Introduction
Hepatitis B virus (HBV) DNA does not integrate at
specific sites of the host genome. Thus, a common cis effect
onto flanking cellular genes can be excluded as a general mechanism
of HBV-associated carcinogenesis. However, integrated HBV DNA can
encode two types of transcriptional activators: The already studied
HBx and the PreS2 activators, including the large HBV surface
protein (LHBs) and the C-terminally truncated middle size surface
proteins (MHBst) (1–3). The
sequence encoding the PreS2 activators is localized on the HBV
surface gene. The surface gene consists of a single open reading
frame (ORF) divided into three coding regions: preS1, preS2 and S,
each starting with an in-frame ATG codon. Through alternate
translation initiation at each of the three AUG codons, a large
(LHB; PreS1 + PreS2 + S), a middle size (MHB; PreS2 + S) and a
small (SHB; S) envelope glycoprotein is able to be synthesized. The
activator function of the surface protein requires the cytoplasmic
orientation of the PreS2 domain (the minimal functional unit) that
occurs in the case of MHBt and in a fraction of LHBs
(4,5). By contrast, full-length MHBs exhibit
no transcriptional activator function: The PreS2 domain is directed
into the lumen of the endoplasmic reticulum (ER). To investigate
the biological importance of MHBs, the present study screened and
identified the proteins interacting with MHBs with the yeast
two-hybrid system 3 to elucidate the biological function of
MHBs.
Materials and methods
Agents and culture media, yeast strains
and plasmids
Taq DNA polymerase, T4 DNA ligase, and
EcoRI and BamHI restriction endonucleases were
purchased from Takara Bio, Inc. (Dalian, China). Lithium acetate,
semi-sulfate adenine, acrylamide and N,N′-bis-acrylamide were
purchased from Sigma (St. Louis, MO, USA). TEMED was obtained from
Boehringer Mannheim GMBH (Mannheim, Germany). Tryptone and yeast
extracts were purchased from Oxoid (Lenexa, KS, USA). X-α-Gal and
Yeast peptone dextrose adenine (YPDA), SD/-Trp SD/-Leu,
SD/-Trp/-Leu, SD/-Trp/-Leu/-His and SD/-Trp/-Leu/-His/-Ade culture
media were obtained from Clontech Laboratories, Inc. (Mountain
View, CA, USA). Protein-G agarose was obtained from Roche
Diagnostics (Indianapolis, IN, USA). The pGEM-T vector was
purchased from Promega Corporation (Madison, WI, USA). Yeast
strains and plasmids for yeast two-hybrid experiments were obtained
from Clontech Laboratories, Inc. as components of the Matchmaker
two hybrid system 3. The yeast strain AH109 (MATa, trp1-901,
leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2:
GAL1UAS-GAL1TATA-HIS3,
GAL2UAS-GAL2TATA-ADE2, URA3:
MEL1UAS-MEL1TATA-LacZ) containing pGBKT7-53,
coding for the DNA binding domain (DNA-BD)/mouse p53 fusion protein
and AH109 used for cloning of bait plasmid, yeast strain Y187 (MATa
ura3-52, his3-200, Ade2-101, trp1-901, leu2-3, 112, gal4Δ, gal80Δ,
met-, URA3: GAL1UAS-GAL1TATA-lacZ MEL1)
containing pTD1-1, in which pACT2 codes for the activation domain
(AD)/SV40 large T antigen fusion protein and Y187 were used for the
cloning of library plasmids. Pretransformed human cDNA liver cell
library yeast strain (Y187). The bacterial strain DH5α was used for
cloning every shuttle plasmid. The yeast Escherichia coli
shuttle plasmids pGBKT7 DNA-BD cloning plasmid, pGADT7 AD cloning
plasmid, pGBKT7-53 control plasmid, pGADT7, pGBKT7-Lam control
plasmid and the pCL1 plasmid were purchased from Clontech
Laboratories, Inc. (K1612-1). The pGEM T vector was obtained from
Promega Corporation.
The A7 plasmid containing the whole sequences of the
adr subtype of HBV, was conserved by the Gene Laboratory of the
Institute of Infectious Diseases in Ditan Hospital (Beijing,
China).
Construction of the bait plasmid and the
expression of the MHBs protein
The MHBs sequences were generated by polymerase
chain reaction (PCR) amplification of the A7 plasmid (HBV strain
adr). The A7 plasmid contains coding sequences for all of the HBV.
The sequences of the primers, which contain the EcoRI and
BamHI restriction enzyme sites, were as follows: Forward:
5′-GAATTCATGGTCACCTTGAGGTGG-3′ and reverse:
5′-GGATCCAGTTTACATATGGGTTTCTG-3′. The PCR conditions were as
follows: 94°C for 4 min, 94°C hot denaturalization for 50 sec, 58°C
annealing for 50 sec and 72°C extension for 1 min for 35 cycles.
The PCR product (6 μl) was cloned with the pGEM-T vector. The
primary structure of the insert was confirmed by direct sequencing.
The fragment encoding MHBs was released from the pGEM-T-MHBs by
digestion with EcoRI and BamHI, and ligated to
pGBKT7. Vector pGBKT7 expressing proteins were fused with amino
acids 1-147 of the GAL4 DNA-BD and pGADT7 expressing proteins were
fused with amino acids 768-881 of the GAL4 AD. The plasmid
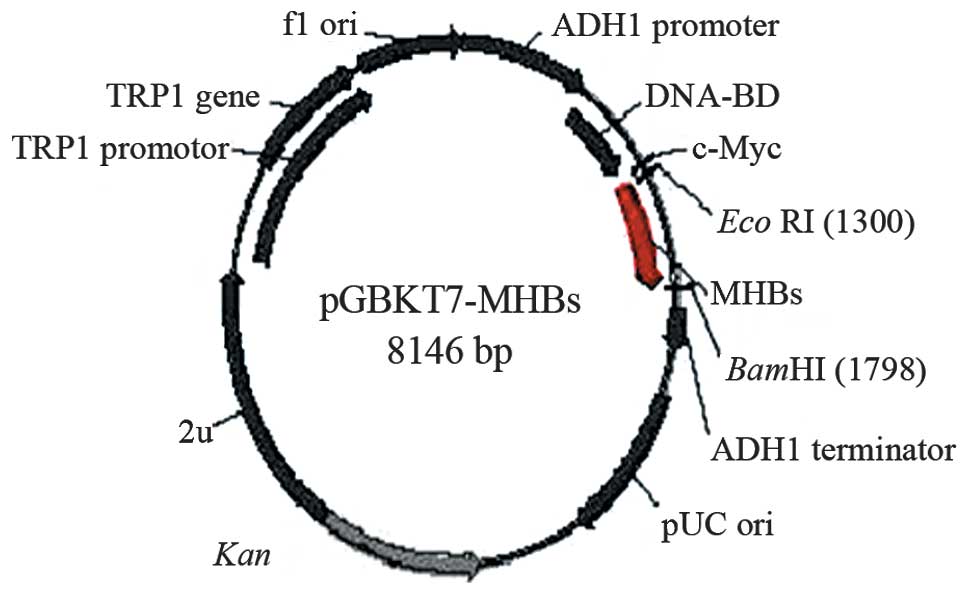
pGBKT7-MHBs (Fig. 1) containing
the full-length MHBs gene was able to direct the expression of the
DNA-BD, c-myc and MHBs fusion protein. The plasmid was transformed
into the AH109 yeast strain using the lithium acetate method
(6). Transformed AH109 (bait) was
cultured on QDO to exclude the auto-activation activity.
Yeast two-hybrid screening of the liver
cell cDNA library
One large (2–3 mm), fresh (<2 months old) colony
of AH109 (bait) was inoculated into 50 ml SD/-Trp and incubated at
30°C overnight (16–24 h) with agitation at 250–270 rpm. Then, the
cells were spun by centrifuging the entire 50 ml culture at 1,000 ×
g for 5 min. Following decanting the supernatant, the cell pellet
was resuspended in the residual liquid by vortexing. A human liver
cDNA library cloned into pACT2 and yeast reporter strain Y187 were
co-cultured. The entire AH109 (bait) culture and the 1 ml human
liver cDNA library (1×106 cfu/ml) were combined and
cultured in a 2-l sterile flask and 45 ml of 2X YPDA/Kan was added
and mixed gently. Following 20 h of mating, the cells were spun
down and resuspended, and then were spread onto 50 large (150 mm)
plates, containing 100 ml of SD/-Ade/-His/-Leu/-Trp (QDO).
Following 6–18 days of growth, the yeast colonies were transferred
onto the plates containing X-α-gal and blue colonies identified the
expression of the MEL1 reporter gene. Approximately
1×106 colonies were screened and positive clones were
identified. The yeast plasmid was isolated from positive yeast
colonies using the Lyticase method (provided by Clontech
Laboratories, Inc.), and transformed into super competent E.
coli DH5α using a chemical method. Transformants were plated on
ampicillin super optimal broth selection media and grown under
selection. Subsequently, pACT2-cDNA constructs were re-isolated, by
BglII restriction enzyme digests and following sequencing of
the positive colonies, the sequences were BLASTed with GenBank
(http://blast.ncbi.nlm.nih.gov/Blast.cgi) to analyze
the function of the genes.
To verify the true protein-protein interaction, the
plasmids of positive colonies were transformed into the Y187 yeast
strain, and false positives were excluded. Next, mating experiments
were performed by mating with the AH109 yeast strain containing
pGBKT7-MHBsor pGBKT7-Lam. Following mating, the diploid yeast were
plated on SD/-Ade-His-Leu-Trp (QDO) covered with X-α-gal to assess
the specificity of interactions. Since plasmid pACT2-cDNA contains
two restriction endonuclease sites of BglII on the two sides
of multiple cloning sites, the gene fragments of different lengths
verified that these screened clones were positive colonies.
Results
Identification of the plasmid and
analysis of the cDNA sequence and homology
The MHBs gene was amplified by PCR from the plasmid
A7 containing the whole fragment of the adr subtype of HBV and the
PCR product was cloned into the pGEM-T vector. Analysis of the PCR
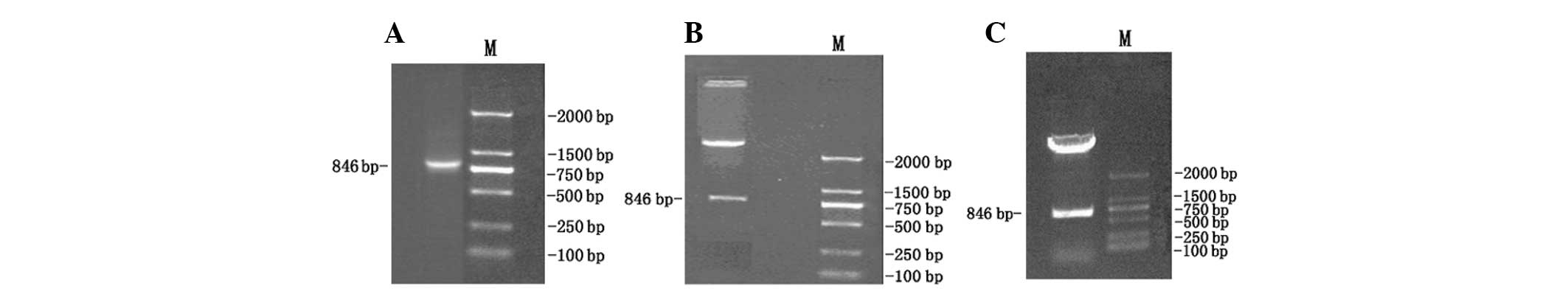
products by agarose gel electrophoresis demonstrated that the clear
bands were the expected size 846 bp of MHB. Following cutting
MHBs-pGEM-T by EcoRI and BamHI restriction enzymes,
MHBs were ligated in-frame into the yeast expression plasmid pGBKT7
EcoRI/BamHI sites using the yeast two-hybrid system
3. Restriction enzyme digestion of the pGBKT7-MHBs plasmid with
EcoRI/BamHI yielded two bands: 7,300 bp empty pGBKT7
and 846 bp MHBs(Fig. 2A, B and C).
The primary structure of the insert was confirmed by direct
sequencing of the pGEM-T-MHBs sequence (Fig. 3).
Screening of the liver cell cDNA library
and identification of the plasmid
The plasmids from the blue colonies containing only
pGBKT7-MHBs were isolated as the bait for screening the human liver
cell cDNA library. Positive clones interact with the MHBs protein
growing on media containing X-α-Gal and lacking leucine,
tryptophan, histidine and adenine (QDO). A total of two positive
colonies were grown on the X-α-gal/QDO medium and expressed (blue
colonies). The two colonies were prescreened by BglII
digestion to ensure that only colonies with different inserts were
subjected to sequencing. One colony was a novel gene with unknown
function and interacted with MHBs in hepatocytes. The protein was
designated MHB-binding protein 1 (MBP1). The MBP1 gene was
amplified by the reverse transcription-polymerase chain reaction
(RT-PCR) technique using HepG2 cDNA as a template and was inserted
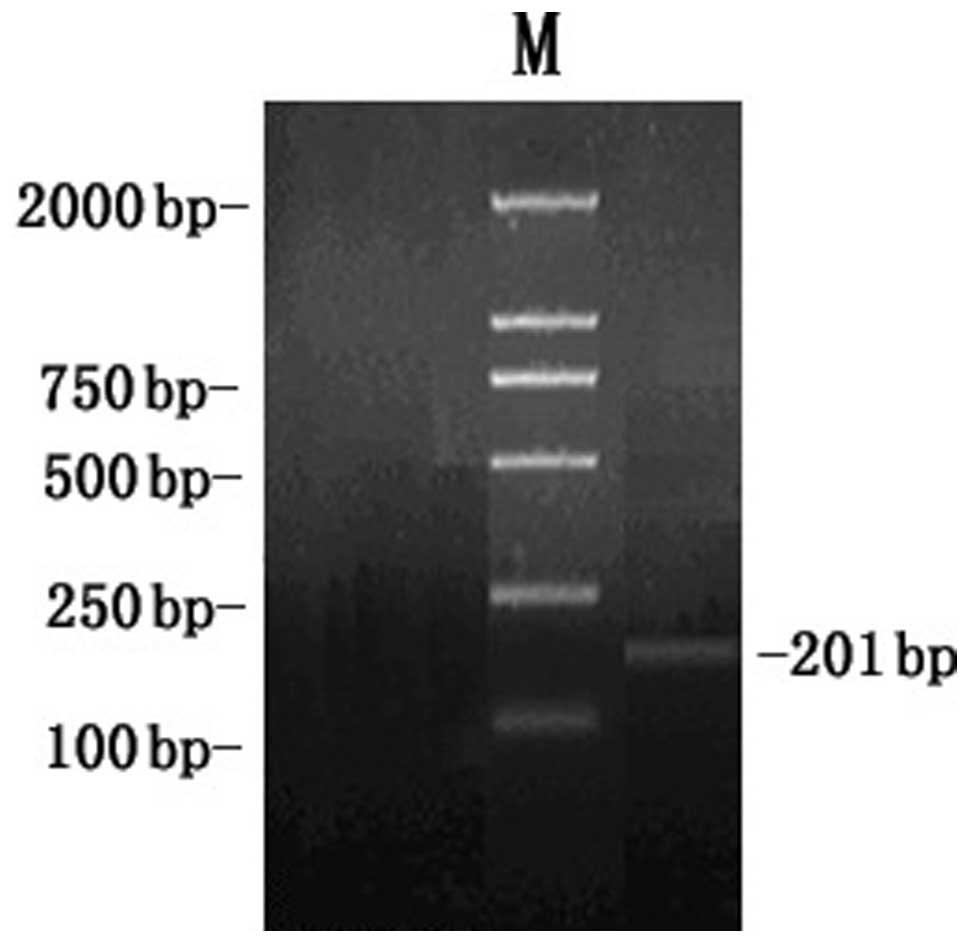
into the pGEM-T vector by TA cloning. In addition, analysis of the
RT-PCR products by agarose gel electrophoresis demonstrated the
clear bands with the expected size 201 bp of MBP1 (Fig. 4).
Analysis of cDNA sequencing and
homology
Two colonies from the hepatocyte cDNA library were
sequenced. Sequence homology searches in GenBank revealed that it
was a novel gene and was designated MBP1. This cDNA was 201 bases
long, which contained an ORF able to encode a protein of 66 amino
acids (Fig. 5). The full-length
sequences accepted by GenBank were obtained using the BLAST program
at the National Center for Biotechnology Information. The GenBank
accession number is DQ307498. A summary of the data is presented in
Table I.
 | Table IComparison between positive clones and
similar sequences in GenBank. |
Table I
Comparison between positive clones and
similar sequences in GenBank.
| High similarity to
known genes | Number of similar
(%) | Homology (%) |
|---|
| Human DNA sequence
from clone RP11-490D19 on chromosome 9 | 1 | 99 |
| Homo sapiens
12 BAC RP11-180M15 (Roswell Park Cancer Institute Human BAC
Library) complete sequence | 1 | 100 |
Discussion
HBV infection is the main factor that induces
hepatocellular carcinoma (HCC) (7). An alternative underlying pathogenic
mechanism of chronic infections and hepatocarcinogenesis may be the
key step to mutual interaction between viral proteins and
hepatocellular proteins, and this action may mediate viral entry
into liver cells and affect the activities and function of these
proteins. Furthermore, the protein from hepatocytes infected with
HBV inversely disturbs viral replication and reduces immunity of
the host, resulting in chronic liver diseases and HCC (8). The length of the nucleotide sequence
of HBV is 3,182 nt and the main serum type is adr in China
(9). The role of MHBs in the
etiology of HCC is not well understood. Screening of hepatocyte
proteins binding with MHBs by the yeast two-hybrid system 3 may aid
in elucidating its role. The present study obtained a total of two
positive colonies grown on the selective
SD/-Trp-Leu-His-Ade/X-α-gal medium, one was the human DNA sequence
from the clone RP11-490D19 on chromosome 9. In total, 15 loci on
chromosome 9p and 17 were analyzed to clarify the involvement of a
loss of heterozygosity (LOH) in HCC in Chinese patients positive
for HBV and/or hepatitis C virus infection. The expression of tumor
suppressor genes, including p53, p16 and the p15 gene was revealed
to be correlated with a deletion of these genes. A high frequency
of LOH was detected on chromosome 9p24 at locus D9S54 (61.8%) and
9p21. No significant association between LOH and HCC
clinico-pathological outcomes was observed. A high frequency of LOH
occurs on chromosomes 9p and 17 in HCC in Chinese patients. Such
sites may contain several putative tumor suppressor genes
critically involved in the development and/or progression of HCC
(10).
The other positive colony was homo sapiens 12
BAC RP11-180M15 (Roswell Park Cancer Institute Human BAC Library,
Buffalo, NY, USA). The end sequences from bacterial artificial
chromosomes (BACs) provide highly specific sequence markers in
large-scale sequencing projects. To date, >300,000 end sequences
have been generated from >186,000 human BAC clones with an
average read length of >460 bp for a total of 141 Mb covering
~4.7% of the genome. Over 60% of the clones have BAC end sequences
(BESs) from the two ends representing >5-fold coverage of the
human genome by the paired-end clones. The quality assessments and
sequence analyses indicate that BESs from human BAC libraries
developed at The California Institute of Technology (CalTech;
Pasadena, CA, USA) and Roswell Park Cancer Institute exhibit
similar properties. The analyses have highlighted differences in
the insert size for different segments of the CalTech library
(11).
The characterization of the interaction pattern of a
protein could provide considerable assistance in the elucidation of
the functions of that protein (12). The interactions between viral
proteins and hepatocellular proteins are important in the
pathogenesis of the virus and may mediate virus entry into
hepatocytes. Yeast two-hybrid 3 is an effective gene analysis
method, which is able to analyze the interactions between protein
and protein, protein and DNA, and protein and RNA in eukaryotic
cells. In addition, it is a novel genetics technique for
investigating the interactions of proteins in physiological
conditions in vivo.
The present study used the yeast two-hybrid system 3
based on the system originally designed by Fields and Song
(13) and took advantage of the
properties of the GAL4 protein of the yeast Saccharomyces
cerevisiae. The GAL4-yeast two-hybrid assay uses two expression
vectors, one uses GAL4-DNA-BD and the other uses GAL4-AD. The
GAL4-DNA-BD fused to a protein ‘X’ and GAL4-AD fused to a protein
‘Y’ to form the bait and the target of the interaction trap,
respectively. A selection of host cells with different reporter
genes and different growth selection markers provide a means to
detect and confirm protein-protein interactions and has
significantly fewer false positives (13–16).
In this way, the bait plasmid pGBKT7-MHBs was
transformed into the AH109 yeast strain. The MHBs gene was
expressed in the yeast cells. Subsequent to the bait plasmid
pGBKT7-MHBsAH109 yeast strain being mated with the liver cDNA
library Y187 yeast strain, the diploid yeast cells were plated on
QDO media containing X-α-gal and two true positive colonies were
obtained. By sequence analysis of isolated library plasmids, two
gene sequences with known functions were obtained; one of them was
termed the Human DNA sequence from clone RP11-490D19 on chromosome
9 (contains the ZNF189 gene for zinc finger protein 189). Notably,
the present study screened Homo sapiens 12 BAC RP11-180M15
(Roswell Park Cancer Institute Human BAC Library) interacting with
MHBs protein from the liver cDNA library.
These interacting proteins screened by yeast
two-hybrid were closely correlated with the occurrence and
development of different types of tumor. Identifying these
interacting proteins may provide novel insights into the biological
functions of the MHBs protein, the pathogenesis of HBV and the
causes of malignancy conversion. Further experiments are required
to elucidate how the interactions between MHBs protein and the
aforementioned interacting proteins affect the occurrence and
development of chronic hepatitis B, hepatic fibrosis and
hepatocarcinoma.
Acknowledgements
The authors would like to thank the Institute of
Infectious Diseases, Ditan Hospital (Beijing, China) for partial
financial and technological assistance in conducting this
study.
Abbreviations:
|
HBV
|
hepatitis B virus
|
|
MHBs
|
middle hepatitis B virus surface
protein
|
|
LHBs
|
large hepatitis B virus surface
protein
|
|
MHBst
|
C-terminally truncated middle size
surface proteins
|
|
ORF
|
open reading frame
|
|
ER
|
endoplasmic reticulum
|
|
PCR
|
polymerase chain reaction
|
|
DNA-BD
|
DNA binding domain
|
|
DNA-AD
|
DNA activation domain
|
|
MBP1
|
MHB-binding protein 1
|
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
|
HCC
|
hepatocellular carcinoma
|
|
QDO
|
quadruple dropout medium lacking
leucine, tryptophan, histidine and adenine
|
References
|
1
|
Murakami S: Hepatitis B virus X protein:
structure, function and biology. Intervirology. 42:81–99. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kekulé AS, Lauer U, Meyer M, Caselmann WH,
Hofschneider PH and Koshy R: The preS2/S region of integrated
hepatitis B virus DNA encodes a transcriptional transactivator.
Nature. 343:457–461. 1990.PubMed/NCBI
|
|
3
|
Hildt E, Saher G, Bruss V and Hofschneider
PH: The hepatitis B virus large surface protein (LHBs) is a
transcriptional activator. Virology. 225:235–239. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hildt E, Urban S and Hofschneider PH:
Characterization of essential domains for the functionality of the
MHBst transcriptional activator and identification of a minimal
MHBst activator. Oncogene. 11:2055–2066. 1995.PubMed/NCBI
|
|
5
|
Bruss V, Lu X, Thomssen R and Gerlich WH:
Post-translational alterations in transmembrane topology of the
hepatitis B virus large envelope protein. EMBO J. 13:2273–2279.
1994.PubMed/NCBI
|
|
6
|
Matsumoto M, Hsieh TY, Zhu N, VanArsdale
T, Hwang SB, Jeng KS, Gorbalenya AE, Lo SY, Ou JH, Ware CF and Lai
MM: Hepatitis C virus core protein interacts with the cytoplasmic
tail of lymphotoxin-beta receptor. J Virol. 71:1301–1309.
1997.PubMed/NCBI
|
|
7
|
Mahoney FJ: Update on diagnosis,
management, and prevention of hepatitis B virus infection. Clin
Microbiol Rev. 12:351–366. 1999.PubMed/NCBI
|
|
8
|
Dong Z and Cheng J: Study on definition of
pre-X region in hepatitis B virus genome. Shijie Huaren Xiaohua
Zazhi. 8:1097–1101. 2003.
|
|
9
|
Tong S, Kim KH, Chante C, Wands J and Li
J: Hepatitis B virus e antigen variants. Int J Med Sci. 2:2–7.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shao J, Li Y, Li H, Wu Q, Hou J and Liew
C: Deletion of chromosomes 9p and 17 associated with abnormal
expression of p53, p16/MTS1 and p15/MTS2 gene protein in
hepatocellular carcinomas. Chin Med J (Engl). 113:817–822.
2000.PubMed/NCBI
|
|
11
|
Zhao S, Malek J, Mahairas G, Fu L, Nierman
W, Venter JC and Adams MD: Human BAC ends quality assessment and
sequence analyses. Genomics. 63:321–332. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Emmert-Buck MR, Gillespie JW, Pawletz CP,
Ornstein DK, Basrur V, Appella E, Wang QH, Huang J, Hu N, Taylor P
and Petricoin EE III: An approach to proteomic analysis of human
tumors. Mol Carcinog. 27:158–165. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fields S and Song O: A novel genetic
system to detect protein-protein interactions. Nature. 340:245–246.
1989. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Osman A: Yeast two-hybrid assay for
studying protein-protein interactions. Methods Mol Biol.
270:403–422. 2004.PubMed/NCBI
|
|
15
|
Gietz RD and Woods RA: Screening for
protein-protein interactions in the yeast two-hybrid system.
Methods Mol Biol. 185:471–486. 2002.PubMed/NCBI
|
|
16
|
Zhen Z: Progress in proteomics. Sheng Wu
Gong Cheng Xue Bao. 17:491–493. 2001.(In Chinese).
|