Introduction
Osteosarcoma is the most common type of primary
malignant bone tumor in children and adolescents. According to a
study in 2005, >400 novel cases of pediatric osteosarcoma are
diagnosed each year in the USA (1). Osteosarcoma is mainly treated with
chemotherapy or surgical excision, but as the prognosis of
unresectable or recurrent cases is poor, novel therapies for the
treatment of this type of tumor are currently being developed
(2). Numerous agents have been
used to treat osteosarcoma over the last 30 years and overall
survival has exceeded 50%. These agents include high-dose
methotrexate, doxorubicin, cisplatin, ifosfamide and etoposide,
which has now been sufficient cumulative experience for patients
with osteosarcoma (3).
Autophagy is a process of selective degradation of
cellular components. There are three major types of autophagy
described, which include macroautophagy, microautophagy and
chaperone-mediated autophagy (4).
At present, the most important type of autophagy is macroautophagy.
Macroautophagy, which is referred to as autophagy in the present
study, is a nonspecific degradation system that mediates the
clearance of long-lived cytoplasmic proteins, including
aggregate-prone proteins, certain pathogens and organelles
(5–8). In mammalian cells, autophagosome
formation begins with a nucleation step, then they expand and fuse
to form complete double membrane vesicles termed autophagosomes.
Autophagosomes fuse with lysosomes and the contents of the
autophagosomes are degraded (9).
In this process, two important ubiquitin-like conjugation processes
are involved. The first is conjugation of autophagy-related protein
(Atg)12 to Atg5 and the second is conjugation of
microtubule-associated protein 1 light chain 3 (LC3; also known as
MAP-LC3 or Atg8) to phosphatidylethanolamine (PE), and the two
processes are essential for autophagosome formation (10). Notably, autophagy contributes to
the maintenance cellular homeostasis and acts as a housekeeping
survival mechanism in different harmful conditions, including
starvation and endoplasmic reticulum stress (11). A study suggested that autophagy may
be important in regulatingof cancer development and progression,
and in determining the response of tumor cells to anticancer
therapy. However, the role of autophagy in these processes is
complicated and depends on the circumstances (12). Autophagy and apoptosis regulate
cell fate and are critical in development, normal physiology and in
numerous diseases. Although there are more marked differences
between these two processes, they can be regulated by the samegene,
such as Bcl- (13). A study
suggested that there is crosstalk between autophagy and apoptosis
in certain situations, for example the autophagy induced by
starvation may be inhibited by caspase-mediated cleavage of beclin1
and the fragment of the cleaved beclin1 translocates to the
mitochondria and induces apoptosis (14). Additionally, autophagic degradation
of active caspase-8 inhibits apoptosis (15). Thus, the regulation of apoptosis
and autophagy by each other is complicated and the mechanism is
unclear.
Dox is a chemotherapeutic agent that activates p53
to induce apoptosis (16). Dox was
widely used for treatment of malignancies and exerts a range of
effects on the structural and functional properties of tumor cells,
ultimately leading to cell death. Although the direct effects of
Dox in the damage of DNA have been well studied, the sequence of
biochemical events that mediate cell death in response to Dox
remains unclear (17).
The present study focused on the role of autophagy
in the Dox-induced apoptosis of osteosarcoma cells. The study also
investigated whether a combination of autophagy inhibitors and Dox
enhanced apoptosis of osteosarcoma cells.
Materials and methods
Reagents
Atg7 small interfering (si)RNA plasmid was purchased
from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA;
sc-29918). Dox and 3-methyladenine (3-MA) were purchased from
Sigma-Aldrich (St. Louis, MO, USA). Fetal bovine serum (FBS) and
Dulbecco’s modified Eagle’s medium (DMEM) were purchased from
Gibco-BRL (Carlsbad, CA, USA). The antibodies anti-nucleoporin p62
antibody (sc-25523; secondary antibody rabbit) anti- cytochrome c
antibody (sc-13156; secondary antibody mouse), anti-MAP LC3β
antibody (sc-376404; secondary antibody mouse), anti-caspase-3
antibody (sc-65496; secondary antibody mouse), cleaved caspase-3
p11 antibody (sc-22171-R; secondary antibody rabbit) and ATG7
antibody (sc-33211; secondary antibody rabbit) were all purchased
from Santa Cruz Biotechnology, Inc.
Cell proliferation assays
U2OS and Saos-2 human cells (ATCC, Rockville, MD,
USA) were grown in DMEM with 10% FBS and cultured at 37°C in 5%
CO2. The cell viability was determined by a MTT [12 μl,
5 mg/ml in phosphate-buffered saline (PBS)] assay. The U2OS and
Saos-2 human cells were cultured at a density of
1–1.5×104 cells/well in 96-well plates. Each group was
replicated in six separate wells. DMSO was used for the
untreated/control cells. Following treatment (0, 100, 250 or 500
nM) Dox, the MTT reagent (Sigma-Aldrich) was added to each well for
4 h. Subsequently, the contents of each well was dissolved in 150
μl dimethylsulfoxide. The absorbance was recorded at a wavelength
of 490 nm using an ELISA reader (BD Biosciences, Franklin Lake, NJ,
USA).
Transfection
Prior to the transfection, the cells were grown to
40% confluence in each dish. According to the manufacturer’s
instructions, the U2OS and Saos-2 cells were transfected with 40
nmol/l control siRNA or Atg7 siRNA using Lipofectamine RNAiMAX
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). After 36
h, the cells were harvested for western blotting.
Western blot analysis
For each assay, the U2OS and Saos-2 cells were
washed with cold PBS twice and then 120 μl radioimmunoprecipitation
assay buffer [50 mM Tris-HCl, pH 6.8; 0.1% SDS, 150 mM NaCl, 1 mM
EDTA, 0.1 mM Na3VO4, 1 mM sodium fluoride
(NaF), 1% Triton X-100, 1% NP-40, 1 mM dithiothreitol, 1 mM PMSF, 1
μg/ml aprotinin, 1 μg/ml leupeptin and 1 μg/ml pepstatin A] was
added to each dish following treatment. The lysates were harvested
into 1.5-ml tubes and agitated in a cold room (4°C) for 20 min.
Subsequently, the cell lysates were centrifuged at 13,000 × g for
15 min, and then the supernatants were harvested. The protein
concentrations were detected by a bicinchoninic acid assay (Sigma
Aldrich). A total of 60 μg protein was used for the western
blotting. The lysates were separated by 10% (w/v)
SDS-polyacrylamide gel electrophoresis. Subsequently, the proteins
were transferred onto polyvinylidene difluoride membranes and they
were blocked with 5% (w/v) skimmed milk in buffer [10 mM Tris-HCl
(pH 7.6), 100 mM NaCl and 0.1% (v/v) Tween-20] for 30 min at room
temperature (25°C). The membranes were incubated in the primary
antibodies overnight in a cold room. The following day, the
membranes were washed three times with Tris-buffered saline and
Tween-20. Subsequently, the membranes were incubated with the
secondary antibodies for 1 h at room temperature. The
semi-quantitation of the proteins was analyzed with a Tanon Gel
Imager system (Tanon, Shanghai, China).
Mitochondrial membrane potential (MMP)
analysis
JC-1 staining to detect the MMP of each group was
conducted by flow cytometry, according to the manufacturer’s
instructions (Molecular Probes, Invitrogen Life Technologies,
Carlsbad, CA, USA). Following treatment, the U2OS and Saos-2 cells
were trypsinized, washed with PBS, and resuspended in PBS at a
concentration of 1×106 cells/ml. The U2OS and Saos-2
cells were then stained with 2.5 μl JC-1 (1 mg/ml) and incubated in
the dark at 37°C for 1.5 h. The JC-1 positive cells were
subsequently detected by a FACSCalibur flow cytometer (BD
Biosciences).
Statistical analysis
Data are representative of three independent
experiments and were analyzed by t-test. P<0.05 was considered
to indicate a statistically significant difference.
Results
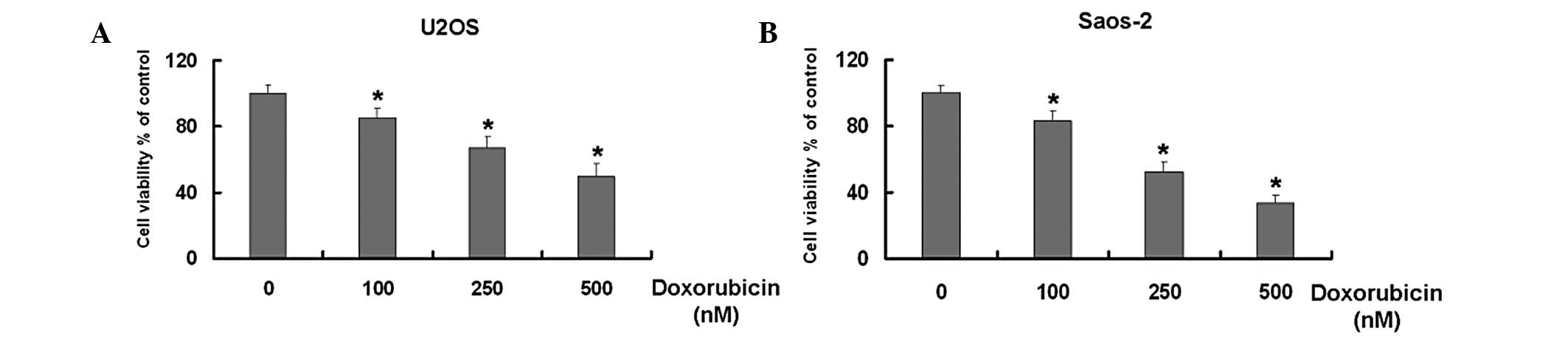
Dox inhibits the growth of U2OS and
Saos-2 osteosarcoma cells in a dose-dependent manner
Dox is widely used for the treatment of numerous
types of tumor (17–21). Initially, the study aimed to
identify whether Dox inhibits the growth of the U2OS and Saos-2
osteosarcoma cell lines. The MTT results demonstrated that Dox
reduced the cell viability of the two U2OS and Saos-2 cell types in
dose-dependent manner compared with that of the untreated cells
(Fig. 1).
Dox induces apoptosis in U2OS and Saos-2
cells
As mentioned previously, Dox inhibited the cell
growth of the two osteosarcoma cell lines. Subsequently, the levels
of apoptosis-associated proteins in the U2OS and Saos-2 cells
treated with Dox were detected to investigate whether growth
inhibition was associated with apoptosis. The levels of the
mitochondrial apoptosis-associated proteins caspase-3 and
cytochrome c were detected.
As shown in Fig. 2A and
B, Dox increased the expression levels of cleaved caspase-3 and
cytochrome c in the U2OS cells compared with those in the
untreated cells. A second osteosarcoma cell line, Saos-2, was used
to confirm the aforementioned results. Similar results were
observed in the Saos-2 cells (Fig. 2D
and E). Furthermore, the MMP was measured in the two cell lines
by flow cytometry. As shown in Fig. 2C
and F, Dox induced a significant loss in the MMP of the two
cells lines.
 | Figure 2Doxorubicin induces apoptosis in U2OS
and Saos-2 cells. (A) Western blot analysis for the expression
levels of caspase-3, cleaved caspase-3 and cytochrome c in
U2OS cells treated with 0, 100, 250 and 500 nM doxorubicin for 24
h. (B) Quantitation of cleaved caspase-3 and cytochrome c
protein expression levels in U2OS cells treated with 0, 100, 250
and 500 nM doxorubicin for 24 h. (C) U2OS cells treated with 0,
100, 250 and 500 nM doxorubicin for 24 h. Following treatment, the
cells were stained with JC-1. (D) Western blot analysis for the
expression levels of caspase-3, cleaved caspase-3 and cytochrome
c in Saos-2 cells treated with 0, 100, 250 and 500 nM
doxorubicin for 24 h. (E) Quantitation of cleaved caspase-3 and
cytochrome c protein expression levels in Saos-2 cells
treated with 0, 100, 250 and 500 nM doxorubicin for 24 h. (F)
Saos-2 cells treated with 0, 100, 250 and 500 nM doxorubicin for 24
h. Following treatment, the cells were stained with JC-1.
*P<0.05 versus the control group. Data are presented
as the mean ± standard deviation, n=3. MMP, mitochondrial membrane
potential. |
These results indicated that Dox induces apoptosis
in osteosarcoma cells through the mitochondrial apoptotic
pathway.
Dox induces autophagy in U2OS and Saos-2
cells
In addition to apoptosis, it has been reported that
Dox induces autophagy in tumor cells (22). Thus, the levels of
autophagy-associated proteins in the U2OS and Saos-2 cells treated
with Dox were subsequently detected. LC3 (the mammalian equivalent
of yeast Atg8) and p62 are the two major markers of autophagy. When
autophagy occurs, the quantity of LC3-II increases and the
expression levels of p62 decrease. Western blotting demonstrated
that the levels of these two proteins in U2OS and Saos-2 cells
treated with Dox were altered compared with those in the untreated
cells. As shown in Fig. 3, Dox
elevated the expression levels of LC3-II and reduced the expression
levels of p62 in U2OS and Saos-2 cells.
 | Figure 3Doxorubicin induces autophagy in U2OS
and Saos-2 cells. (A) Western bloting for the expression levels of
LC3 and p62 in U2OS cells treated with 0, 100, 250 and 500 nM
doxorubicin for 24 h. (B) Quantitation of LC3-II and p62 protein
expression levels in U2OS cells treated with 0, 100, 250 and 500 nM
doxorubicin for 24 h. (C) Western blot analysis for the expression
levels of LC3 and p62 in Saos-2 cells treated with 0, 100, 250 and
500 nM doxorubicin for 24 h. (D) Quantitation of LC3-II and p62
protein expression levels in Saos-2 cells treated with 0, 100, 250
and 500 nM doxorubicin for 24 h. *P<0.05 versus the
control group. Data are presented as the mean ± standard deviation,
n=3. LC3, microtubule-associated protein 1 light chain 3. |
These results indicated that Dox induces autophagy
in U2OS and Saos-2 cells.
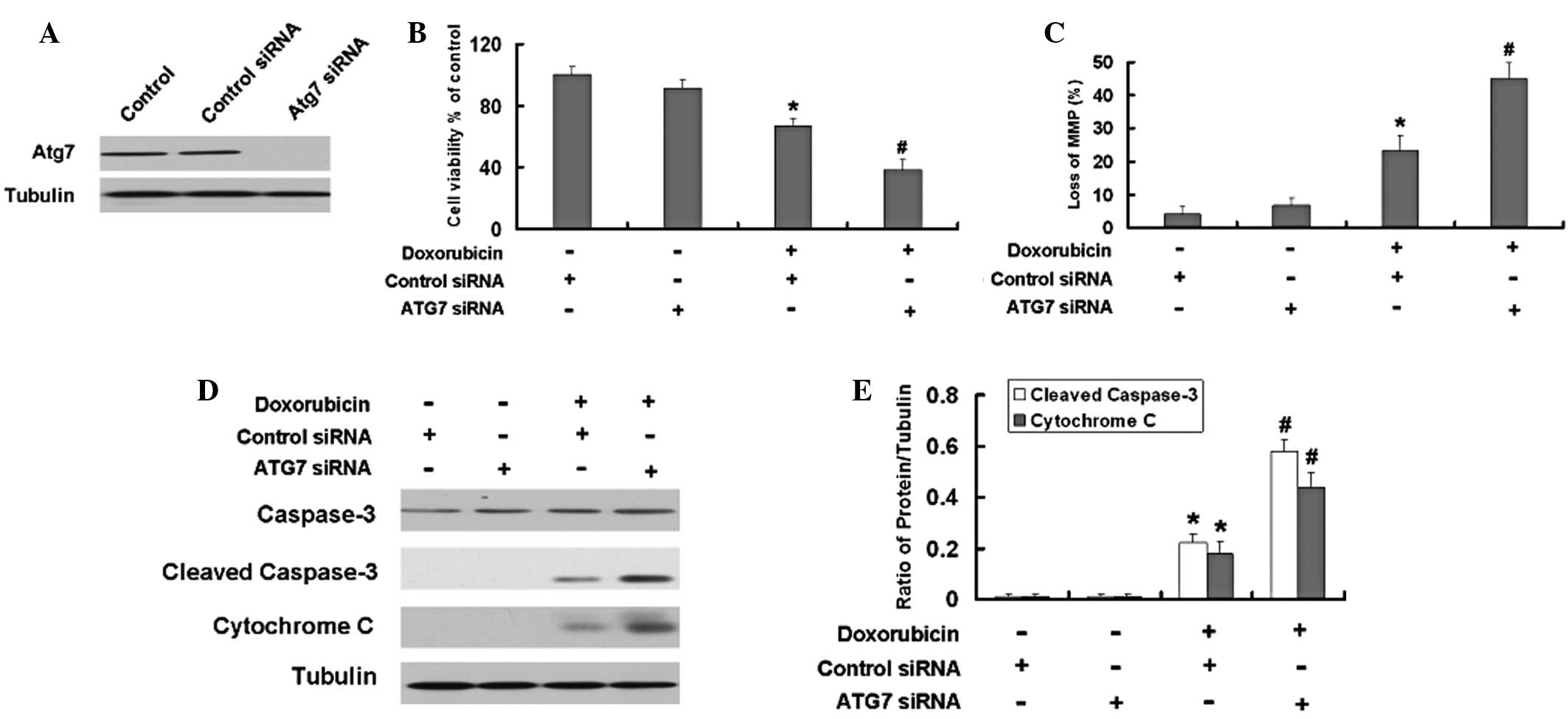
Inhibition of autophagy by Atg7 siRNA or
an autophagy inhibitor enhances the apoptosis induced by Dox in
U2OS and Saos-2 cells
Numerous studies have shown that autophagy is
important in the apoptosis induced by antitumor agents, but the
role varies with different agents and types of tumor (12,23).
Atg7 siRNA and an autophagy inhibitor were used to investigate the
role of autophagy in Dox-induced apoptosis. As shown the Fig. 4A, siAtg7 reduced the expression
levels of Atg7 in the U2OS cells compared with those in the control
cells. The MTT results demonstrated that Atg7 siRNA intensifies the
growth inhibition induced by Dox in the U2OS cells (Fig. 4B). Furthermore, the Atg7 siRNA
aggravated the loss of MMP in the U2OS cells (Fig. 4C). Subsequently, the levels of the
apoptosis-associated proteins, cleaved capase-3 and cytochrome
c, were detected. The Atg7 siRNA further increased the
expression levels of the apoptosis-associated proteins, cleaved
capase-3 and cytochrome c, induced by Dox in the U2OS cells
(Fig. 4D and E).
Subsequently, the autophagy inhibitor 3-MA was used
to confirm the role of autophagy in Dox-induced apoptosis. The
combination of 3-MA and Dox also intensified the growth inhibition
of the U2OS cells induced by Dox (Fig.
5A). As shown in Fig. 5B, C and
D, the combination of 3-MA and Dox further increased the
expression levels of the apoptosis-associated proteins cleaved
capase-3 and cytochrome c and loss of MMP induced by Dox in
the U2OS cells.
 | Figure 5Inhibition of autophagy by the
autophagy inhibitor 3-MA enhances apoptosis induced by doxorubicin
in U2OS cells. (A) Cell viability was determined by an MTT assay.
U2OS cells were treated by doxorubicin (250 nM), 3-MA (10 mM) or
doxorubicin and 3-MA for 24 h. Data are presented as the mean ±
standard deviation, n=6. (B) U2OS cells were treated by doxorubicin
(250 nM), 3-MA (10 mM) or doxorubicin and 3-MA for 24 h. Following
the treatment, the cells were stained with JC-1. (C) Western blot
analysis for the expression levels of caspase-3, cleaved caspase-3
and cytochrome c in the cells treated with doxorubicin (250
nM), 3-MA (10 mM) or doxorubicin and 3-MA for 24 h. (D)
Quantitation of cleaved caspase-3 and cytochrome c protein
expression levels. *P<0.05 versus the control group;
#P<0.05 versus the doxorubicin group. Data are
presented as the mean ± standard deviation, n=3. DMSO,
dimethylsulfoxide; 3-MA, 3-methyladenine; MMP, mitochondrial
membrane potential. |
Saos-2 cells were used to confirm these results and
similar results were observed (Figs.
6 and 7).
 | Figure 7Inhibition of autophagy by the
autophagy inhibitor 3-MA enhances apoptosis induced by doxorubicin
in Saos-2 cells. (A) Cell viability was determined by an MTT assay.
Saos-2 cells were treated with doxorubicin (250 nM), 3-MA (10 mM)
or doxorubicin and 3-MA for 24 h. Data are presented as the mean ±
standard deviation, n=6. (B) Saos-2 cells were treated with
doxorubicin (250 nM), 3-MA (10 mM) or doxorubicin and 3-MA for 24
h. Following the treatment, the cells were stained with JC-1. (C)
Western blot analysis for the expression levels of caspase-3,
cleaved caspase-3 and cytochrome c treated with doxorubicin
(250 nM), 3-MA (10 mM) or doxorubicin and 3-MA for 24 h. (D)
Quantitation of cleaved caspase-3 and cytochrome c protein
expression levels. *P<0.05 versus the control group;
#P<0.05 versus the doxorubicin group. Data are
presented as the mean ± standard deviation, n=3. DMSO,
dimethylsulfoxide; 3-MA, 3-methyladenine; MMP, mitochondrial
membrane potential. |
These results in the U2OS and Saos-2 cells indicated
that autophagy is important in Dox-induced apoptosis and the
inhibition of autophagy further increases the cytotoxicity of Dox
in osteosarcoma cells.
Discussion
Osteosarcoma is common type of primary bone tumor.
Due to the high metastatic potential and the common acquisition of
chemotherapeutic resistance in osteosarcoma, the clinical outcome
is poor (18). Chemotherapy for
osteosarcoma is administered in neoadjuvant and adjuvant settings.
Clinical treatment commonly uses chemotherapy for the treatment of
osteosarcoma, in order to avoid amputation. Dox is a common
conventional chemotherapeutic drug used for the treatment of
osteosarcoma (19). Studies have
indicated that Dox inhibits the cell growth of human leukemia cells
(17) and breast cancer cell lines
(20). In order to investigate the
effect of Dox on osteosarcoma, the present study investigated the
effect of Dox on the cell viability of two human osteosarcoma cell
lines, U2OS and Saos-2, by MTT assay. The results demonstrated that
Dox inhibited the cell proliferation of the U2OS and Saos-2 cells
in a dose-dependent manner.
A previous study reported that Dox induced apoptosis
by increasing the expression levels of cleaved caspase-3 (18). Cleaved caspase-3 is an activated
form of caspase-3, and it leads to cell death. Therefore, in order
to detect whether Dox induces the apoptosis of the two osteosarcoma
cell lines, the expression levels of cleaved caspase-3 and
cytochrome c were detected by western blot analysis of the
U2OS and Saos-2 cells. The results showed that Dox increased the
expression levels of cleaved caspase-3 and cytochrome c in a
dose-dependent manner in the U2OS and Saos-2 cells. Additionally,
the MMP loss was measured in the two cell lines using flow
cytometry. Dox evidently induced the loss of the MMP in the two
cells lines.
Although the present study demonstrated that Dox
induces apoptosis of U2OS and Saos-2 cells, the effect of the
treatment requires enhancement. Therefore, the other functions of
Dox were investigated. Fong et al (22) identified that Dox induced increased
expression levels of LC3 in the A2780 epithelial ovarian cancer
cell line. LC3, a homologue of Apg8p that is essential for
autophagy in yeast, is a classical marker of autophagy. In
particular, LC3-II is the first mammalian protein identified that
specifically associates with autophagsome membranes (24). In order to determine whether Dox
has an effect on autophagy in U2OS and Saos-2 cells, the expression
levels of LC3 were detected by western blotting and it was
demonstrated that Dox induced the increased expression levels of
LC3 in the U2OS and Saos-2 cells. In addition, Dox reduced the
levels of another autophagy marker, p62. All the results showed
that Dox induced autophagy in the U2OS and Saos-2 cells.
A growing number of studies have reported crosstalk
between apoptosis and autophagy (25). The correlation between them is
complicated. A number of studies have provided evidence that
autophagy serves as a survival pathway in tumor cells treated with
anticancer drugs and proposed a rationale for the use of autophagy
inhibitors in combination with therapies designed to induce
apoptosis in human cancers (26,27).
Therefore, autophagy inhibition represents a major therapeutic
target for chemosensitization (28). Although the present study
demonstrated that Dox induced apoptosis and autophagy in U2OS and
Saos-2 cells, the correlation between apoptosis and autophagy
induced by Dox is unknown. In order to elucidate the correlation,
Dox was used in combination with the autophagy inhibitor 3-MA and
Atg7 siRNA to detect the cell proliferation and the expression
levels of apoptosis-associated proteins induced by Dox in the U2OS
and Saos-2 cells. As expected, the results showed that the
autophagy inhibitor 3-MA and Atg7 siRNA significantly enhanced the
cell proliferation inhibition and notably increased the expression
levels of the apoptosis proteins cleaved caspase-3 and cytochrome
c induced by Dox in the U2OS and Saos-2 cells. These results
demonstrated that autophagy is preventative against the apoptosis
induced by Dox in U2OS and Saos-2 cells.
The present study provides significant data
indicating that inhibition of autophagy may enhance the tumor cell
proliferation inhibition and apoptosis of U2OS and Saos-2 cells
induced by Dox. Thus, activation of autophagy may be involved in
the resistance to apoptosis of U2OS and Saos-2 cells. Autophagy may
enable tumor cells adapt to metabolic stress and promote cell
survival during apoptosis. In conclusion, the present study showed
that inhibition of autophagy may be a novel strategy to increase
the efficacy of anticancer drugs in the treatment of U2OS and
Saos-2 cells.
References
|
1
|
Chou AJ, Merola PR, Wexler LH, et al:
Treatment of osteosarcoma at first recurrence after contemporary
therapy: the Memorial Sloan-Kettering Cancer Center experience.
Cancer. 104:2214–2221. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nakase M, Inui M, Okumura K, Kamei T,
Nakamura S and Tagawa T: p53 gene therapy of human osteosarcoma
using a transferrin-modified cationic liposome. Mol Cancer Ther.
4:625–631. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Janeway KA and Grier HE: Sequelae of
osteosarcoma medical therapy: a review of rare acute toxicities and
late effects. Lancet Oncol. 11:670–678. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang Z and Klionsky DJ: Eaten alive: a
history of macroautophagy. Nat Cell Biol. 12:814–822. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Klionsky DJ: The molecular machinery of
autophagy: unanswered questions. J Cell Sci. 118:7–18. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Levine B and Klionsky DJ: Development by
self-digestion: molecular mechanisms and biological functions of
autophagy. Dev Cell. 6:463–477. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mizushima N: The pleiotropic role of
autophagy: from protein metabolism to bactericide. Cell Death
Differ. 12(Suppl 2): 1535–1541. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rubinsztein DC, Gestwicki JE, Murphy LO
and Klionsky DJ: Potential therapeutic applications of autophagy.
Nat Rev Drug Discov. 6:304–312. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang WP and Klionsky DJ: Autophagy in
yeast: a review of the molecular machinery. Cell Struct Funct.
27:409–420. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Luo S and Rubinsztein DC: Apoptosis blocks
Beclin 1-dependent autophagosome synthesis: an effect rescued by
Bcl-xL. Cell Death Differ. 17:268–277. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Del Bello B, Toscano M, Moretti D and
Maellaro E: Cisplatin-induced apoptosis inhibits autophagy, which
acts as a pro-survival mechanism in human melanoma cells. PLoS One.
8:e572362013.PubMed/NCBI
|
|
12
|
Hippert MM, O’Toole PS and Thorburn A:
Autophagy in cancer: good, bad, or both? Cancer Res. 66:9349–9351.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Thorburn A: Apoptosis and autophagy:
regulatory connections between two supposedly different processes.
Apoptosis. 13:1–9. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Djavaheri-Mergny M, Maiuri MC and Kroemer
G: Cross talk between apoptosis and autophagy by caspase-mediated
cleavage of Beclin 1. Oncogene. 29:1717–1719. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hou W, Han J, Lu C, Goldstein LA and
Rabinowich H: Autophagic degradation of active caspase-8: a
crosstalk mechanism between autophagy and apoptosis. Autophagy.
6:891–900. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lowe SW, Ruley HE, Jacks T and Housman DE:
p53-dependent apoptosis modulates the cytotoxicity of anticancer
agents. Cell. 74:957–967. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu R, Shtil AA, Tan TH, Roninson IB and
Kong AN: Adriamycin activates c-jun N-terminal kinase in human
leukemia cells: a relevance to apoptosis. Cancer Lett. 107:73–81.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Spina A, Sorvillo L, Di Maiolo F, et al:
Inorganic phosphate enhances sensitivity of human osteosarcoma U2OS
cells to doxorubicin via a p53-dependent pathway. J Cell Physiol.
228:198–206. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gill J, Ahluwalia MK, Geller D and Gorlick
R: New targets and approaches in osteosarcoma. Pharmacol Ther.
137:89–99. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Radetzki S, Köhne CH, von Haefen C, et al:
The apoptosis promoting Bcl-2 homologues Bak and Nbk/Bik overcome
drug resistance in Mdr-1-negative and Mdr-1-overexpressing breast
cancer cell lines. Oncogene. 21:227–238. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tarquini F, Tiribuzi R, Crispoltoni L, et
al: Caspase 3 activation and PARP cleavage in lymphocytes from
newborn babies of diabetic mothers with unbalanced glycaemic
control. Cell Biochem Funct. 32:87–95. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fong MY, Jin S, Rane M, Singh RK, Gupta R
and Kakar SS: Withaferin A synergizes the therapeutic effect of
doxorubicin through ROS-mediated autophagy in ovarian cancer. PLoS
One. 7:e422652012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li H, Jin X, Zhang Z, Xing Y and Kong X:
Inhibition of autophagy enhances apoptosis induced by the
PI3K/AKT/mTor inhibitor NVP-BEZ235 in renal cell carcinoma cells.
Cell Biochem Funct. 31:427–433. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kabeya Y, Mizushima N, Ueno T, et al: LC3,
a mammalian homologue of yeast Apg8p, is localized in autophagosome
membranes after processing. EMBO J. 19:5720–5728. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kang R, Zeh HJ, Lotze MT and Tang D: The
Beclin 1 network regulates autophagy and apoptosis. Cell Death
Differ. 18:571–580. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhong JT, Xu Y, Yi HW, et al: The BH3
mimetic S1 induces autophagy through ER stress and disruption of
Bcl-2/Beclin 1 interaction in human glioma U251 cells. Cancer Lett.
323:180–187. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chang Z, Shi G, Jin J, et al: Dual
PI3K/mTOR inhibitor NVP-BEZ235-induced apoptosis of hepatocellular
carcinoma cell lines is enhanced by inhibitors of autophagy. Int J
Mol Med. 31:1449–1456. 2013.PubMed/NCBI
|
|
28
|
Fimia GM and Piacentini M: Regulation of
autophagy in mammals and its interplay with apoptosis. Cell Mol
Life Sci. 67:1581–1588. 2010. View Article : Google Scholar : PubMed/NCBI
|