Introduction
Malignant gliomas, the most common primary brain
malignant tumor, are aggressive, highly invasive and neurologically
destructive types of cancer. Glioblastoma multiforme (GBM), the
most aggressive manifestation of all gliomas, typically affects
adults between 45–60 years of age. Despite therapeutic advances in
surgical techniques, radiotherapy and chemotherapy, the prognosis
of patients with GBM remains discouraging (1–4),
largely due to recurrences as a result of tumor growth into
adjacent brain regions.
Establishing the molecular basis of tumorigenesis
and the progression of malignant gliomas is crucial to improve
current therapies and develop novel treatment strategies. It has
been suggested that gene expression profiles from glioma specimens
may predict patient outcome more accurately than pathological
criteria (5,6). One potential candidate gene for
therapeutic targeting in glioma is the gene encoding forkhead box
protein P1 (FOXP1), a transcription factor that is widely expressed
and important in the development of various human tissues (7–11).
Of note, FOXP1 has been suggested to be both a tumor suppressor
candidate and potential oncogene, due to its differential
expression levels in distinctive types of tumors. Several authors
have suggested that FOXP1 is an oncogene with high expression
levels and the protein overexpression is often identified in many
types of B-cell lymphomas associated with a poor outcome (12–15).
By contrast, loss of FoxP1 has been observed in endometrial
(16), prostate (17,18)
and renal cell carcinoma (19),
and the loss of FOXP1 in breast cancer has been associated with
lower survival rates (17,20).
To the best of our knowledge, associations between
FoxP1 expression and the clinical features of gliomas, to determine
its clinicopathological significance, have not been
investigated. Therefore, the present study investigated the role of
FOXP1 in patients with gliomas and whether it may be a potential
target for novel therapeutic strategies. FOXP1 gene and protein
expression in glioma samples were investigated and compared with
those in tissue adjacent to the tumors. Then, an exogenous
expression vector was transfected into the glioma cell line U251,
in an attempt to elucidate whether its overexpression alters the
biological characteristics of glioma cells.
Materials and methods
Patients
The patient population consisted of 25 adults (16
male, 9 female; mean age at sampling, 54.3 years). Written informed
consent was obtained from all patients, and the study was approved
by and performed according to the guidelines of the Institutional
Review Board of the General Hospital of Tianjin Medical University
(Tianjin, China; approval no. 85–188). GBM was verified in the
histological specimens between July 2008 and July 2012 by a
neuropathologist according to the World Health Organization
criteria. All 25 cases were classified as grade 4, with 18 cases of
GBM and 7 cases of glioblastoma with oligodendroglioma.
Region-specific specimen collection
Deep-seated tumors were removed using an
intraoperative navigation system (Brainlab, Feldkirchen, Germany)
that minimized invasiveness and maximized patient safety and
accurate tumor resection. Brain tissue samples were collected from
the resection zone, categorized as peripheral normal brain, tumor
marginal tissue or tumor core, and stored in liquid nitrogen as
described previously (21).
Quantitative polymerase chain reaction
(qPCR)
Fresh-frozen tumor and peritumoral tissue samples
were collected from 25 patients with glioma at our hospital. Total
RNA from a subset of these fresh frozen tissue samples was
extracted using TRIzol reagent (Invitrogen Life Technologies,
Carlsbad, CA, USA). Total RNA extraction, quality control and
1-step qPCR were performed as previously described (22,23).
FOXP1-specific oligonucleotide primers: forward,
5′-TGCAGAGCAGCCACGCCTAC-3′ and reverse,
5′-CCGTTCAGCTCTTCCCGTATT-3′, were designed to give a 148-base pair
(bp) PCR product. The levels of GAPDH mRNA were used to standardize
the mRNA data, using GAPDH primers obtained from Invitrogen Life
Technologies, and the melting curves were analyzed. Amplification
conditions consisted of 30 min at 42°C for reverse transcription
and 2 min at 94°C for Taq activation, followed by 35 cycles at 94°C
for 20 sec, 58°C for 20 sec and elongation at 72°C for 30 sec.
Expression of FOXP1 protein in tissue
samples
Western blot analysis was used to determine protein
levels in the cancerous tissues. Briefly, frozen tissues were
homogenated with lysis buffer, centrifuged at 4°C for 30 min (9,000
× g). The supernatant was collected and the bicinchoninic acid
assay was used to determine protein concentration. A 10%
polyacrylamide gel was prepared to load protein samples and 5%
non-fat dry milk was added to block the non-specific antigen. The
primary antibodies (Rabbit anti-human ATP binding cassette E1
(ABCE1) polyclonal antibody; Abcam, Cambridge, MA, USA) and the
secondary antibody HRP-conjugated goat anti-rabbit IgG (H&L)
antibody (Cell Signaling Technology, Beverly, MA, USA) were
applied.
Cell lines and culture conditions
The human glioma cell line (U251) was purchased from
American Type Culture Collection (ATCC; Mannassas, VA, USA). Cells
were cultured in RPMI-1640 supplemented with 10% fetal bovine
serum, 10 U/l penicillin G and 100 mg/l streptomycin at 37°C in a
humidified atmosphere containing 5% CO2.
PCR Amplification and cloning of the
FOXP1 cDNA
A cDNA encoding full-length FOXP1 was amplified by
PCR in a 50 μl reaction containing 1 μl human fetal brain cDNA
library (1:10 dilution; Clontech Laboratories, Mountain View, CA,
USA), oligonucleotide primers (forward, 5′-CTCGGATTCATGATG
CAGGAATCTGCGACAGAGAC AATAAGC-3′ and reverse,
5′-CTCGAATTCTCATTCCAGATCTTCAGATAAAGG CTCTTCTTC-3′) at 0.5 μmol/l,
deoxynucleotide triphosphates at 200 μmol/l, 1X reaction buffer and
2.5 units Pfu Turbo polymerase (Stratagene Inc., La Jolla, CA,
USA). The thermal cycler was programmed as follows: 95°C for 1 min,
followed by 26 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C
for 3 min, followed by a final 10 min extension at 72°C. The
PCR-generated full-length FOXP1 was digested with BamHI and EcoRI,
ligated together and cloned to create the eukaryotic expression
construct p-enhanced green fluorescence protein (EGFP)-N1-FOXP1.
Plasmid DNA was prepared and the construct insert was fully
sequenced.
Lipofectamine 2000 was used for transfection of
2×105 cells/well (six-well plate) and 5 μg of
recombinant plasmids. Empty vector was used as a control. An
inverted fluorescence microscope (Axiovert 200; Carl Zeiss,
Göttingen, Germany) was used for observation and obtaining
images.
MTT assay
A total of 1×104 U251 cells were cultured
in 96-well tissue-culture plates overnight and then transfected
with plasmids as described above. Following 1–5 days, glioma cells
were incubated for 3 h in 100 μl MTT (0.1 mg/ml; Sigma, St. Louis,
MO, USA). Cells were resuspended in 200 μl isopropanol (to dissolve
the formazan) and the optical density (OD) of the solution was
determined using a spectrometer (Fluostar Optima; BMG Labtech,
Ortenberg, Germany) at an incident wavelength of 570 nm. Cell
viabilities in experimental wells were expressed as a percentage of
the viability in the control well.
Invasion assay
Invasion assay was performed using a Boyden chamber
system (Neuro Probe) with a fibronectin-precoated (0.5 mg/ml)
polycarbonate membrane (8 μm pore size). The lighter side of the
polycarbonate membrane was precoated with 250 μg/ml
Matrigel® (BD Biosciences, Franklin Lakes, NJ, USA). The
bottom chambers were filled with 30 μl RPMI-1640 medium containing
2% bovine serum albumin (BSA), while the top chambers were filled
with 50 μl RPMI-1640 serum-free medium containing 0.2% BSA. Cells
(5×104/well) were added to the top chamber and incubated
for 15 h in an incubator at 37°C with 5% CO2. Three
independent experiments were performed in triplicate. The cells
were fixed in methanol and stained with haematoxylin. The top
surface of the membrane was gently scrubbed with a cotton bud, the
cells that had migrated to the lower side of the membrane were
counted under the microscope (Carl Zeiss) and the numbers of
migrated cells were calculated as the mean ± standard
deviation.
Adhesion assay
Matched quantities of pEGFP-N1-FOXP1- and control
empty vector-transfected U251 cells (3×104) were plated
onto a Matrigel-precoated (50 μg/ml) 96-well plate in triplicate.
The cells were washed at 30, 60 and 120 min to remove non-adherent
cells. Following washing, the adhered cells were quantified using
an MTT assay. The relative OD was determined at 570 nm using
Wellscan MK3 ELISA (Ani Labsystems Ltd. Helsinki, Finland) and a
450 nm reference filter. The OD values reflected the proportion of
cells in the Matrigel-coated 96-well plate.
Wound healing assay
The wound healing assay was conducted as previously
described (24). The distance of
wound closure (compared with the control at 0 h) was measured at
three independent wound sites per group. The relative cell motility
was calculated as the wound width at t = 0 h minus the wound width
at t = 24 h, as indicated. Values from at least three independent
experiments were pooled and expressed as the mean ± standard
deviation.
Statistical analysis
All experiments were repeated at least three times.
Student’s t-test was used to evaluate the differences between
experimental and control groups. The data were analyzed by one-way
analysis of variance with SPSS 13.0 (SPSS, Inc., Chicago, IL, USA).
The frequencies of FOXP1 expression among cancer samples were
analyzed by the χ2 test with modification by the
Fisher’s exact test to account for frequency values <5. All of
the P-values reported are two-sided. P<0.05 was considered to
indicate a statistically significant difference between values.
Results
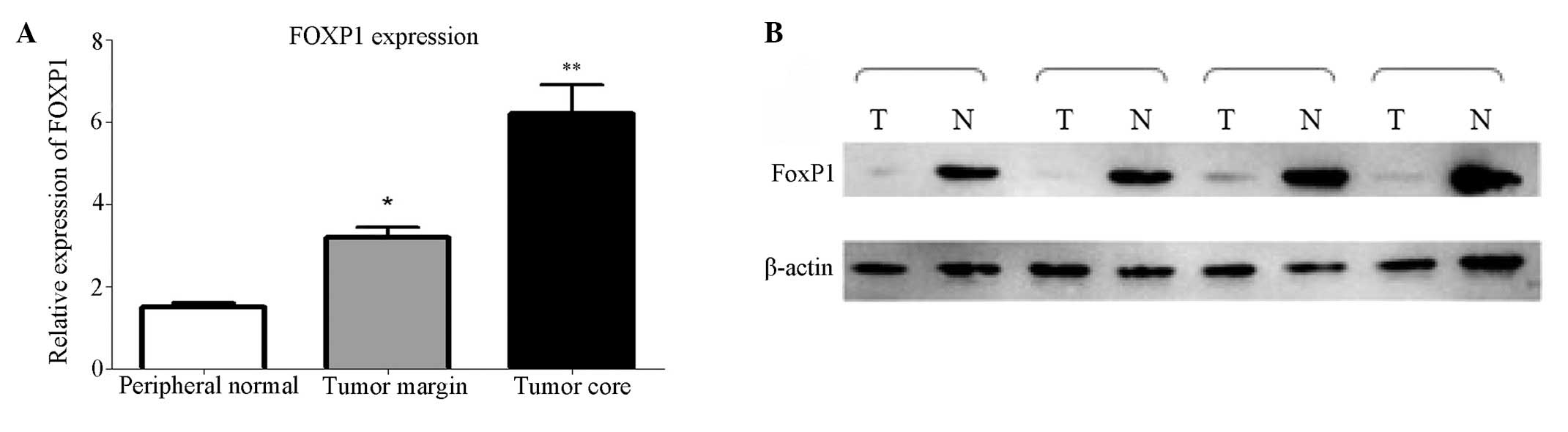
Expression of FoxP1 in human glioma
tissues
In 25 sets of tissue samples collected from
different brain regions, expression of the FOXP1 gene was
significantly downregulated in tumor core samples (P<0.05)
compared with peripheral normal brain tissue, as determined by
qPCR, although expression decreased progressively in samples
obtained from more distal areas (Fig.
1A). Immunoblotting confirmed the reduction in FOXP1 protein
expression in malignant gliomas was consistent with the reduced
transcript levels (Fig. 1B).
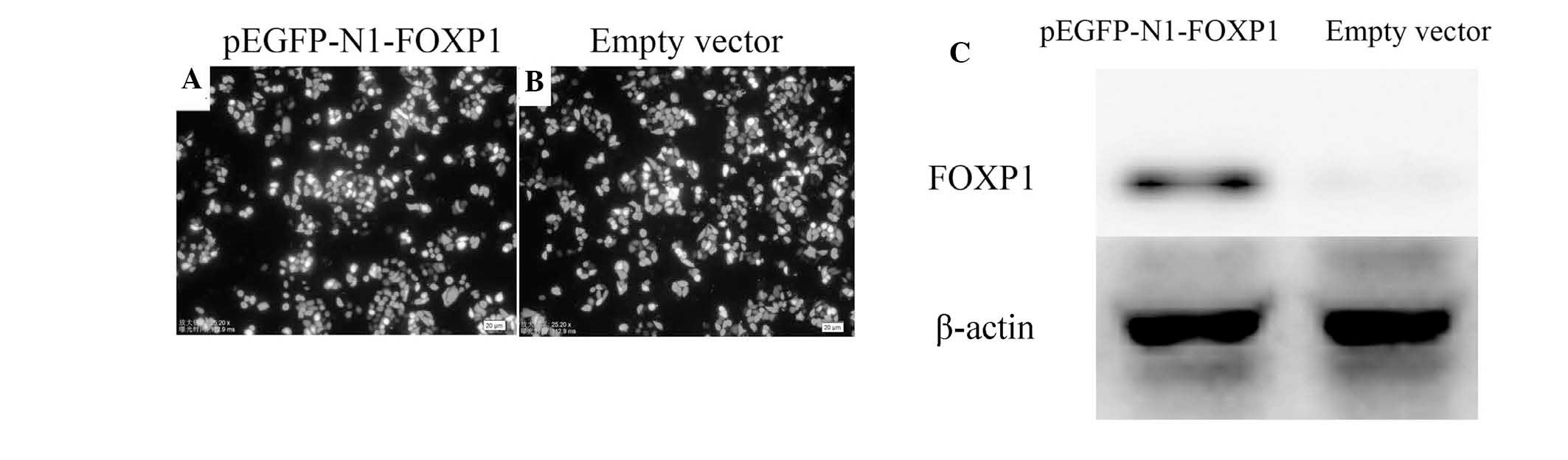
Transfection efficiency
As demonstrated in Fig.
2A and B, under fluorescence microscopy observation, the
transfection efficiency of the two groups (pEGFP-N1-FOXP1 and empty
vector) was satisfactory, with all exceeding 80%.
Western blot analysis demonstrated that transfected
pEGFP-N1-FOXP1 significantly increased exogenous expression of
FOXP1 in U251 cells as compared with that in the empty vector
control (Fig. 2C).
FOXP1 inhibited proliferation and
increased apoptosis in U251 cells
The MTT assay revealed that the proliferation of
cells in the empty vector group increased almost 4-fold from 24 to
96 h following transfection, while the proliferation in the
pEGFP-N1-FOXP1 group was relatively slow, which increased only
1-fold in 72 h (P<0.01; Fig.
3A). Flow cytometric analysis demonstrated that the rate of
apoptosis increased significantly (P<0.01) in
pEGFP-N1-FOXP1-transfected cells as compared with that of the empty
vector control (Fig. 3B and
C).
Effect of exogenous expression of FOXP1
on the invasive, motive and adhesive abilities and proliferation of
U251 cells
To study the role of FOXP1 on cell invasion,
motility and adhesion, the major characteristics of metastasis, all
the assays were performed 48 h following transfection. In an
invasion assay, the number of cells that migrated to the bottom
side of the membrane on a chamber, where the cells were seeded, was
calculated (Fig. 4A). The data
demonstrated that the number of migrated cells in the transfected
FOXP1 group decreased dramatically compared with that in the empty
vector control group (Fig. 4B).
These results suggested that the overexpression of FOXP1
significantly suppressed the migration ability of U251 cells.
To examine whether the depletion of FOXP1 had any
effect on the motive ability of the cells, a wound-healing
experiment was performed using U251 cells transfected with FOXP1 or
the empty vector. The data revealed that there was a significant
difference in the relative wound closure at 15 and 24 h for the two
groups (P<0.01, Fig 4C and
D).
The elevated FOXP1 levels that correlated with
glioma metastasis in the tumor patients prompted us to examine
whether FOXP1 had an effect on cell adhesion to the extracellular
matrix (ECM). For this purpose, a cell adhesion assay was
performed. The results demonstrated that the cells, when
transfected with FOXP1, had a characteristically low adhesion
ability as compared with the empty vector control (Fig. 4E). The results were consistent at
different time-points, including at 30, 60 and 120 min,
respectively. These results indicated that FOXP1 may inhibit tumor
cell adhesion to the ECM. In conclusion, the data suggested that
FOXP1 exerts its effects on metastasis by inhibiting invasion,
migration and adhesion in U251 cells.
Discussion
FOXP1 was first identified in a study screening for
glutamine-rich transcription factors in B cells (25). The full length of the human FOXP1
gene was initially cloned with a monoclonal antibody (JC12) that
recognized a differentially expressed protein in malignant and
normal B cells (26). As a member
of the FOXP subset of ‘forkhead’ (Fox) transcription factors, FOXP1
has multiple functions, including the regulation of B-cell
development (10), lung and
esophagus development (11),
monocyte and macrophage differentiation (27) and cardiac development (28). Of note, separate studies
investigating different tumor types have suggested that FOXP1 may
act as either a tumor suppressor or an oncogene (25). Therefore, the present study aimed
to verify the association between FOXP1 expression and the clinical
parameters of malignant gliomas.
In the present study, it was demonstrated for the
first time, to the best of our knowledge, that FOXP1 mRNA and
protein expression in glioma tumors is significantly elevated
compared with matched peripheral normal tissue. The immunostaining
results are consistent with the qPCR results, which suggested that
the expression of FOXP1 may be important in the tumorigenesis and
progression of gliomas. Previous studies demonstrated that the
positive expression of FOXP1 in malignant endometrium was linked
with deep myometrial invasion and poor differentiation (16). Furthermore, FOXP1 expression was
associated with the postoperative Gleason score of prostate cancer
(29) and with tumor grading of
renal cancer (19), whereas no
significant correlations were identified between FOXP1 expression
and other clinical characteristics in these patients. Of note, the
loss of FOXP1 protein expression in breast carcinoma is closely
associated with poor patient outcome (17). This accumulative evidence suggests
that FOXP1 may act as a tumor suppressor gene in human cancer. The
results of the present study support these findings, demonstrating
that low expression levels of FOXP1 protein are more common in
glioma tissues than in normal tissues.
Establishing the factors responsible for the
development, proliferation, metastasis and recurrence of brain
tumors may facilitate the identification of potential diagnostic
markers or targets for new therapeutic regimens. In the present
study, the aim was to to investigate the potential role of FOXP1
and its possible regulatory mechanism in glioblastoma. Therefore,
an exogenous expression vector of FOXP1 was established and
transfected it into the glioma cell line U251 in order to identify
its role in the proliferation, migration and invasion of U251
cells. The results further confirm that FOXP1 acts as a tumor
suppressor gene in glioma and is crucial in controlling glioma cell
invasion, migration and motility. These results are consistent with
evidence from earlier studies, which suggested that targeting FOXP1
may be a strategy for the development of novel glioma
therapies.
To the best of our knowledge, the present study was
the first report of the differential expression of FOXP1 in
gliomas, indicating that FOXP1 may be used as a novel therapeutic
target for glioma treatment. The results demonstrated elevated
expression levels of FOXP1 in malignant glioma tissue, and that
this exogenous high expression significantly reduced the
proliferation, migration and invasion activity of glioma cells.
Further studies are required to elucidate the underlying mechanisms
of the involvement of FOXP1 in glioma carcinoma.
References
|
1
|
Komotar RJ, Otten ML, Moise G and Connolly
ES Jr: Radiotherapy plus concomitant and adjuvant temozolomide for
glioblastoma-a critical review. Clin Med Oncol. 2:421–422.
2008.PubMed/NCBI
|
|
2
|
Minniti G, De Sanctis V, Muni R, et al:
Radiotherapy plus concomitant and adjuvant temozolomide for
glioblastoma in elderly patients. J Neurooncol. 88:97–103. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stupp R, Mason WP, van den Bent MJ, et al:
Radiotherapy plus concomitant and adjuvant temozolomide for
glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen SM, Chen HC, Chen SJ, et al:
MicroRNA-495 inhibits proliferation of glioblastoma multiforme
cells by downregulating cyclin-dependent kinase 6. World J Surg
Oncol. 11:872013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Furnari FB, Fenton T, Bachoo RM, et al:
Malignant astrocytic glioma: genetics, biology, and paths to
treatment. Genes Dev. 21:2683–2710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rich JN, Hans C, Jones B, et al: Gene
expression profiling and genetic markers in glioblastoma survival.
Cancer Res. 65:4051–4058. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Feng J, Zhang X, Zhu H, Wang X, Ni S and
Huang J: High expression of FoxP1 is associated with improved
survival in patients with non-small cell lung cancer. Am J Clin
Pathol. 138:230–235. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Koon HB, Ippolito GC, Banham AH and Tucker
PW: FOXP1: a potential therapeutic target in cancer. Expert Opin
Ther Targets. 11:955–965. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Green MR, Gandhi MK, Courtney MJ, Marlton
P and Griffiths L: Relative abundance of full-length and truncated
FOXP1 isoforms is associated with differential NFkappaB activity in
Follicular Lymphoma. Leuk Res. 33:1699–1702. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu H, Wang B, Borde M, et al: Foxp1 is an
essential transcriptional regulator of B cell development. Nat
Immunol. 7:819–826. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shu W, Lu MM, Zhang Y, Tucker PW, Zhou D
and Morrisey EE: Foxp2 and Foxp1 cooperatively regulate lung and
esophagus development. Development. 134:1991–2000. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wlodarska I, Veyt E, De Paepe P, et al:
FOXP1, a gene highly expressed in a subset of diffuse large B-cell
lymphoma, is recurrently targeted by genomic aberrations. Leukemia.
19:1299–1305. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lenz G, Wright GW, Emre NC, et al:
Molecular subtypes of diffuse large B-cell lymphoma arise by
distinct genetic pathways. Proc Natl Acad Sci USA. 105:13520–13525.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Adams H, Tzankov A, Lugli A and Zlobec I:
New time-dependent approach to analyse the prognostic significance
of immunohistochemical biomarkers in colon cancer and diffuse large
B-cell lymphoma. J Clin Pathol. 62:986–997. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hoeller S, Schneider A, Haralambieva E,
Dirnhofer S and Tzankov A: FOXP1 protein overexpression is
associated with inferior outcome in nodal diffuse large B-cell
lymphomas with non-germinal centre phenotype, independent of gains
and structural aberrations at 3p14.1. Histopathology. 57:73–80.
2010. View Article : Google Scholar
|
|
16
|
Giatromanolaki A, Koukourakis MI, Sivridis
E, Gatter KC, Harris AL and Banham AH: Loss of expression and
nuclear/cytoplasmic localization of the FOXP1 forkhead
transcription factor are common events in early endometrial cancer:
relationship with estrogen receptors and HIF-1alpha expression. Mod
Pathol. 19:9–16. 2006. View Article : Google Scholar
|
|
17
|
Bates GJ, Fox SB, Han C, et al: Expression
of the forkhead transcription factor FOXP1 is associated with that
of estrogen receptor-beta in primary invasive breast carcinomas.
Breast Cancer Res Treat. 111:453–459. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takayama K, Horie-Inoue K, Ikeda K, et al:
FOXP1 is an androgen-responsive transcription factor that
negatively regulates androgen receptor signaling in prostate cancer
cells. Biochem Biophys Res Commun. 374:388–393. 2008. View Article : Google Scholar
|
|
19
|
Toma MI, Weber T, Meinhardt M, et al:
Expression of the Forkhead transcription factor FOXP1 is associated
with tumor grade and Ki67 expression in clear cell renal cell
carcinoma. Cancer Invest. 29:123–129. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fox SB, Brown P, Han C, et al: Expression
of the forkhead transcription factor FOXP1 is associated with
estrogen receptor alpha and improved survival in primary human
breast carcinomas. Clin Cancer Res. 10:3521–3527. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei KC, Huang CY, Chen PY, et al:
Evaluation of the prognostic value of CD44 in glioblastoma
multiforme. Anticancer Res. 30:253–259. 2010.PubMed/NCBI
|
|
22
|
Barbaric D, Dalla-Pozza L and Byrne JA: A
reliable method for total RNA extraction from frozen human bone
marrow samples taken at diagnosis of acute leukaemia. J Clin
Pathol. 55:865–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kelleher KL, Leck KJ, Hendry IA and
Matthaei KI: A one-step quantitative reverse transcription
polymerase chain reaction procedure. Brain Res Brain Res Protoc.
6:100–107. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Su F, Li H, Yan C, Jia B, Zhang Y and Chen
X: Depleting MEKK1 expression inhibits the ability of invasion and
migration of human pancreatic cancer cells. J Cancer Res Clin
Oncol. 135:1655–1663. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li C and Tucker PW: DNA-binding properties
and secondary structural model of the hepatocyte nuclear factor
3/fork head domain. Proc Natl Acad Sci USA. 90:11583–11587. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Banham AH, Beasley N, Campo E, et al: The
FOXP1 winged helix transcription factor is a novel candidate tumor
suppressor gene on chromosome 3p. Cancer Res. 61:8820–8829.
2001.PubMed/NCBI
|
|
27
|
Shi C, Sakuma M, Mooroka T, et al:
Down-regulation of the forkhead transcription factor Foxp1 is
required for monocyte differentiation and macrophage function.
Blood. 112:4699–4711. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Li S, Yuan L, et al: Foxp1
coordinates cardiomyocyte proliferation through both
cell-autonomous and nonautonomous mechanisms. Gen Dev.
24:1746–1757. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Banham AH, Boddy J, Launchbury R, et al:
Expression of the forkhead transcription factor FOXP1 is associated
both with hypoxia inducible factors (HIFs) and the androgen
receptor in prostate cancer but is not directly regulated by
androgens or hypoxia. Prostate. 67:1091–1098. 2007. View Article : Google Scholar
|