Introduction
Renal cell carcinoma (RCC) is a kidney cancer that
originates from the proximal convoluted tubule. It represents the
leading cause of mortality among urological malignancies (1,2). It
is the 10th most common type of cancer in Europe (3). In the United States, there are
~65,000 novel cases and almost 14,000 mortalities from RCC annually
(4). RCC includes several
different histological subtypes that possess distinct biological
behaviors and prognoses. The most common type of RCC is clear cell
RCC (ccRCC), which originates in the lining of the proximal renal
tubule, representing >75–80% of all cases of RCC (5). Approximately 30% of patients have
metastatic disease when diagnosed with RCC and radical nephrectomy
remains the main treatment for RCC patients due to resistance to
radiation and chemotherapy (6).
Although the overall survival rate is >60% over 5 years, ~30% of
patients with a diagnosis of localized RCC develop metastatic
recurrence (7,8). Patients with metastatic RCC face a
poor prognosis and have limited therapeutic options. The median
survival rate in a recent cohort was only 1.5 years with <10% of
patients surviving to 5 years (9).
The histological grade combined with clinical stage, which is
considered to be the gold standard method for the prediction of
patient prognosis, is not accurate when used alone (10). Thus, molecular markers and novel
treatments are required in order to improve the prognosis for
patients with RCC.
MicroRNAs (miRNAs) are a class of naturally
occurring, endogenous small non-coding RNA, in the size range of
19–25 nt. miRNAs regulate gene expression at the
post-transcriptional level by binding through partial sequence
homology, to the 3′ untranslated region (3′UTR) of mammalian target
mRNAs and causing translational inhibition and/or mRNA degradation
(11). Since they were initially
described almost 20 years ago, there has been a steady increase in
their identification and the latest Release 20 of miRBase has
24,521 entries of miRNAs from various species, including 734 mature
miRNAs from Gallus gallus (12). It has been predicted that as many
as 30% of protein-encoding genes may be regulated by miRNAs and
they may function as oncogenes and tumor suppressors (13). Upregulated miRNAs in cancer may
function as oncogenes by negatively regulating tumor suppressors.
By contrast, downregulated miRNAs may normally function as tumor
suppressor genes and inhibit cancer by regulating oncogenes
(14,15). It has demonstrated the important
roles of miRNAs in regulating various cellular functions, including
cell apoptosis, cell proliferation, neural development and stem
cell differentiation (16,17). Recently, miRNAs have also been
revealed to be important in tumorigenesis, cancer invasion and
metastasis (18).
miR-145 has been reported to be frequently
downregulated in various types of cancer, including renal,
prostate, bladder, lung and colon cancer, as well as B-cell
malignancies. However, the functions of miR-145 are yet to be
investigated in RCC. The present study focused on the functions and
direct target of miR-145 in RCC.
Materials and methods
Cells and culture conditions
The human ccRCC-derived cell lines 786-O and A498
were obtained from the Shanghai Institute of Cell Biology, Chinese
Academy of Sciences (Shanghai, China). The cells were incubated in
RPMI-1640 (HyClone Laboratories, Inc., Logan, UT, USA) or
Dulbecco’s modified Eagle’s medium (DMEM; Gibco-BRL, Carlsbad, CA,
USA) supplemented with 10% heat-inactivated fetal calf serum, 100
U/ml penicillin and 100 mg/l streptomycin at 37°C in a humidified
atmosphere containing 5% CO2.
Transfection of miR-145 mimics, scrambled
control (NC) and luciferase reporter plasmid
Mature miR-145 mimics, the NC and the luciferase
reporter plasmid were designed and synthesized by GenePharma
(Shanghai, China). The sequence of miR-145 mimics was
5′-GUCCAGUUUUCC CAGGAAUCCCU-3′. The sequence of NC mimics was
5′-UUCUCCGAACGUGUCACGUTT-3′. The insertion fragment was confirmed
by DNA sequencing. Cell transfection and cotransfection were
performed using Lipofectamine 2000 (Invitrogen Life Technologies)
according to the manufacturer’s instructions.
Cell viability assay
The cell proliferation was determined by the MTT
assay. The cells transfected with miR-145 mimics or the NC were
seeded in 96-well plates at a density of 3,000 cells per well. Cell
proliferation was documented every 24 h for five days according to
the manufacturer’s instructions. Briefly, MTT solution was added
into each well and incubated at 37°C for 4 h. The plates were spun
(200 × g for 10 min), and the purple colored precipitates of
formazan were dissolved in 200 μl dimethylsulfoxide. Absorbance
(optical density, OD) was measured at 490 nm using an automatic
multi-well spectrophotometer (Bio-Rad, Richmond, CA, USA). There
were six replicate wells for every time point in each group. The
suppression rate was calculated using the formula: Suppression rate
= (1 - ODmiR-145 / ODmiR-NC) × 100%. All the
experiments were performed in triplicate.
Cell migration and invasion assay
Cell motility was measured using 8 μm-pore
polycarbonate membrane Boyden chambers inserted in a transwell
apparatus (Costar, Cambridge, MA, USA). The transfected cells
(miR-145 mimics and NC) growing in the log phase were treated with
trypsin/EDTA solution, washed once with serum-containing RPMI-1640
medium, centrifuged (200 × g for 10 mins), and re-suspended as
single-cell solutions. A total of 1×105 cells in 0.2 ml
serum-free RPMI-1640 medium were seeded on a transwell apparatus
(Costar). RPMI-1640 (600 μl) containing 20% fetal bovine serum was
added to the lower chamber. An invasion assay was performed by the
same procedure except that the filters of the transwell chambers
were coated with 30 μg Matrigel (BD Biosciences, San Jose, CA,
USA). After the cells were incubated for 12–24 h at 37°C in a 5%
CO2 incubator, the cells on the top surface of the
insert were removed by wiping with a cotton swab. The cells that
migrated to the bottom surface of the insert were fixed in 100%
methanol for 2 min, stained in 0.5% crystal violet for 2 min,
rinsed in phosphate-buffered saline and then subjected to
microscopic inspection (magnification, ×200; BX51WI-DPMC; Olympus,
Tokyo, Japan). The values for invasion and migration were obtained
by counting five fields per membrane and represent the average of
three independent experiments.
Western blot analysis
The primary antibodies used in the present study,
including anti-MMP-11 and anti-β-actin were products of Bioworld
Technology (St. Louis Park, MN, USA). The total protein of cells
was prepared using RIPA lysis buffer. The protein concentration in
the resulting lysate was determined using the bicinchoninic acid
protein assay (Beyotime Institute of Biotechnology, Shanghai,
China). Equal quantities of protein were loaded onto SDS-PAGE and
transferred onto polyvinylidene difluoride membranes. Following
inhibition with 5% degreased milk in Tris-buffered saline with
Tween-20 (TBST; containing 0.1% Tween-20), the membranes were
incubated overnight with the appropriate primary antibody. The
membranes were then washed and incubated with the corresponding
horseradish peroxidase-conjugated secondary antibody (goat
anti-rabbit) at 1:1,000 dilution in TBST. The blot was developed
with enhanced chemiluminescence solution (Pierce Biotechnology,
Inc., Rockford, IL, USA) and images were captured by a FluorChem
imaging system (Alpha Innotech, San Leandro, CA, USA). The
intensity of each spot was read and analyzed using AlphaEaseFC
software (Alpha Innotech, San Leandro, CA, USA). β-actin was used
as a loading control.
Luciferase assay
The cells were plated in a 12-well plate at ~90%
confluence and transfected with 0.5 μg reporter plasmid, 40 nmol
miR-145 mimics or their negative control by Lipofectamine 2000.
Each sample was also cotransfected with 0.05 μg pRL-CMV plasmid
expressing Renilla luciferase (Promega Corporation, Madison,
WI, USA) as an internal control for transfection efficiency. The
cells were harvested with passive lysis buffer, a component of the
Dual-Luciferase Reporter Assay system (Tecan, Theale, UK), 48 h
after transfection according to the manufacturer’s instructions. An
appropriate volume of cell lysate was added to a well of the F96
MicroWell Plates, followed by 25 μl Luciferase Assay Reagent II
(Tecan). Firefly luciferase activities and Renilla
luciferase activities were measured using a luminometer (Tecan).
Firefly luciferase activity was normalized to Renilla
luciferase activity for each transfected well. Each assay was
replicated three times.
Statistical analysis
Data are presented as the mean ± standard deviation
and compared using Student’s t-test in Stata 10.0 (College Station,
TX, USA). Double-tailed P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-145 suppresses cell proliferation in
RCC cell lines
To measure the effect of miR-145 on cell
proliferation, an MTT assay was used. As shown in Fig. 1, the upregulation of miR-145
significantly inhibited cell proliferation. MTT assays revealed
that following 144 h of treatment, the suppression rate of miR-145
reached 35.81%±4.1% in 786-O cells and 41.15±3.5% in A498 cells.
These results indicated that miR-145 may be important in 786-O and
A498 cells.
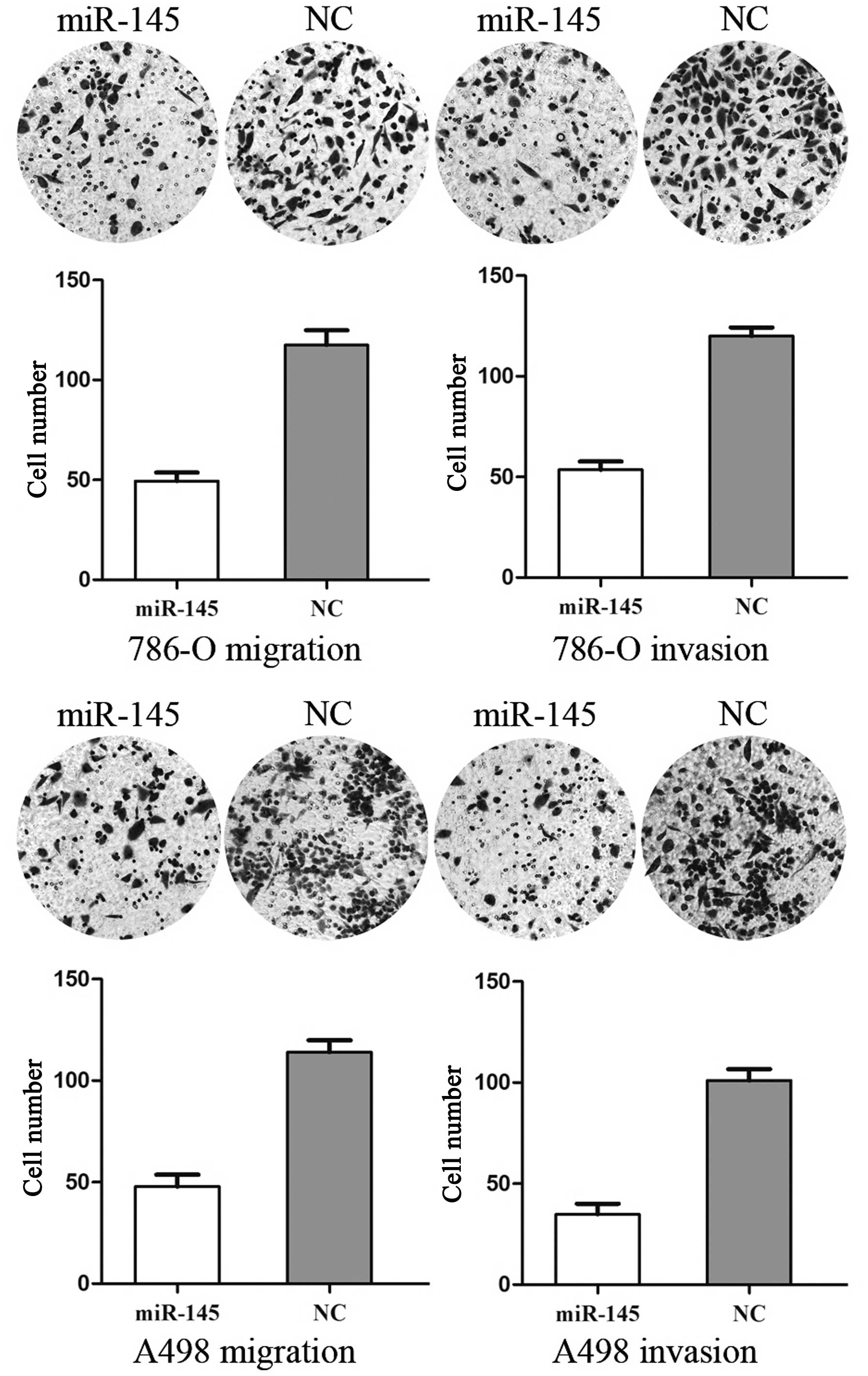
miR-145 inhibits cell migration and
invasion in RCC cell lines
To measure the effect of miR-145 on tumor cell
migration and invasion, the transwell apparatus assay was used
(Fig. 2). The transfected cells
(miR-145 mimics and NC mimics) growing in the log phase were
collected and cultured on transwell apparatus. Following 12 h
incubation, cell migration was significantly decreased in the
miR-145 groups compared with the control group (P<0.05). Using
transwell apparatus pre-coated with Matrigel, the effects of
miR-145 on cell invasiveness were examined. Following 24 h
incubation, miR-145 transfected cells demonstrated significantly
decreased invasiveness compared with the control cells (P<0.05).
These results indicated that miR-145 inhibited cell migration and
invasion in RCC cell lines.
miR-145 suppresses the expression of
MMP-11 in RCC cell lines
Sachdeva et al revealed that miR-145 was able
to target multiple metastasis related genes, including MMP-11 and
ADAM-17 (19). Western blot
analysis was performed to examine whether the protein level of
MMP-11 was decreased following the transfection of miR-145. As
shown in Fig. 3, MMP-11 was
significantly decreased in 786-O and A498 cell lines 72 h after
transfection of miR-145. Thus, miR-145 reduces the protein level of
MMP-11 in RCC cells.
MMP-11 is a direct target of miR-145
Luciferase reporter assays were performed to
evaluate whether the site was able to directly mediate the
inhibition of expression. As shown in Fig. 4, the overexpression of miR-145 was
able to suppress MMP-11 3′UTR-luciferase activity by 57% in 786-O
cells and 45% in A498 cells (P<0.05). Therefore, MMP-11 may be a
direct target of miR-145 in vitro.
Discussion
miR-145 was initially identified in mice from heart
tissues using small RNA cloning techniques and then later reported
in humans, revealing a unique seed sequence that is conserved in
Xenopus and mammals (20).
Human miR-145 (hsa-miR-145) is enriched in germline and
mesoderm-derived tissues, including in the uterus, ovary, testis,
prostate, spleen and heart (21).
It is located on chromosome 5 (5q32–33) within a 4.09 kb region.
The first study regarding the downregulation of miR-145 in various
types of tumor was conducted by Michael et al (22). The authors demonstrated that the
total number of clones sequenced for miR-145 was two from patients
with colon adenocarcinomas compared with eight from normal tissue
using the small RNA cloning approach. The results were confirmed by
northern blot analysis. Notably, the authors also found a decreased
level of miR-145 in precancerous adenomatous polyps, suggesting a
possible role in tumor initiation.
miR-145 has been reported to be frequently
downregulated in various types of cancer, including renal,
prostate, bladder, lung and colon cancer, as well as B-cell
malignancies (23–27). The identification of miR-145 target
genes is critical for understanding its role in tumorigenesis and
is important for defining novel therapeutic targets. miR-145
suppresses tumor cell growth by targeting insulin receptor
substrate-1 (28), c-Myc (29) and several other genes associated
with carcinogenesis (19). miR-145
impacts migration, invasion and metastasis by targeting Fli 1
(30) and mucin 1 (31), respectively, and also affects
p53-mediated cell cycle arrest by targeting p21 (32). In addition, the downregulation of
miR-145 is associated with an aggressive phenotype and poor
prognosis in prostate cancer (33,34).
Therefore, upregulating miR-145 or providing analogous
pharmaceutical compounds exogenously, may be effective cancer
therapies for several types of tumor resulting from the activation
or overexpression of these oncogenes. The present study revealed
that miR-145 reduced cell migration and invasion by downregulating
the expression of MMP-11. The results suggested that miR-145 may be
used for the development of novel molecular markers and therapeutic
approaches to inhibit the metastasis of RCC.
Metastasis is a multi-step process that requires
cancer cells to detach from the main tumor, migrate and invade
through the stroma, intravasate, survive in the circulatory system,
arrive at a secondary site and extravasate, invade and grow at the
secondary site (35). MMPs have
been previously shown to contribute to migration and invasion in
various types of cancer. They are a family of zinc-dependent
extracellular endoproteinases that are collectively capable of
degrading essentially all components of the extracellular matrix
and basement membrane (36). They
regulate and shape the tumor microenvironment, and are synthesized
and secreted by multiple cell types, including corneal epithelial
cells and fibroblasts (37). To
date, at least 24 different human MMPs have been identified and
they can be classified into five groups on the basis of substrate
specificity, including interstitial collagenases, gelatinases,
stromelysins, matrilysins and membrane-type MMPs (38,39).
MMPs are found in normal and pathological tissues in
which matrix remodeling is involved, including embryonic
development, wound healing, arthritis and angiogenesis, as well as
tumor invasion and metastasis (40). Therefore, elevated levels of MMPs
have been detected in the serum and urine of patients with several
different types of cancer, including cancer of the bladder, breast,
lung, colon, head and neck as well as melanoma (41). These MMPs are secreted as inactive
zymogens (pro-MMPs) requiring extracellular activation and their
activity is tightly regulated by specific tissue inhibitors
(42). In view of the important
role in tumor invasion and metastasis, inhibitors of MMP activity
have been investigated as a method of preventing/decreasing tumor
spread.
MMP-11, also termed stromelysin-3, is encoded by the
MMP11 gene located on chromosome 22 q11.23 (43). It was originally identified by
screening a breast cancer cDNA library for genes that were
expressed at higher levels in invasive carcinomas compared with
breast fibroadenomas (44).
Additional investigation has demonstrated that MMP-11 is usually
overexpressed in numerous types of human carcinoma, including
breast, non-small cell lung and colorectal carcinomas, however, is
rarely expressed in normal tissue, including the normal tissue
surrounding the tumor. Notably, only adults have been shown to
express MMP-11 in tumors and regenerating or healing tissues
(45–47). The functions of MMP-11 in cancer
progression have been demonstrated by several preclinical
observations. The upregulated expression of MMP-11 enhances tumor
incidence in mice (48), homing of
malignant epithelial cells (49),
cancer progression by remodeling extracellular matrix (50) and has antiapoptotic and
antinecrotic effects on tumor cells (51,52).
MMP-11 deficiency increases tumor-free survival rate and modulates
local or distant invasion (53).
The knockdown of MMP-11 mRNA in gastric cancer cells suppresses
tumor growth in vitro and in vivo and inhibits the
spread of murine hepatocarcinoma cells to lymph nodes (53). The levels of MMP-11 expression may
serve as a marker for transformation and invasion in several types
of cancer, otherwise, it may be a target for cancer therapy in
order to inhibit metastasis. The results from the present study
suggested that miR-145 suppressed the migration and invasion of RCC
cells through the downregulation of MMP-11. It may be investigated
as a predictive marker for the early detection of tumor metastasis
and for targeting therapeutic drugs to inhibit the invasion of
RCC.
In conclusion, to the best of our knowledge this is
the first study to demonstrate that miR-145 inhibited RCC cell
migration and invasion by downregulating the expression of MMP-11.
These findings have therapeutic implications and may be exploited
for further treatment of RCC.
Future investigation is required to address whether
the potential of miR-145 may be fully realized in cancer treatment.
If so, it may be beneficial for the treatment of RCC.
References
|
1
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: the impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar
|
|
5
|
Yan BC, Mackinnon AC and Al-Ahmadie HA:
Recent developments in the pathology of renal tumors: morphology
and molecular characteristics of select entities. Arch Pathol Lab
Med. 133:1026–1032. 2009.PubMed/NCBI
|
|
6
|
Yu ZH, Zhang Q, Wang YD, Chen J, Jiang ZM,
Shi M, Guo X, Qin J, Cui GH, Cai ZM, et al: Overexpression of
cyclooxygenase-1 correlates with poor prognosis in renal cell
carcinoma. Asian Pac J Cancer Prev. 14:3729–3734. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar
|
|
8
|
Zisman A, Pantuck AJ, Wieder J, Chao DH,
Dorey F, Said JW, deKernion JB, Figlin RA and Belldegrun AS: Risk
group assessment and clinical outcome algorithm to predict the
natural history of patients with surgically resected renal cell
carcinoma. J Clin Oncol. 20:4559–4566. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xue YJ, Xiao RH, Long DZ, Zou XF, Wang XN,
Zhang GX, Yuan YH, Wu GQ, Yang J, Wu YT, et al: Overexpression of
FoxM1 is associated with tumor progression in patients with clear
cell renal cell carcinoma. J Transl Med. 10:2002012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Choi YD, Kim KS, Ryu S, Park Y, Cho NH,
Rha SH, Jang JJ, Ro JY, Juhng SW and Choi C: Claudin-7 is highly
expressed in chromophobe renal cell carcinoma and renal oncocytoma.
J Korean Med Sci. 22:305–310. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Garzon R, Pichiorri F, Palumbo T,
Visentini M, Aqeilan R, Cimmino A, Wang H, Sun H, Volinia S, Alder
H, et al: MicroRNA gene expression during retinoic acid-induced
differentiation of human acute promyelocytic leukemia. Oncogene.
26:4148–4157. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yao Y, Charlesworth J, Nair V and Watson
M: MicroRNA expression profiles in avian haemopoietic cells. Front
Genet. 4:1532013.PubMed/NCBI
|
|
13
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
15
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yoo AS, Staahl BT, Chen L and Crabtree GR:
MicroRNA-mediated switching of chromatin-remodelling complexes in
neural development. Nature. 460:642–646. 2009.PubMed/NCBI
|
|
17
|
le Sage C, Nagel R, Egan DA, Schrier M,
Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA, et
al: Regulation of the p27(Kip1) tumor suppressor by miR-221 and
miR-222 promotes cancer cell proliferation. EMBO J. 26:3699–3708.
2007.PubMed/NCBI
|
|
18
|
Zhang H, Li Y and Lai M: The microRNA
network and tumor metastasis. Oncogene. 29:937–948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sachdeva M and Mo YY: miR-145-mediated
suppression of cell growth, invasion and metastasis. Am J Transl
Res. 2:170–180. 2010.PubMed/NCBI
|
|
20
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lakshmipathy U, Love B, Adams C,
Thyagarajan B and Chesnut JD: Micro RNA profiling: an easy and
rapid method to screen and characterize stem cell populations.
Methods Mol Biol. 407:97–114. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Michael MZ, O’ Connor SM, van Holst
Pellekaan NG, Young GP and James RJ: Reduced accumulation of
specific microRNAs in colorectal neoplasia. Mol Cancer Res.
1:882–891. 2003.PubMed/NCBI
|
|
23
|
Doberstein K, Steinmeyer N, Hartmetz AK,
Eberhardt W, Mittelbronn M, Harter PN, Juengel E, Blaheta R,
Pfeilschifter J and Gutwein P: MicroRNA-145 targets the
metalloprotease ADAM17 and is suppressed in renal cell carcinoma
patients. Neoplasia. 15:218–230. 2013.PubMed/NCBI
|
|
24
|
Ozen M, Creighton CJ, Ozdemir M and
Ittmann M: Widespread deregulation of microRNA expression in human
prostate cancer. Oncogene. 27:1788–1793. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ichimi T, Enokida H, Okuno Y, Kunimoto R,
Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama
K, et al: Identification of novel microRNA targets based on
microRNA signatures in bladder cancer. Int J Cancer. 125:345–352.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Akao Y, Nakagawa Y and Naoe T:
MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 26:311–320.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Akao Y, Nakagawa Y, Kitade Y, Kinoshita T
and Naoe T: Downregulation of microRNAs-143 and -145 in B-cell
malignancies. Cancer Sci. 98:1914–1920. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi B, Sepp-Lorenzino L, Prisco M, Linsley
P, deAngelis T and Baserga R: Micro RNA 145 targets the insulin
receptor substrate-1 and inhibits the growth of colon cancer cells.
J Biol Chem. 282:32582–32590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sachdeva M, Zhu S, Wu F, Wu H, Walia V,
Kumar S, Elble R, Watabe K and Mo YY: p53 represses c-Myc through
induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA.
106:3207–3212. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Larsson E, Fredlund Fuchs P, Heldin J,
Barkefors I, Bondjers C, Genové G, Arrondel C, Gerwins P, Kurschat
C, Schermer B, et al: Discovery of microvascular miRNAs using
public gene expression data: miR-145 is expressed in pericytes and
is a regulator of Fli1. Genome Med. 1:1082009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sachdeva M and Mo YY: MicroRNA-145
suppresses cell invasion and metastasis by directly targeting mucin
1. Cancer Res. 70:378–387. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Seoane J, Le HV and Massagué J: Myc
suppression of the p21(Cip1) Cdk inhibitor influences the outcome
of the p53 response to DNA damage. Nature. 419:729–734. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang L, Tang H, Thayanithy V, Subramanian
S, Oberg AL, Cunningham JM, Cerhan JR, Steer CJ and Thibodeau SN:
Gene networks and microRNAs implicated in aggressive prostate
cancer. Cancer Res. 69:9490–9497. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen X, Gong J, Zeng H, Chen N, Huang R,
Huang Y, Nie L, Xu M, Xia J, Zhao F, et al: MicroRNA145 targets
BNIP3 and suppresses prostate cancer progression. Cancer Res.
70:2728–2738. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kwon YJ, Hurst DR, Steg AD, Yuan K, Vaidya
KS, Welch DR and Frost AR: Gli1 enhances migration and invasion via
up-regulation of MMP-11 and promotes metastasis in ERalpha negative
breast cancer cell lines. Clin Exp Metastasis. 28:437–449. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yan D, Dai H and Liu JW: Serum levels of
MMP-11 correlate with clinical outcome in Chinese patients with
advanced gastric adenocarcinoma. BMC Cancer. 11:1512011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li DQ, Shang TY, Kim HS, Solomon A,
Lokeshwar BL and Pflugfelder SC: Regulated expression of
collagenases MMP-1, -8, and -13 and stromelysins MMP-3, -10, and
-11 by human corneal epithelial cells. Invest Ophthalmol Vis Sci.
44:2928–2936. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kähäri VM and Saarialho-Kere U: Matrix
metalloproteinases and their inhibitors in tumour growth and
invasion. Ann Med. 31:34–45. 1999.PubMed/NCBI
|
|
39
|
Clark AF: New discoveries on the roles of
matrix metalloproteinases in ocular cell biology and pathology.
Invest Ophthalmol Vis Sci. 39:2514–2516. 1998.PubMed/NCBI
|
|
40
|
Wu D, Ding J, Wang L, Pan H, Zhou Z, Zhou
J and Qu P: microRNA-125b inhibits cell migration and invasion by
targeting matrix metallopeptidase 13 in bladder cancer. Oncol Lett.
5:829–834. 2013.PubMed/NCBI
|
|
41
|
Moses MA, Wiederschain D, Loughlin KR,
Zurakowski D, Lamb CC and Freeman MR: Increased incidence of matrix
metalloproteinases in urine of cancer patients. Cancer Res.
58:1395–1399. 1998.PubMed/NCBI
|
|
42
|
Egeblad M and Werb Z: New functions for
the matrix metalloproteinases in cancer progression. Nat Rev
Cancer. 2:161–174. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
43
|
Valdivia A, Peralta R, Matute-González M,
Garcia Cebada JM, Casasola I, Jiménez-Medrano C, Aguado-Pérez R,
Villegas V, Gonzalez-Bonilla C, Manuel-Apolinar L, et al:
Co-expression of metalloproteinases 11 and 12 in cervical scrapes
cells from cervical precursor lesions. Int J Clin Exp Pathol.
4:674–682. 2011.PubMed/NCBI
|
|
44
|
Pan W, Arnone M, Kendall M, Grafstrom RH,
Seitz SP, Wasserman ZR and Albright CF: Identification of peptide
substrates for human MMP-11 (stromelysin-3) using phage display. J
Biol Chem. 278:27820–27827. 2003. View Article : Google Scholar
|
|
45
|
Rouyer N, Wolf C, Chenard MP, Rio MC,
Chambon P, Bellocq JP and Basset P: Stromelysin-3 gene expression
in human cancer: an overview. Invasion Metastasis. 14:269–275.
1994.PubMed/NCBI
|
|
46
|
Kossakowska AE, Huchcroft SA, Urbanski SJ
and Edwards DR: Comparative analysis of the expression patterns of
metalloproteinases and their inhibitors in breast neoplasia,
sporadic colorectal neoplasia, pulmonary carcinomas and malignant
non-Hodgkin’s lymphomas in humans. Br J Cancer. 73:1401–1408.
1996.PubMed/NCBI
|
|
47
|
Têtu B, Brisson J, Lapointe H and Bernard
P: Prognostic significance of stromelysin 3, gelatinase A, and
urokinase expression in breast cancer. Hum Pathol. 29:979–985.
1998.PubMed/NCBI
|
|
48
|
Nöel AC, Lefebvre O, Maquoi E, VanHoorde
L, Chenard MP, Mareel M, Foidart JM, Basset P and Rio MC:
Stromelysin-3 expression promotes tumor take in nude mice. J Clin
Invest. 97:1924–1930. 1996.PubMed/NCBI
|
|
49
|
Masson R, Lefebvre O, Noël A, Fahime ME,
Chenard MP, Wendling C, Kebers F, LeMeur M, Dierich A, Foidart JM,
et al: In vivo evidence that the stromelysin-3 metalloproteinase
contributes in a paracrine manner to epithelial cell malignancy. J
Cell Biol. 140:1535–1541. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Noël A, Boulay A, Kebers F, Kannan R,
Hajitou A, Calberg-Bacq CM, Basset P, Rio MC and Foidart JM:
Demonstration in vivo that stromelysin-3 functions through its
proteolytic activity. Oncogene. 19:1605–1612. 2000.PubMed/NCBI
|
|
51
|
Boulay A, Masson R, Chenard MP, El Fahime
M, Cassard L, Bellocq JP, Sautès-Fridman C, Basset P and Rio MC:
High cancer cell death in syngeneic tumors developed in host mice
deficient for the stromelysin-3 matrix metalloproteinase. Cancer
Res. 61:2189–2193. 2001.PubMed/NCBI
|
|
52
|
Wu E, Mari BP, Wang F, Anderson IC, Sunday
ME and Shipp MA: Stromelysin-3 suppresses tumor cell apoptosis in a
murine model. J Cell Biochem. 82:549–555. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Peruzzi D, Mori F, Conforti A, Lazzaro D,
De Rinaldis E, Ciliberto G, La Monica N and Aurisicchio L: MMP11: a
novel target antigen for cancer immunotherapy. Clin Cancer Res.
15:4104–4113. 2009. View Article : Google Scholar : PubMed/NCBI
|