Introduction
At present, there are a number of different
types/subtypes of human lymphomas and leukemias. These were
originally classified according to histology and disease course,
but are now being re-grouped based on genetic aberrations,
including chromosomal translocations, oncogene mutations, and
expression profiles (1).
The Philadelphia (Ph) chromosome, which results from
translocation t(9:22), is one of the most characterized genetic
abnormalities associated with leukemia. The association was
initially described by Nowell and Hungerford (2) ~50 years ago, rendering it the first
chromosomal rearrangement to be associated with a specific
malignancy. It is now established that t(9;22) is observed in
>90% of patients with chronic myelogenous leukemia (CML), which
is a clonal bone marrow stem cell disorder characterized by the
slow progression of proliferation of mature and immature
granulocytes (neutrophils, eosinophils and basophils) that may
eventually lead to acceleration towards a blast crisis (3). The Ph chromosome is also detected in
~20% of adult acute lymphoblastic leukemia (ALL), 5% of pediatric
ALL, and rare cases of acute myelogenous leukemia (4).
The BCR-ABL fusion protein is the molecular
consequence of the Ph chromosome translocation and is an active
cytoplasmic tyrosine kinase (5).
This fusion protein varies in size from 190 to 230 kDa, depending
on the site of the breakpoint within the bcr gene. The
majority of patients with CML express a 210-kDa BCR-ABL protein,
while patients with Ph+ ALL commonly express a 190-kDa
BCR-ABL protein (6). A larger,
230-kDa BCR-ABL fusion protein is only rarely identified in a
subgroup of patients with chronic neutrophilic leukemia.
It is widely believed that the breakpoint in the BCR
sequence is correlated with the leukemic phenotype (7). In CML, the break on chromosome 22 is
predominantly within a BCR gene region termed M-bcr. The
majority of breaks occur immediately downstream of exon 2 or 3 of
the M-bcr region and result in b2a2 (e13a2) or
b3a2 (e14a2) fusion transcripts (8). In acute leukemia, however, the
breakage also occurs outside M-bcr (in approximately half the
cases), usually in intron 1 of the BCR gene (26,141),
resulting in an e1a2 fusion transcript (9). The difference in length of
BCR-ABL transcripts is not only a reflection of the site of
breakage but may also be a result of alternative splicing (10).
ABL is a non-receptor tyrosine kinase expressed in
the majority of tissues. It transduces signals from cell-surface
receptors for growth factors and adhesion receptors to regulate the
cytoskeletal structure (11). BCR
is also a signaling protein that contains multiple modular domains
and its fusion to ABL is known to increase the tyrosine-kinase
activity of ABL, and bring novel regulatory domains/motifs to ABL,
such as the growth factor receptor-binding protein 2 and Src
homolgy 2 domain-binding sites (12).
The development of leukemia is a complex process and
the detailed involvement of BCR-ABL proteins in this process has
been the subject of numerous studies. Apart from its diagnostic
value, those studies focused on whether there is a prior genetic
alteration to the t(9;22) translocation, its exact involvement in
pathogenesis as well as progression of related leukemias and its
involvement in cancer stem cells (13). However, there is controversy as
numerous leukemia-specific genetic rearrangements, including
bcr-abl are also observed in the peripheral blood and bone
marrow of healthy individuals (14). The biological explanation of this
observation remains largely unexplained (15).
The present study aimed to investigate the presence
of bcr-abl fusion transcripts in the peripheral blood of
healthy individuals. Nested reverse transcription polymerase chain
reaction (RT-PCR) amplification was used to assure maximal
sensitivity, and identify primers that distinguish the t(9:22)
common variants including, b2a2, b3a2 and
e1a2. The results of the present study are discussed with
regard to previous studies in a comprehensive overview focusing on
the significance of these results.
Materials and methods
Sample collection
A single donation of 5 ml EDTA peripheral blood was
drawn from 189 healthy subjects, of which 145 were adults and 44
were children. Informed consent was obtained from healthy
volunteers. The study protocol was approved by the Institutional
Review Board (IRB) of the University of Jordan. The adult group was
composed of 73 males (50.3%) and 72 females (49.7%) aged between 20
to 86 years (mean ± SD=50.0±17.7), while the group of children was
composed of 25 males (56.8%) and 19 females (43.2%) aged between 2
and 16 years (mean ± SD=8.4±3.9). The details of the age and gender
distribution of the study population are listed in Table I. The subjects were recruited
randomly from volunteers representing the different geographical
and ethnic backgrounds of the Jordanian population. All the
subjects were healthy and free of malignancy as demonstrated by a
short medical history taken by qualified clinical
professionals.
 | Table IIncidence of bcr-abl
transcripts (p210 and p190) in leukocytes of healthy individuals,
grouped according to age and gender. |
Table I
Incidence of bcr-abl
transcripts (p210 and p190) in leukocytes of healthy individuals,
grouped according to age and gender.
| Age group | No. of p210
positive samples (%) | No. of p190
positive samples (%) |
|---|
|
|---|
| M | F |
|---|
| Children | 2/25 (8.0) | 2/19 (10.5) | 0 (0.0) |
| Adults | 10/73 (13.7) | 5/72 (6.9) | 0 (0.0) |
| 20–39 years | 4/22 | 1/24 | |
| 40–59 years | 2/24 | 2/23 | |
| 60–86 years | 4/27 | 2/25 | |
| Total | 12/98 (12.2) | 7/91 (7.7) | 0 (0.0) |
RNA extraction
Total RNA was extracted from peripheral blood
samples collected in EDTA tubes using the cell lysis and RNA
isolation reagent TRIzol (Invitrogen Life Technologies, Carlsbad,
CA, USA). RNA was extracted within a few hours of blood collection.
The quality of the RNA was analyzed by electrophoresis gels and the
concentration was determined spectrophotometrically (Bio-Rad,
Hercules, CA, USA). RNA was stored at −70°C until use.
cDNA synthesis
Approximately 1 μg total RNA was converted into cDNA
using an RT system kit according to the manufacturer’s instructions
(Promega, Madison, WI, USA). This kit utilized random hexamers and
the MuLV RT enzyme.
Nested PCR amplification and transcript
detection
Following cDNA synthesis, nested PCR was conducted
to detect the presence of the three predominant bcr-abl
fusion transcripts (b2a2, b3a2 and e1a2).
Primers and conditions used in this study have been adopted from
previous studies with minor modifications (16,17).
In this study, uniplex PCR was used instead of multiplex PCR. The
RNA integrity, cDNA synthesis and PCR amplification were checked
using internal control primers for the glyceraldehyde 3-phosphate
dehydrogenase gene (18). A known
positive CML sample for the bcr-abl transcript was used as a
positive control. A non-template negative control was included in
each PCR run to check for contamination. Nested PCR products (5 μl)
were electrophoresed in 2% ethidium bromide stained agarose gels,
visualized and images were captured (UVP, Cambridge, UK) under UV
light.
Estimation of the minimal level of
detection of the nested PCR
The minimal level of detection of the nested PCR was
estimated using two methods. In the first method, cells from the
K562 BCR-ABL-positive cell line were serially diluted in MCF-7
cells, which are BCR-ABL negative. Dilutions were prepared in
10-fold steps, from undiluted to a dilution of 10−7. In
the second method, BCR-ABL-positive RNA obtained from K562 cells
was diluted in negative RNA from MCF-7 cells. The dilutions were
also prepared in 10-fold steps, from undiluted to
10−7.
The two methods showed a sensitivity level of
10−6 (1 in 1,000,000 dilution) using primers and
conditions used in this study.
Statistical analysis
Stepwise logistic regression (SLR) was utilized to
analyze gender and age as potential predictors of the presence of
the t(9;22) translocation. Backward and forward SLR were
investigated. The Hosmer-Lemeshow test was used to assess model
fitness. P<0.05 was considered to indicate a statistically
significant difference. Neither model analyzed contained a
constant. Two types of statistical analysis, −2 log-likelihood and
Wald statistics, were conducted with the first one being more
reliable. If discrepancies were noted between the two analyses with
regard to whether a predictor was useful to the model, the change
was adopted in the −2 log-likelihood model. Cox and Snell’s
R2 and Nagelkerke’s R2 were used to estimate
the coefficient of determination and the strength of the prediction
model.
Results
Amplification results for p190 and p210
fusion transcripts
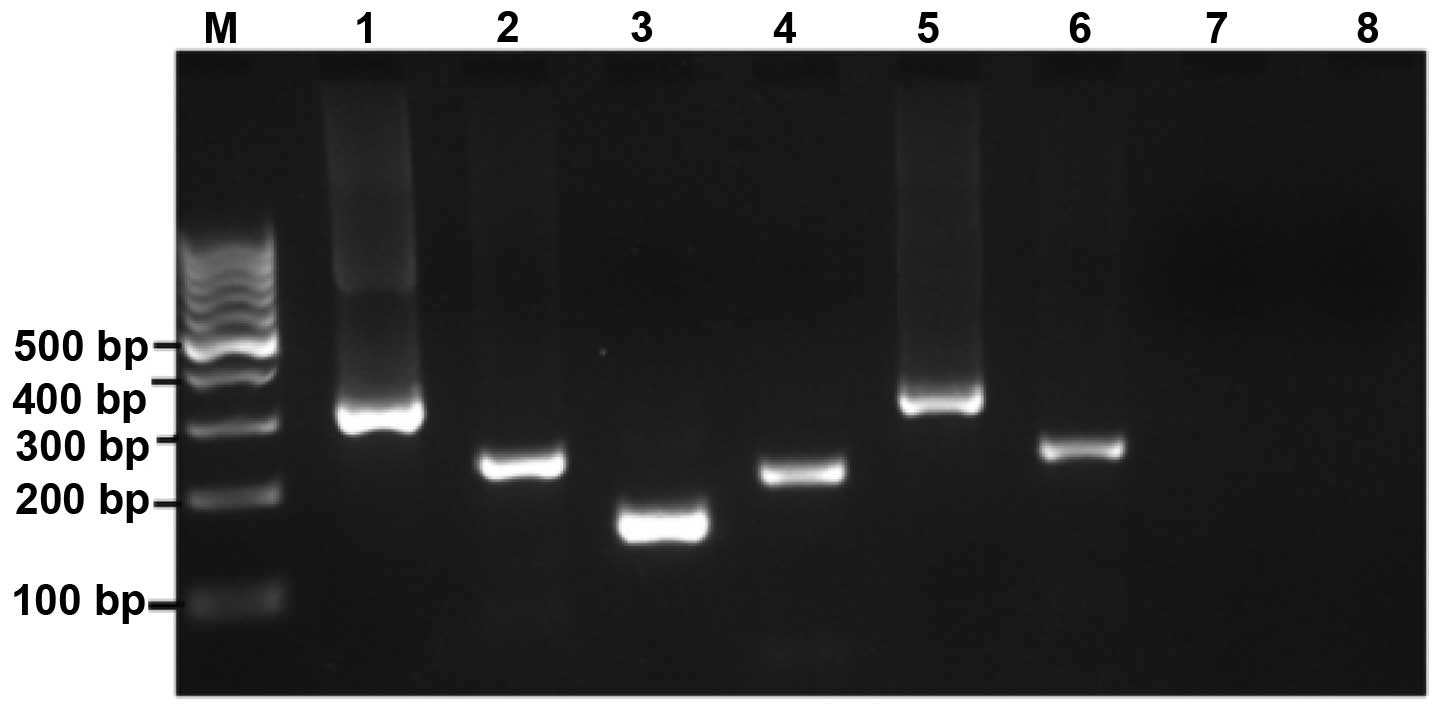
The frequency of bcr-abl (p210 and p190)
fusion transcripts in the peripheral blood samples obtained from a
total of 189 healthy individuals, grouped according to age and
gender, was analyzed using a highly sensitive nested RT-PCR assay
(Fig. 1). The reactions were
handled with care, and controls were included in every run to avoid
false positive results due to contamination.
The amplification results are summarized in Table I. No expression of the
bcr-abl p190 RNA transcript was identified in any of the
samples tested. However, ~10% of healthy individuals were positive
for the bcr-abl p210 transcripts. These p210 fusion
transcripts were detected in 10.3% of adults and 9.1% of children
(Table II). As regards, the
incidence of translocation was higher in males (12.2%) compared
with females (7.7%).
 | Table IIComparison between studies concerning
the occurrence of bcr-abl fusion transcripts in samples
obtained from healthy individuals. |
Table II
Comparison between studies concerning
the occurrence of bcr-abl fusion transcripts in samples
obtained from healthy individuals.
| Study (year) | Sample type
(no.) | Positive results
(%) | Method | Ref no. |
|---|
| Biernaux et
al (1995) | Adult PB (73) | 30.0 (p210) | RT-PCR (N) | (21) |
| Children PB
(22) | 4.5 (p210) | | |
| Cord blood
(22) | 0.0 (p210) | | |
| Bose et al
(1998) | Adult PB (16) | 25.0 (p210) | RT-PCR (N) | (22) |
| | 69.0 (p190) | | |
| Non-CML cell
linesa (7) | 43.0 (p210) | | |
| | 100.0 (p190) | | |
| NIH3T3 murine
fibroblasts | 0.0 | | |
| Song et al
(2011) | Adult PB (46,
p210) | 54.0 (p210) | RT-PCR (N) | (15) |
| (53, p190) | 77.0 (p190) | | |
| Child PB (28,
p210) | 32.0 (p210) | | |
| (27, p190) | 67.0 (p190) | | |
| Cord blood
(50) | 16.0 (p210) | | |
| | 42.0 (p190) | | |
| Current study | Adult PB (145) | 10.3 (p210) | RT-PCR (N) | |
| | 0.0 (p190) | | |
| Child PB (44) | 9.1 (p210) | | |
| | 0.0 (p190) | | |
The bcr-abl p210 fusion transcripts detected
in 19 individuals were approximately equally distributed between
the b3a2 (10) and b2a2
(9) subtypes. None of the tested
samples contained the two transcripts together (Table III). This distribution is
comparable to that observed in CML patients where the majority have
either the b3a2 (55.0%) or the b2a2 (40.0%)
transcripts, and in 5% of the cases b3a2 and b2a2
transcripts coexist as a result of alternative splicing (19,20).
Furthermore, although it may not be statistically significant, 3
out of 4 children had the b2a2 transcripts, and although the
b3a2 transcripts were detected equally in males (5) and females (5), the b2a2 transcripts were
detected in 7 males vs. 2 females (Table III).
 | Table IIIGender and age distribution of
b3a2 and b2a2 bcr-abl p210 fusion transcripts. |
Table III
Gender and age distribution of
b3a2 and b2a2 bcr-abl p210 fusion transcripts.
| Sample no. | Age group
(years) | Fusion
transcript | Gender |
|---|
| 1 | Child (8) | b3a2 | F |
| 2 | Child (9) | b2a2 | F |
| 3 | Child (10) | b2a2 | M |
| 4 | Child (12) | b2a2 | M |
| 5 | Adult (20) | b3a2 | M |
| 6 | Adult (22) | b3a2 | F |
| 7 | Adult (46) | b3a2 | F |
| 8 | Adult (49) | b3a2 | M |
| 9 | Adult (55) | b3a2 | F |
| 10 | Adult (61) | b3a2 | M |
| 11 | Adult (63) | b3a2 | M |
| 12 | Adult (71) | b3a2 | F |
| 13 | Adult (73) | b3a2 | M |
| 14 | Adult (23) | b2a2 | F |
| 15 | Adult (25) | b2a2 | M |
| 16 | Adult (39) | b2a2 | M |
| 17 | Adult (49) | b2a2 | M |
| 18 | Adult (65) | b2a2 | M |
| 19 | Adults (70) | b2a2 | M |
Statistical analysis
Logistic regression analysis was conducted utilizing
stepwise analysis in the forward and backward directions and
without a constant in the model. Gender and age were consistently
presented as a potential predictor of the t(9;22) translocation,
irrespective of whether Wald or −2 Log likelihood statistics were
used. As stated previously, the incidence of translocation was
higher in males (12.2%) compared with females (7.7%). Males were
2.4 times [adjusted OR=2.4 95% confidence interval (CI), 1.2–4.8;
P=0.008] more likely to have translocations. A significantly
increased risk was also observed in adults compared with children,
in which adults were 6 times (adjusted OR=6 95% CI, 3.3–10.7;
P<0.001) more likely to have translocations. The model for
gender and age was strong with Cox and Snell’s R2 and
Nagelkerke’s R2 of 0.45 and 0.59, respectively.
Discussion
There are few studies concerning the the occurrence
of bcr-abl fusion transcripts in healthy individuals (21,22,15).
The results of these studies as well as the present study are
summarized in Table II, which
also shows the number and type of samples studied and the detection
method used. Due to its superior sensitivity, all listed studies
used a nested RT-PCR assay to detect the bcr-abl transcripts
and primers specific for the p210 transcripts. In addition, certain
studies also included primers for the p190 transcripts.
In regard to the detection of p210 fusion
transcripts in the peripheral blood of healthy adults, Table II shows that the detection rate
ranges from 10.3 (current study) to ≤54% (15) with two studies demonstrating
results within this range, of 25 (22) and 30% (21). Notably, the current study recruited
significantly more healthy volunteers (145 samples) than the three
other studies (16, 46 and 73 samples). As for the occurrence of
p190 transcripts in the peripheral blood of healthy adults, the
variation was more significant and ranged from 0.0 (current study)
to ≤69 (22) and 77% (15). The present study also used more
samples for the p210 analysis (145 samples) compared with the two
other studies (16 and 53 samples, respectively).
Regarding the bcr-abl p210 transcripts in the
peripheral blood of healthy children, the results show a rate of
9.1% which is higher than that reported by Biernaux et al
(21) in 1995 (4.5%) but much
lower than that of Song et al (15) in 2011 (32%). With the p190
transcripts in children, while it was not possible to detect them
in any of our 44 samples, the only other study which observed this
rate detected transcripts in the blood of ~67% of the 27 children
tested (15).
Contradictory results have been demonstrated in two
studies that observed the bcr-abl transcripts in umbilical
cord blood (CB) samples. Although Biernaux et al (21) failed to detect these transcripts in
CB samples, Song et al (15) observed p210 and p190 transcripts at
high frequencies (16 and 42%, respectively) in CB.
The abovementioned results of the three previous
studies as well as the present study, are not consistent and show
significant variation in the rate of bcr-abl transcript
detection in healthy individuals.
The present study aimed to analyze the presence of
the most common variants of the bcr-abl fusion transcripts
in the peripheral blood of 189 healthy individuals. Highly
sensitive nested PCR assays were used to separately detect each of
the p190 and p210 fusion transcripts. The results demonstrated the
presence of p210 fusion transcripts in ~10% of the tested samples,
with gender and age associations, where older males appeared more
likely to have the transcripts in the peripheral blood. This
correlation with advancing age was also demonstrated in two
previous studies (15,21). For example in one of these studies
(21), the bcr-abl
transcripts were not detected in CB and were only detected in 1 of
22 children, but were detected in 18 of 52 adults. This increase
may be due to the fact that, in adults an increased number of cell
divisions had occurred and therefore, there was an increased chance
of the accumulation of DNA rearrangements (21).
A number of other studies have demonstrated the
occurrence of bcr-abl leukemic transcripts in the blood of
seemingly healthy individuals (15,21,22).
However, as demonstrated in Table
II, there is considerable variation in the results. This
variation was observed in the detection frequencies of p210 and
p190 bcr-abl fusion transcription in peripheral and CB
samples, as well as the frequency between children and adults. It
is not easy to explain this discrepancy but it is suggested that
the following factors may contribute. Only a small number of
studies are available, and a number of these studies used a small
number of samples. By comparison, the current study recruited the
largest number of healthy volunteers. Ethnic, geographical or
environmental elements may also contribute to the variation. In
addition, another source of variation may be due to the methodology
implemented in each study. As all studies shown in Table II used nested RT-PCR, this factor
may hold less weight. However, there may still be variation due to
laboratory to laboratory variation, assay sensitivity and
experimental errors, of which false positives may be a predominant
issue, particularly with a highly sensitive method, such as nested
RT-PCR. One comparison that may highlight the importance of
false-positive results concerns the occurrence of p190 transcripts
in the peripheral blood of healthy adults. Although the present
study failed to detect any such transcripts in the samples tested,
other authors have demonstrated high frequencies of ~69 (22) and 77% (15). Notably, the present study used more
samples (145 samples) compared with the two other studies (16 and
53 samples, respectively). Concordant with the results of the
present study, no such high frequencies of bcr-abl p190
transcripts have been demonstrated even in ALL patients, where
these transcripts are supposed to be a diagnostic hallmark for 20%
of patients (4). In addition, our
laboratory, which offers a diagnostic service to the Jordan
University Hospital and numerous other nearby hospitals, has not
detected p190 transcripts in such high frequencies in the hundreds
of suspected ALL cases that have been screened, using the same
highly sensitive nested RT-PCR assay (unpublished data). Therefore,
there is a requirement for more studies recruiting larger numbers
of healthy volunteers of all ages, from different geographical
regions and ethnicities and preferably using different detection
techniques.
Regardless of the variation between studies on the
frequency of bcr-abl transcripts in normal individuals, the
presence of the transcripts has been confirmed. This may suggest
the requirement for diagnostic laboratories and clinicians
worldwide to consider the presence of transcripts in healthy
individuals when interpreting bcr-abl or t(9;22) positive
results.
Although there have not been enough studies
concerning the clinical value of the occurrence of bcr-abl
transcripts in healthy individuals, numerous reports have focused
on the involvement of t(9;22) and the resultant BCR-ABL fusion
proteins on the oncogenesis of CML. One theory states that CML is,
as the name indicates, a chronic disease of the hematopoietic stem
cell where such cells, which harbor the p210 bcr-abl
transcripts, remain quiescent for years prior to transforming into
a malignant colony subsequent to acquiring extra mutations
(15). Thus, in this theory, the
presence of p210 bcr-abl transcripts in seemingly healthy
individuals may be considered a risk factor for CML, which is
consistent with the observation that the occurrence of p210
bcr-abl transcripts in healthy individuals and the incidence
of CML is more commonly observed in older individuals. Furthermore,
in line with this hypothesis, there are numerous reports that have
demonstrated the development of CML-like conditions in mouse models
following inoculation with bcr-abl transduced hematopoietic
progenitor cells (23,24,25).
These CML-like conditions developed with a certain latency which
may also be due to the accumulation of additional mutations,
required for clonal expansion (26).
However, the presence of bcr-abl transcripts
in healthy individuals as a risk factor for CML is not a
straightforward conclusion. The incidence of these transcripts is
higher than that for CML, suggesting that a number of those healthy
individuals are not likely to develop CML. One explanation for this
is that the bcr-abl transcripts detected in healthy
individuals may be generated in non-self-renewing terminally
differentiated cells, and not in hematopoietic stem cells or early
progenitors, which are known to be more capable of developing
malignant clones (27). A recent
study has suggested that patients who eventually develop CML may be
suffering from a specific cytotoxic T-cell immune response deficit
with respect to the BCR-ABL antigen (28). In addition, certain HLA types have
been shown to be associated with reduced risk of CML (29). Such studies hypothesized that the
immuno-competency of an individual is a determining protective
factor against developing CML.
In conclusion, this study was consistent with
previous studies concerning the occurrence of bcr-abl fusion
transcripts in the peripheral blood of healthy individuals, and
that this was more likely in adults compared with children.
However, there are significant discrepancies in the reported
frequencies of the various types of these transcripts between
different studies. It is therefore recommended that further studies
be conducted to reach a consensus on this issue, where larger
numbers of healthy individuals are recruited, preferably from
different geographical and ethnic backgrounds, and where different
detection techniques are used to assess methodological variation.
The collected samples may cover peripheral blood, bone marrow and
CB. Moreover, a close and long-term follow-up of
bcr-abl-positive cases is essential to assess the
implication of these results on the development of related
leukemias.
Acknowledgements
This study was supported by a student grant from the
Deanship of Scientific Research, University of Jordan.
References
|
1
|
Avery A: Molecular diagnostics of
hematologic malignancies. Top Companion Anim Med. 24:144–150. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nowell PC and Hungerford DA: Chromosome
studies in human leukemia. IV Myeloproliferative syndrome and other
atypical myeloid disorders. J Natl Cancer Inst. 29:911–931.
1962.PubMed/NCBI
|
|
3
|
Chen Y, Peng C, Li D and Li S: Molecular
and cellular bases of chronic myeloid leukemia. Protein Cell.
1:124–132. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Laurent E, Talpaz M, Kantarjian H and
Kurzrock R: The BCR gene and philadelphia chromosome-positive
leukemogenesis. Cancer Res. 61:2343–2355. 2001.PubMed/NCBI
|
|
5
|
Kurzrock R, Kantarjian HM, Druker BJ and
Talpaz M: Philadelphia chromosome-positive leukemias: from basic
mechanisms to molecular therapeutics. Ann Intern Med. 138:819–830.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Melo JV: The diversity of BCR-ABL fusion
proteins and their relationship to leukemia phenotype. Blood.
88:2375–2384. 1996.PubMed/NCBI
|
|
7
|
Pane F, Frigeri F, Sindona M, Luciano L,
Ferrara F, Cimino R, Meloni G, Saglio G, Salvatore F and Rotoli B:
Neutrophilic-chronic myeloid leukemia: a distinct disease with a
specific molecular marker (BCR/ABL with C3/A2 junction). Blood.
88:2410–2414. 1996.PubMed/NCBI
|
|
8
|
Groffen J, Stephenson JR, Heisterkamp N,
de Klein A, Bartram CR and Grosveld G: Philadelphia chromosomal
breakpoints are clustered within a limited region, bcr, on
chromosome 22. Cell. 36:93–99. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Heisterkamp N, Knoppel E and Groffen J:
The first BCR gene intron contains breakpoints in Philadelphia
chromosome positive leukemia. Nucleic Acids Res. 16:10069–10081.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shtivelman E, Lifshitz B, Gale RP, Roe BA
and Canaani E: Alternative splicing of RNAs transcribed from the
human abl gene and from the bcr-abl fused gene. Cell. 47:277–284.
1986. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Woodring PJ, Hunter T and Wang JY:
Regulation of F-actin-dependent processes by the Abl family of
tyrosine kinases. J Cell Sci. 116:2613–2626. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li S, Couvillon AD, Brasher BB and Van
Etten RA: Tyrosine phosphorylation of Grb2 by Bcr/Abl and epidermal
growth factor receptor: a novel regulatory mechanism for tyrosine
kinase signaling. EMBO J. 20:6793–6804. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rumpold H and Webersinke G: Molecular
pathogenesis of philadelphia-positive chronic myeloid leukemia - is
it all BCR-ABL? Curr Cancer Drug Targets. 11:3–19. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Janz S, Potter M and Rabkin CS: Lymphoma-
and leukemia-associated chromosomal translocations in healthy
individuals. Genes Chromosomes Cancer. 36:211–223. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song J, Mercer D, Hu X, Liu H and Li MM:
Common leukemia- and lymphoma-associated genetic aberrations in
healthy individuals. J Mol Diagn. 13:213–219. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cross NC, Melo JV, Feng L and Goldman JM:
An optimized multiplex polymerase chain reaction (PCR) for
detection of BCR-ABL fusion mRNAs in haematological disorders.
Leukemia. 8:186–189. 1994.PubMed/NCBI
|
|
17
|
Nogva HK, Evensen SA and Madshus IH:
One-tube multiplex RT-PCR of BCR-ABL transcripts in analysis of
patients with chronic myeloid leukaemia and acute lymphoblastic
leukaemia. Scand J Clin Lab Invest. 58:647–654. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kaneda R, Toyota M, Yamashita Y, Koinuma
K, Choi YL, Ota J, Kisanuki H, Ishikawa M, Takada S, Shimada K and
Mano H: High-throughput screening of genome fragments bound to
differentially acetylated histones. Genes Cells. 9:1167–1174. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Okamoto K, Karasawa M, Sakai H, Ogura H,
Morita K and Naruse T: A novel acute lymphoid leukemia type BCR/ABL
transcript in chronic myelogenous leukemia. Br J Haematol.
96:611–613. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lichty B, Keating A, Callum J, Yee K,
Croxford R, Corpus G, Nwachukwu B, Kim P, Guo J and Kamel-Reid S:
Expression of p210 and p190 BCR-ABL due to alternative splicing in
chronic myelogenous leukemia. Br J Haematol. 103:711–715. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Biernaux C, Loos M, Sels A, Huez G and
Stryckmans P: Detection of major bcr-abl gene expression at a very
low level in blood cells of some healthy individuals. Blood.
86:3118–3122. 1995.PubMed/NCBI
|
|
22
|
Bose S, Deininger M, Gora-Tybor J, Goldman
JM and Melo JV: The presence of typical and atypical BCR-ABL fusion
genes in leukocytes of normal individuals: biologic significance
and implications for the assessment of minimal residual disease.
Blood. 92:3362–3367. 1998.
|
|
23
|
Daley GQ, Van Etten RA and Baltimore D:
Induction of chronic myelogenous leukemia in mice by the
P210bcr/abl gene of the Philadelphia chromosome. Science.
247:824–830. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kelliher MA, McLaughlin J, Witte ON and
Rosenberg N: Induction of a chronic myelogenous leukemia-like
syndrome in mice with v-abl and BCR/ABL. Proc Natl Acad Sci USA.
87:6649–6653. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Elefanty AG and Cory S: Hematologic
disease induced in BALB/c mice by a bcr-abl retrovirus is
influenced by the infection conditions. Mol Cell Biol.
12:1755–1763. 1992.PubMed/NCBI
|
|
26
|
Bäsecke J, Griesinger F, Trümper L and
Brittinger G: Leukemia- and lymphoma-associated genetic aberrations
in healthy individuals. Ann Hematol. 81:64–75. 2002.
|
|
27
|
Ren R: Mechanisms of BCR-ABL in the
pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer.
5:172–183. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rusakiewicz S, Madrigal A, Travers P and
Dodi AI: BCR/ABL- specific CD8+ T cells can be detected
from CML patients, but are only expanded from healthy donors.
Cancer Immunol Immunother. 58:1449–1457. 2009.PubMed/NCBI
|
|
29
|
Posthuma EF, Falkenburg JH, Apperley JF,
Gratwohl A, Roosnek E, Hertenstein B, Schipper RF, Schreuder GM,
D’Amaro J, Oudshoorn M, et al: HLA-B8 and HLA-A3 coexpressed with
HLA-B8 are associated with a reduced risk of the development of
chronic myeloid leukemia. The Chronic Leukemia Working Party of the
EBMT. Blood. 93:3863–3865. 1999.PubMed/NCBI
|