipoma. Br J Cancer. 82:1271–1275. 2000.PubMed/NCBI
|
|
29
|
Dong Y, Sui L, Sugimoto K, Tai Y and
Tokuda M: Cyclin D1-CDK4 complex, a possible critical factor for
cell proliferation and prognosis in laryngeal squamous cell
carcinomas. Int J Cancer. 95:209–215. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Weber BL, Westin EH and Clarke MF:
Differentiation of mouse erythroleukemia cells enhanced by
alternatively spliced c-MYB mRNA. Science. 249:1291–1293. 1990.
View Article : Google Scholar : PubMed/NCBI
|