Introduction
Hepatitis B virus (HBV) infection is a predominant
global health concern with >350 million individuals chronically
infected worldwide. Approximately one million mortalities occur
annually due to the long-term complications associated with HBV
infection (1). Following infection
of hepatoctyes, HBV relaxed-circle DNA (rcDNA) is transferred to
the nucleus where it forms covalently closed circular DNA (cccDNA),
which represents the intracellular template of the virus. Within
infected cells, the pregenomic RNA (pgRNA) is then transcribed from
the cccDNA and transported to the cytoplasm, where the mature
capsids of the rcDNA are reverse transcribed and either secreted
from the cells or returned to the nucleus to form the cccDNA pool.
While HBV is a non-cytopathic virus, the immune response against
virus-infected liver cells and the production of inflammatory
cytokines have been proposed to be responsible for liver disease
and viral clearance (2).
HBV infections are predominantly divided into either
self-limited acute hepatitis B (AHB), inactive hepatitis B surface
antigen (HBsAg) carriers, chronic hepatitis B (CHB) and occult HBV
infection (OBI) (3). AHB is
characterized by transient liver inflammation and virusemia. CHB
consists of four phases: Immune-tolerant (IT), immune-clearance
(IC), non/low-replicative (LR) and hepatitis B e antigen
(HBeAg)-negative hepatitis (ENH) (4). Patients are classified as having the
OBI form of the disease if they have undetectable HBsAg, but
detectable HBV DNA in the sera or liver tissues (5). Anti-viral drug treatment for patients
with CHB predominantly involves interferon-α (INF-α) and nucleoside
analogues (NUC), including lamivudine, adefovir and entecavir.
However, such drugs are not capable of directly eradicating cccDNA,
which has been reported to have a significant role in HBV infection
relapse (6), the pathogenesis of
hepatocellular carcinoma and cirrhosis (5), and the infection of liver transplant
cases (7). Monitoring intrahepatic
(IH) cccDNA levels in patients with CHB may have potential as a
predictor of therapeutic response or disease progression (8). However, at present, IH HBV cccDNA and
total DNA (tDNA) persistence, and the correlation between IH cccDNA
and other biochemical, virological and serological parameters in
patients with AHB and those with CHB who are receiving anti-viral
treatment have yet to be elucidated.
This study aimed to investigate the persistence of
IH cccDNA and tDNA and their correlation in patients with AHB and
those with CHB receiving anti-viral treatment. Furthermore, the
present study aimed to obtain and compare the thresholds of IH
cccDNA and tDNA levels in order to assess successful therapeutic
outcome for patients with CHB, which may be beneficial in
determining an anti-viral treatment endpoint.
Patients and methods
Patients and samples
Liver biopsy specimens were collected from a total
of 60 patients, including 11 patients with AHB who achieved
spontaneous virological seroclearance, 46 patients with CHB in
phase III antiviral clinical trials receiving NUC (adefovir, 10
mg/day or lamivudine, 100 mg/day), either as a monotherapy or with
pegylated-INF-α (100 μg/week) and three patients with autoimmune
hepatitis (AIH) as negative controls. Patients who were coinfected
with hepatitis D, hepatitis C or human immunodeficiency virus, or
those with Wilson’s disease, primary biliary cirrhosis or with a
substantial daily alcohol intake (20 g/day for females; 30 g/day
for males) were excluded from the study. Among the patients with
CHB, 21 had failed primary treatment, 11 had achieved virological
response (VR) and 14 had achieved VR and HBsAg seroclearance and
had been relapse-free for >6 months. Each patient signed an
informed consent document approved by the Ethics Committee of
Shenzhen Third People’s Hospital (Shenzhen, China). Biopsy
specimens were frozen in liquid nitrogen and stored at −80°C until
experimental analysis.
IH HBV cccDNA quantification
DNA was extracted from biopsy specimens using the
QIAamp® DNA Mini kit (Qiagen, Hilden, Germany). To
enhance the specificity of cccDNA detection, the Plasmid-Safe™
ATP-Dependent DNase (PSAD; Epicentre Biotechnologies, Madison, WI,
USA) was used to degrade rcDNA and single-stranded DNA (ssDNA)
prior to quantitative polymerase chain reaction (qPCR) analysis,
according to the manufacturer’s instructions (9). IH cccDNA levels were measured using
qPCR analysis as described previously (9). Two forward primers, [CCC1,
5′-GCGGWCTCC CCGTCTGTGCC-3′; diaphanous-related formin 1 (DRF1),
5′-GTCTGTGCCTTCTCATCTGC-3′], and one reverse primer, (CCC2, 5′-GTC
ATGCCCCAAAGCCACC-3′), were used for cccDNA amplification using the
LightCycler® system (Roche Diagnostics, Mannheim,
Germany) and SYBR Premix Ex Taq (Takara Bio Inc., Otsu, Japan).
Serial dilutions of a plasmid containing an HBV monomer (pHBVEcoRI)
served as a quantification standard. β-globin DNA (house keeping
gene) was detected using the LightCycler® Control kit
DNA (Roche Diagnostics) in order to count the cell number in the
biopsies and calculate copies/cell.
Serum and IH HBV tDNA quantification
Blood was collected on the day of the liver biopsy
and serum was immediately stored at -20°C. DNA was extracted from
200 μl serum using the QIAamp® DNA Blood Mini kit
(Qiagen). Prior to degradation using PSAD, serum and IH tDNA levels
were measured using the Cobas® TaqMan® test
as described previously (Roche Diagnostics) (10).
Serum HBsAg and HBeAg quantification
Serum HBsAg and HBeAg levels were quantified by
enzyme immunoassay using the Abbott ARCHITECT platform (Abbott
Laboratories, Chicago, IL, USA) according to the manufacturer’s
instructions. HBsAg >0.05 IU/ml and HBeAg >1 IU/ml were
considered to be positive results.
Serum alanine aminotransferase (ALT)
quantification
Serum ALT levels were measured using blood chemistry
analyses (DiaSys Diagnostic Systems, Holzheim, Germany), according
to the manufacturer’s instructions.
Statistical analysis
Statistical analyses were performed using the SPSS
13.0 statistical software (SPSS Inc., Chicago, IL, USA). Continuous
variables are expressed as the mean with the range, and were
analyzed using the Mann-Whitney rank sum test or one-way analysis
of variance. Serum HBsAg and HBeAg (IU/ml) levels were
logarithmically transformed prior to analysis. Categorical
variables were compared using Pearson’s χ2 test.
Correlations were analyzed using Pearson’s correlation coefficient.
Area under the receiver operating characteristic (ROC) curve
analysis was performed to predict the likelihood of achieving VR in
combination with HBsAg seroclearance in the patients with CHB.
Two-sided P<0.05 was considered to indicate a statistically
significant difference.
Results
Baseline characteristics
The clinical, biochemical and serological
characteristics of the patient groups used in this study are shown
in Table I. Among the 11 patients
with AHB, all patients were HBeAg-negative (HBeAg−), and
one patient was also anti-hepatitis B core antibody-positive
(anti-HBcAb+). Among the patients with CHB who had
failed primary treatment, three patients were HBeAg− and
anti-HBcAb−, and the remaining 18 patients were
HBeAg+ and anti-HBcAb+. In the group of
patients with CHB who had achieved VR, six patients were
HBeAg−, of whom two patients were
anti-HBcAb+, and five patients were HBeAg+
and anti-HBcAb+. In the group of patients with CHB who
had achieved VR and HBsAg seroclearance, 11 patients were
HBeAg− and anti-HBcAb− and three patients
were HBeAg+ and anti-HBcAb+
(P<0.0001).
 | Table IClinical, serological and biochemical
parameters of patients. |
Table I
Clinical, serological and biochemical
parameters of patients.
| Patient groups | |
|---|
|
| |
|---|
| | CHB receiving
anti-viral treatment | |
|---|
| |
| |
|---|
| Parameters | AHB | 1 | 2 | 3 | P-value |
|---|
| Age (years) | 28.18 (21–41) | 32.86 (22–42) | 37.18 (27–73) | 36.93 (21–55) | 0.062 |
|
HBeAg+/HBeAg− | 0/11 | 18/3 | 5/6 | 3/11 | <0.0001 |
| Gender (M/F) | 10/1 | 19/2 | 9/2 | 11/3 | 0.713 |
| ALT (IU/ml) | 34.27 (12–83) | 76.86 (29–262) | 36.27 (14–102) | 25.57 (15–36) | 0.001 |
| HBeAg
(log10 IU/ml) | −0.42
(−1–0.009) | 1.42 (−1–3.10) | 0.59
(−0.77–2.49) | −0.44
(−2–0.74) | <0.0001 |
| HBsAg
(log10 IU/ml) | 0.61
(−0.19–3.45) | 3.01
(1.49–4.76) | 3.41
(1.54–5.06) | −1.43
(−2.52–0.10) | <0.0001 |
| HBV DNA
(copies/ml) | 0 | 1.33×107
(2.32×104–9.07×107) | 0 | 0 | 0.120 |
IH cccDNA and tDNA quantification
The lower limits of detection for IH cccDNA and tDNA
quantification were 0.00024 copies/cell and 0.0001 copies/cell,
respectively. IH cccDNA levels were detectable in four patients
with AHB, 21 patients with CHB who had failed primary treatment, 10
patients who had achieved VR and 11 patients who had achieved VR
and HBsAg seroclearance. IH tDNA levels were detectable in 10
patients with AHB, 21 patients with CHB who had failed primary
treatment, 11 patients who had achieved VR, 13 patients who had
achieved VR and HBsAg seroclearance and one patient with AIH. The
median IH cccDNA level in patients with AHB (0.002 copies/cell;
range, <0.00024–0.013) was identified to be significantly lower
than that in patients with CHB who had failed primary treatment
(4.18 copies/cell; range, 0.078–42.4; P<0.0001) as well as that
in patients with CHB who had achieved VR (0.039 copies/cell; range,
<0.00024–0.17; P=0.005) but not that in patients with CHB who
had achieved VR and HBsAg seroclearance (0.012 copies/cell; range,
<0.00024–0.093; P=0.076). The median IH tDNA level in patients
with AHB (0.04 copies/cell; range, <0.0001–0.15) was observed to
be significantly lower than that in patients with CHB who had
failed primary treatment (371 copies/cell; range, 3.94–2,940;
P<0.0001), that in patients with CHB who had achieved VR (1.62
copies/cell; range, 0.0039 to 5.50; P=0.001) but not that in
patients with CHB who had achieved VR and HBsAg seroclearance
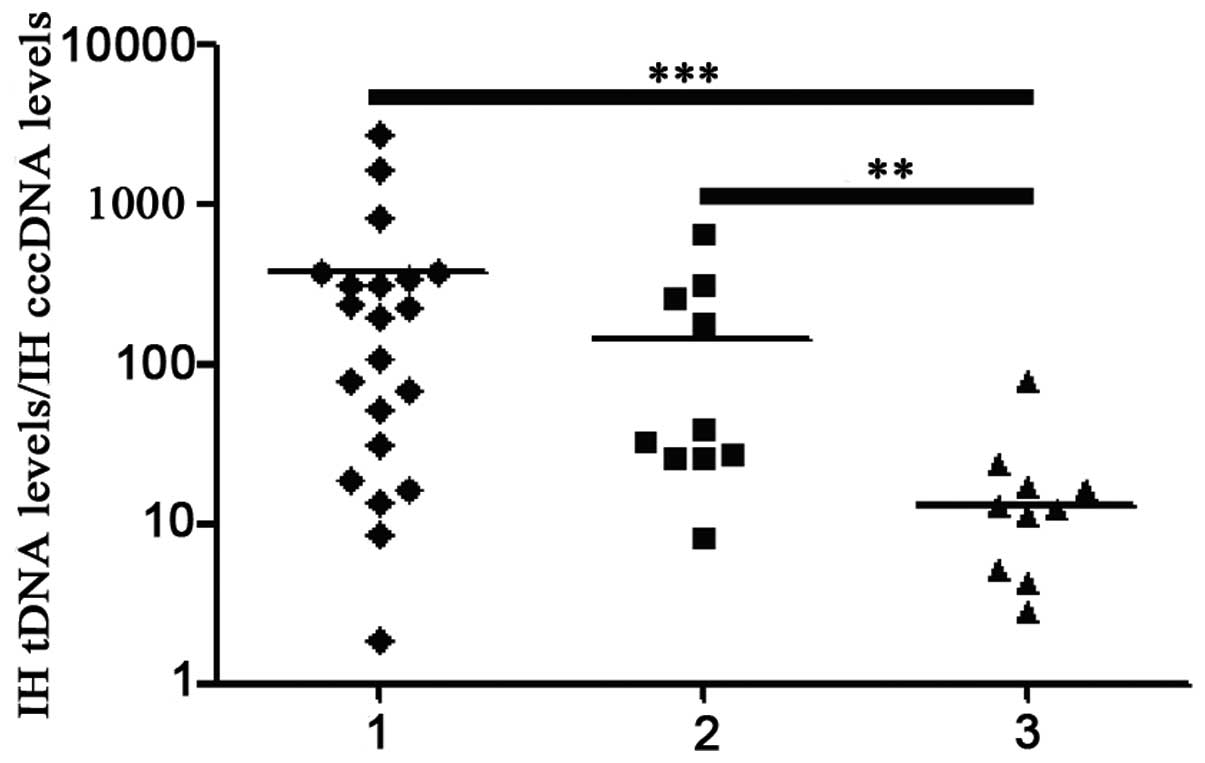
(0.096 copies/cell; range, <0.0001–0.38; P=0.443) (Fig. 1).
 | Figure 1IH cccDNA and tDNA levels in patients
with AHB and patients with CHB receiving anti-viral treatment:
Group 1, patients with AHB; group 2, patients with CHB who failed
primary treatment; group 3, patients with CHB who achieved VR;
group 4, patients with CHB who achieved VR and HBsAg seroclearance.
**P<0.01; ***P<0.001. AHB, acute
hepatitis B; CHB; chronic hepatitis B; IH, intrahepatic; cccDNA,
covalently closed circular DNA; tDNA, total DNA; VR, virological
response; HBsAg, hepatitis B surface antigen; HBeAg, hepatitis B e
antigen. |
Among patients with CHB, the median IH cccDNA level
in those who had achieved VR and HBsAg seroclearance was observed
to be significantly lower than that in those who had failed primary
treatment (P<0.0001), but not that in those who had achieved VR
(P=0.169). Furthermore, the median IH tDNA level in patients with
CHB who had achieved VR and HBsAg seroclearance was detected to be
significantly lower than that in patients with CHB who had failed
primary treatment (P<0.0001) and those who had achieved VR
(P=0.001). As shown in Fig. 1, the
median IH cccDNA and tDNA levels in patients with CHB who had
achieved VR were significantly lower than those who had failed
primary treatment (P<0.0001 and P<0.0001, respectively).
Furthermore, Fig. 2 shows that the
IH tDNA:cccDNA ratio in patients with CHB who had achieved VR and
HBsAg seroclearance (20.33; range, 0–79.25) was significantly lower
that in those who had failed primary treatment (380.29; range,
1.89–2,753.93; P<0.0001) and those who had achieved VR (155.52;
range, 0–643.11; P=0.004). The median IH cccDNA level in the
patients who were HBeAg− (0.14 copies/cell; range,
<0.00024–2.08) was significantly lower than in those who were
HBeAg+ (3.24 copies/cell; range, 0.00042–42.4;
P<0.0001). As shown in Fig. 1,
the median IH tDNA level (1.38 copies/cell; range, <0.0001–17.2)
in the former was significantly lower than that in the latter (299
copies/cell; range, 0.070–2940; P<0.0001).
Correlation analysis
IH cccDNA and tDNA levels were positively correlated
with serum ALT (P=0.024 and P=0.0048, respectively) and serum HBeAg
levels (P=0.0001 and P=0.015, respectively); however, no
correlation was observed with HBV DNA levels (P=0.121 and P=0.437,
respectively). IH cccDNA levels were not observed to be correlated
with serum HBsAg levels in either patients who were
HBeAg+ (P=0.84) or in patients who were
HBeAg− (P=0.146). As shown in Table II, IH tDNA levels
were positively correlated with serum HBsAg levels in patients who
were HBeAg− (P=0.007), but not in those who were
HBeAg+ (P=0.247). IH cccDNA levels were observed to be
positively correlated with IH tDNA levels (P<0.0001) but not
with the IH tDNA:cccDNA ratio (P=0.701). However, as demonstrated
in Fig. 3, IH tDNA levels were
positively correlated with the IH tDNA:cccDNA ratio (P=0.0037).
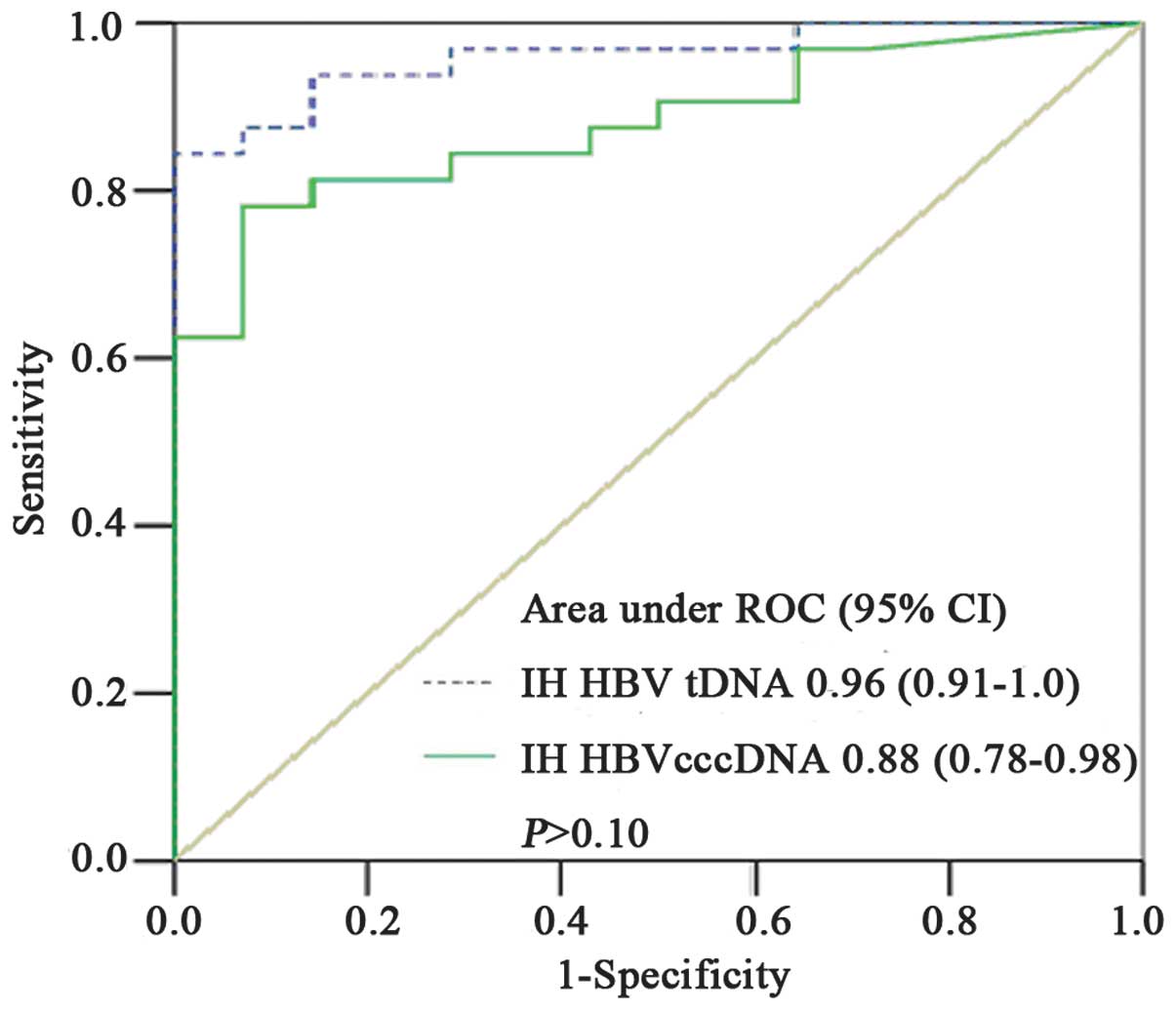
Prediction for achieving VR in
combination with HBsAg seroclearance
Areas under the ROC curve were used to predict the
likelihood of achieving VR and HBsAg seroclearance in patients with
CHB who received anti-viral treatment. The areas for IH cccDNA and
tDNA levels were observed to be 0.88 [95% confidence interval (CI):
0.78–0.98] and 0.96 (95% CI: 0.91–1.00), respectively. As shown in
Fig. 4, the cutoff value for IH
cccDNA levels was 0.015 copies/cell with a sensitivity of 81.3%,
specificity of 85.7%, positive predictive value (PPV) of 85.71% and
negative predictive value (NPV) of 88.37%. The cutoff value for IH
tDNA levels was 0.23 copies/cell with a sensitivity of 93.8%,
specificity of 85.7%, PPV of 85.71% and NPV of 95.35%
(P>0.10).
Discussion
Approximately two billion individuals are exposed to
HBV worldwide, of whom >350 million are chronically infected. It
is well established that cellular immune responses, particularly T
cells, contribute to HBV clearance. CD4+ T cells are
classified into type 1 and type 2 T helper cells (Th1 and Th2,
respectively), which differ in their patterns of secreted cytokines
(11). Host immune reactions that
are biased towards Th1 have been associated with AHB, with Th1-type
cytokines, including IFN-γ and tumor necrosis factor-β (TNF-β),
potentially controlling HBV replication by activating
CD8+ T-cell, natural killer (NK) cell and macrophage
responses, consequently stimulating a cascade of inflammatory
cytokines and degrading HBV RNA directly (12). Inefficiencies in the innate and
adaptive immune responses may result in CHB, in which high levels
of transforming growth factor-β1, predominantly secreted by
regulatory T cells, may impair NK cell function by reducing natural
killer group 2 member D/DNAX activation protein 10 and natural
killer cell receptor 2B4/SLAM-associated protein expression, and
suppress innate antiviral immunity by blocking the cell cycle in G1
(13) as well as inhibit the
secretion of IFN-γ and TNF-α from HBV-specific T cells (12).
In the present study, the median IH cccDNA level in
patients with AHB was observed to be significantly lower than that
of the patients with CHB who were receiving anti-viral treatment.
This finding suggests that in patients with AHB, the immune system
may be capable of reducing IH cccDNA to markedly low levels. In
patients with CHB, the median IH cccDNA level in those who had
achieved VR in combination with HBsAg seroclearance, was observed
to be significantly lower than that in those who had failed primary
treatment and those that had achieved VR. The outcome of anti-viral
treatment in patients with CHB is highly variable, ranging from
primary treatment failure to VR, serological response, biochemical
response and virological breakthrough (14). While it has been hypothesized that
clinical resolution of HBV infection occurs when HBV replication is
completely controlled by the host’s immune system (15), it has been suggested that achieving
VR in combination with HBsAg seroclearance may be the most
effective therapeutic response (16). Furthermore, the present study
demonstrated that cccDNA production may be attenuated in patients
with CHB who achieve VR and HBsAg seroclearance following
anti-viral treatment.
To predict the likelihood of achieving VR and HBsAg
seroclearance following anti-viral treatment in patients with CHB,
the areas under the ROC curves of IH tDNA and cccDNA levels were
compared. The area under the ROC curve of IH tDNA levels was
observed to be higher than that of IH cccDNA levels (0.96 vs.
0.88); however, the difference was not statistically significant
(P>0.10). This may be explained by changes in virion
productivity, which was defined as the ratio between IH HBV tDNA
and cccDNA levels and reflected the level of cccDNA transcription
(17). In the present study, while
the IH tDNA:cccDNA ratio was significantly lower in patients with
CHB who achieved VR and HBsAg seroclearance, it was revealed to be
positively correlated with IH tDNA, but not cccDNA levels. IH
cccDNA transcription is tightly controlled by various intrinsic
factors, including methylation and acetylation of H3 and H4
histones in cccDNA minichromosomes (18,19)
and recruitment of transcription factors, including histone
deacetylase 1, sirtuin 1, enhancer of zeste homolog 2 and yin yang
1 (20). Effective anti-viral
treatment may be beneficial to control cccDNA transcription and
consequently reduce virion productivity. In addition, it has been
reported that amplification of the cccDNA pool may be suppressed by
a negative-feedback mechanism involving the viral large surface
protein (15), and HBeAg loss may
significantly impair virion productivity (21). Such reports are verified by the
present study, in which the proportion of HBeAg−
patients was observed to be significantly higher in the group of
patients with CHB who had achieved VR and HBsAg seroclearance. A
previous report indicated that in patients with CHB who had
achieved therapeutic response with adefovir, the reduction rate of
IH tDNA levels was significantly higher than that of cccDNA levels
(22). However, it has also been
reported that IH tDNA levels may be as sensitive as IH cccDNA
levels for predicting the likelihood of achieving sustained
virological response (SVR) (8),
although achieving VR in combination with HBsAg seroclearance is
likely to be more effective than achieving SVR in anti-viral
treatment. The findings of the present study reveal a potentially
novel therapeutic strategy for suppressing virion productivity
through reducing the size of the cccDNA pool rather than through
eradicating IH cccDNA completely.
IH cccDNA was not detected in the negative controls
employed in the present study, however tDNA persistence was
detected in one patient, being diagnosed as OBI. This finding may
be attributed to the complex composition of IH tDNA, which aside
from rcDNA, consists of linear DNA, including double linear DNA
(dlDNA), as well as single-stranded DNA (ssDNA). Certain linear DNA
molecules, produced through illegitimate replication and deficient
HBV transcription (23), may be
present in hepatocytes but not secreted into the blood (24). Alternatively, IH tDNA may be
detected by means of integrated HBV (25). The findings of the present study
indicate that IH tDNA detection may be valuable in the diagnosis of
OBI in patients with hepatitis without HBV virusemia.
While cryo-preserved liver tissue is not always
available clinically, serum HBsAg has been regarded as a surrogate
marker for assessing IH cccDNA levels (21). Previous reports observed that serum
HBsAg levels were positively correlated with IH cccDNA levels in
patients with CHB who were HBeAg+, but not in those who
were HBeAg− (26,27).
However, in the present study, no significant correlation was
observed between serum HBsAg and IH cccDNA levels in either
HBeAg+ or HBeAg− patients. This may be a
consequence of HBsAg production, which is complex, abundant and far
exceeding that required for virion assembly, being independent of
HBV replication (28), or the
occurrence of increased HBsAg translation in HBeAg−
patients as a compensatory mechanism to adapt to the increased
immunological pressure (17).
Alternatively, HBsAg may be produced from integrated HBV (26), or IH cccDNA copies may be reduced
by anti-viral treatment (29).
It was observed in the present study that IH cccDNA
levels were positively correlated with serum HBeAg levels and that
IH cccDNA and tDNA levels were significantly lower in
HBeAg− patients than in HBeAg+ patients.
HBeAg has been reported to have an important role in HBV
persistence, and HBeAg seroconversion has been suggested to
increase Th1 cytokine secretion to inhibit HBV replication
(12). The findings of the present
study are in accordance with a previous study that demonstrated
that pre-core mRNA was solely transcribed from the cccDNA template
and not the integrated HBV, and that serum HBeAg detection may
serve as a cccDNA surrogate in anti-viral screening assays
(30).
In the present study, IH cccDNA levels were observed
to be positively correlated with IH tDNA but not serum HBV DNA
levels. This poor correlation between IH cccDNA and serum HBV DNA
levels may be partially attributed to the fact that a number of
patients were virusemia negative and indicates that serum HBV DNA
levels may not represent an ideal biomarker for evaluating IH HBV
production. CHB is a necroinflammatory liver disease and it has
been suggested that raised serum ALT levels may predict activation
of the host immune system, hepatocyte damage and increased cell
proliferation (31). The positive
correlation between serum ALT levels and IH cccDNA and tDNA levels
observed in the present study indicate that cccDNA replication may
have a role in the progression of HBV inflammation.
In conclusion, the present study showed that
extremely low levels of IH cccDNA may be present in patients with
AHB and patients with CHB who achieve VR and HBsAg seroclearance
following anti-viral treatment. IH cccDNA levels were identified to
be positively correlated with IH tDNA, serum HBeAg and ALT levels,
and not correlated with serum HBV DNA and HBsAg levels. Therefore,
IH cccDNA and tDNA have potential value for the prediction of a
successful therapeutic response in patients with CHB receiving
anti-viral treatment, and the latter may also be beneficial in OBI
diagnosis.
Acknowledgements
The authors would like to thank XC Chen and M Wang
from the Institute of Hepatology, Shenzhen Third People’s Hospital
for their technical assistance.
References
|
1
|
Lok AS and McMahon BJ: Chronic Hepatitis
B. Hepatology. 45:507–539. 2007. View Article : Google Scholar
|
|
2
|
Jiang YF, Ma ZH, Zhao PW, Pan Y, Liu YY,
Feng JY and Niu JQ: Effect of thymosin-α(1) on T-helper 1 cell and
T-helper 2 cell cytokine synthesis in patients with hepatitis B
virus e antigen-positive chronic hepatitis B. J Int Med Res.
38:2053–2062. 2010.
|
|
3
|
European association for the study of the
liver. EASL Clinical Practice Guidelines: management of chronic
hepatitis B. J Hepatol. 50:227–242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nguyen T, Thompson AJ, Bowden S, et al:
Hepatitis B surface antigen levels during the natural history of
chronic hepatitis B: a perspective on Asia. J Hepatol. 52:508–513.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wong DK, Huang FY, Lai CL, et al: Occult
hepatitis B infection and HBV replicative activity in patients with
cryptogenic cause of hepatocellular carcinoma. Hepatology.
54:829–836. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lau GK, Liang R, Chiu EK, Lee CK and Lam
SK: Hepatic events after bone marrow transplantation in patients
with hepatitis B infection: a case controlled study. Bone Marrow
Transplant. 19:795–799. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chaiteerakij R, Komolmit P, Sa-nguanmoo P
and Poovorawan Y: Intrahepatic HBV DNA and covalently closed
circular DNA (cccDNA) levels in patients positive for anti-HBc and
negative for HBsAg. Southeast Asian J Trop Med Public Health.
41:867–875. 2010.PubMed/NCBI
|
|
8
|
Sung JJ, Wong ML, Bowden S, et al:
Intrahepatic hepatitis B virus covalently closed circular DNA can
be a predictor of sustained response to therapy. Gastroenterology.
128:1890–1897. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bowden S, Jackson K, Littlejohn M and
Locarnini S: Quantification of HBV covalently closed circular DNA
from liver tissue by real-time PCR. Methods Mol Med. 95:41–50.
2004.PubMed/NCBI
|
|
10
|
Weinberger KM, Wiedenmann E, Böhm S and
Jilg W: Sensitive and accurate quantitation of hepatitis B virus
DNA using a kinetic fluorescence detection system (TaqMan PCR). J
Virol Methods. 85:75–82. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ramezani A, Banifazl M, Mamishi S, Sofian
M, Eslamifar A and Aghakhani A: The influence of human leukocyte
antigen and IL-10 gene polymorphisms on hepatitis B virus outcome.
Hepat Mon. 12:320–325. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chang JJ and Lewin SR: Immunopathogenesis
of hepatitis B virus infection. Immunol Cell Biol. 85:16–23. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun C, Fu B, Gao Y, Liao X, Sun R, Tian Z
and Wei H: TGF-β1 down-regulation of NKG2D/DAP10 and 2B4/SAP
expression on human NK cells contributes to HBV persistence. PLoS
Pathog. 8:e10025942012.
|
|
14
|
Chinese Society of Hepatology and Chinese
Society of Infectious Diseases; Chinese Medical Association. The
guideline of prevention and treatment for chronic hepatitis B (2010
version). Zhonghua Gan Zang Bing Za Zhi. 19:13–24. 2011.(In
Chinese).
|
|
15
|
Levrero M, Pollicino T, Petersen J,
Belloni L, Raimondo G and Dandri M: Control of cccDNA function in
hepatitis B virus infection. J Hepatol. 51:581–592. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang M, Qiu N, Lu S, et al: Serum
hepatitis B surface antigen is correlated with intrahepatic total
HBV DNA and cccDNA in treatment-naïve patients with chronic
hepatitis B but not in patients with HBV related hepatocellular
carcinoma. J Med Virol. 85:219–227. 2013.PubMed/NCBI
|
|
17
|
Manesis EK, Papatheodoridis GV, Tiniakos
DG, et al: Hepatitis B surface antigen: relation to hepatitis B
replication parameters in HBeAg-negative chronic hepatitis B. J
Hepatol. 55:61–68. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pollicino T, Belloni L, Raffa G, Pediconi
N, Squadrito G, Raimondo G and Levrero M: Hepatitis B virus
replication is regulated by the acetylation status of hepatitis B
virus cccDNA-bound H3 and H4 histones. Gastroenterology.
130:823–837. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vivekanandan P, Thomas D and Torbenson M:
Methylation regulates hepatitis B viral protein expression. J
Infect Dis. 199:1286–1291. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Belloni L, Allweiss L, Guerrieri F, et al:
IFN-α inhibits HBV transcription and replication in cell culture
and in humanized mice by targeting the epigenetic regulation of the
nuclear cccDNA minichromosome. J Clin Invest. 122:529–537.
2012.
|
|
21
|
Volz T, Lutgehetmann M, Wachtler P, et al:
Impaired intrahepatic hepatitis B virus productivity contributes to
low viremia in most HBeAg-negative patients. Gastroenterology.
133:843–852. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Werle-Lapostolle B, Bowden S, Locarnini S,
et al: Persistence of cccDNA during the natural history of chronic
hepatitis B and decline during adefovir dipivoxil therapy.
Gastroenterology. 126:1750–1758. 2004. View Article : Google Scholar
|
|
23
|
Chou YC, Jeng KS, Chen ML, et al:
Evaluation of transcriptional efficiency of hepatitis B virus
covalently closed circular DNA by reverse transcription-PCR
combined with the restriction enzyme digestion method. J Virol.
79:1813–1823. 2005. View Article : Google Scholar
|
|
24
|
Xu WS, Zhao KK, Miao XH, Ni W, Cai X,
Zhang RQ and Wang JX: Effect of oxymatrine on the replication cycle
of hepatitis B virus in vitro. World J Gastroenterol. 16:2028–2037.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang Z, Jhunjhunwala S, Liu J, et al: The
effects of hepatitis B virus integration into the genomes of
hepatocellular carcinoma patients. Genome Res. 22:593–601. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Thompson AJ, Nguyen T, Iser D, et al:
Serum hepatitis B surface antigen and hepatitis B e antigen titers:
disease phase influences correlation with viral load and
intrahepatic hepatitis B virus markers. Hepatology. 51:1933–1944.
2010. View Article : Google Scholar
|
|
27
|
Jaroszewicz J, Calle Serrano B, Wursthorn
K, et al: Hepatitis B surface antigen (HBsAg) levels in the natural
history of hepatitis B virus (HBV)-infection: A European
perspective. J Hepatol. 52:514–522. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chan HL, Wong VW, Tse AM, et al: Serum
hepatitis B surface antigen quantitation can reflect hepatitis B
virus in the liver and predict treatment response. Clin
Gastroenterol Hepatol. 5:1462–1468. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Takkenberg RB, Zaaijer HL, Menting S, et
al: Detection of hepatitis B virus covalently closed circular DNA
in paraffin-embedded and cryo-preserved liver biopsies of chronic
hepatitis B patients. Eur J Gastroenterol Hepatol. 22:952–960.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou T, Guo H, Guo JT, Cuconati A, Mehta A
and Block TM: Hepatitis B virus e antigen production is dependent
upon covalently closed circular (ccc) DNA in HepAD38 cell cultures
and may serve as a cccDNA surrogate in antiviral screening assays.
Antiviral Res. 72:116–124. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moucari R, Mackiewicz V, Lada O, et al:
Early serum HBsAg drop: a strong predictor of sustained virological
response to pegylated interferon alfa-2a in HBeAg-negative
patients. Hepatology. 49:1151–1157. 2009. View Article : Google Scholar : PubMed/NCBI
|